Abstract
The tetrameric paramyxovirus hemagglutinin-neuraminidase (HN) protein mediates attachment to sialic acid-containing receptors as well as cleavage of the same moiety via its neuraminidase (NA) activity. The X-ray crystallographic structure of an HN dimer from Newcastle disease virus (NDV) suggests that a single site in two different conformations mediates both of these activities. This conformational change is predicted to involve an alteration in the association between monomers in each HN dimer and to be part of a series of changes in the structure of HN that link its recognition of receptors to the activation of the other viral surface glycoprotein, the fusion protein. To explore the importance of the dimer interface to HN function, we performed a site-directed mutational analysis of residues in a domain defined by residues 218 to 226 at the most membrane-proximal part of the dimer interface in the globular head. Proteins carrying substitutions for residues F220, S222, and L224 in this domain were fusion deficient. However, this fusion deficiency was not due to a direct effect of the mutations on fusion. Rather, the fusion defect was due to a severely impaired ability to mediate receptor recognition at 37°C, a phenotype that is not attributable to a change in NA activity. Since each of these mutated proteins efficiently mediated attachment in the cold, it was also not due to an inherent inability of the mutated proteins to recognize receptors. Instead, the interface mutations acted by weakening the interaction between HN and its receptor(s). The phenotype of these mutants correlates with the disruption of intermonomer subunit interactions.
The paramyxoviruses are enveloped, negative-stranded RNA viruses. The group includes Newcastle disease virus (NDV), Sendai virus, simian virus 5, mumps virus, and the various parainfluenza viruses. The surfaces of paramyxovirions and paramyxovirus-infected cells display two types of glycoprotein spike structures, which mediate early virion-cell and cell-cell interactions (20). The hemagglutinin-neuraminidase (HN) protein is a type II homotetramer, which mediates both attachment to sialic acid-containing receptors and cleavage of the same moiety from both soluble and membrane-bound glycoconjugates (13). The latter is catalyzed by the neuraminidase (NA) activity associated with the protein.
All paramyxovirus HN proteins are pairs of dimers. For some viruses, e.g., the Australia-Victoria isolate of NDV (NDV-AV) (22), the HN monomers are linked by disulfide bonds mediated by cysteines in the membrane-proximal stalk region. The stalk supports a terminal globular domain in which reside the attachment and NA activities and all the antigenic sites (16, 25). The X-ray crystallographic structures of the globular head of the non-disulfide-linked dimer of the HN of NDV-L-Kansas as well as forms of the protein complexed with an inhibitor and the β-anomer of sialic acid have been solved (2). These structures indicate that receptor recognition and NA activity are mediated by a single sialic acid binding site in two different conformations. Loss of both activities by mutagenesis of active-site residues was cited as supportive of the single-site hypothesis (1, 2). On the other hand, we have shown that similarly mutated attachment- and NA-deficient NDV HN proteins can be rendered somewhat competent to mediate attachment by NA activity contributed either endogenously or exogenously (9). These findings raise the possibility that the lack of attachment activity exhibited by active-site mutants may be secondary to their NA deficiency.
The second paramyxovirus surface glycoprotein is the fusion (F) protein, which, as its name implies, mediates both virus-cell and cell-cell fusion (13, 20). F is produced as a precursor, called Fo, which requires both proteolytic cleavage and a conformational change to become fusogenic (13, 21). However, for most paramyxoviruses, including NDV, expression in transfected cells of wild-type F in the absence of HN does not result in syncytium formation (reviewed in reference 13). The promotion of fusion by these F proteins requires coexpression of the homologous HN protein. Combinations of HN and F proteins derived from heterologous viruses fail to induce fusion.
The mechanism of HN-dependent, F-mediated fusion has still not been completely elucidated. By the construction of HN chimeras composed of domains from heterologous HN proteins, it has been established that the stalk region of HN defines the domain that determines fusion specificity for the homologous F protein, with various contributions from the transmembrane domain (5, 24, 26). However, it is not clear how attachment, mediated by the receptor recognition site in the globular domain of HN, is related to the HN-F interaction and, in turn, to the fusion activation of the F protein.
The X-ray crystallographic structures (2) suggest that, upon binding sialic acid, the globular domain of HN undergoes a conformational change that leads to an alteration in the association between monomers in each dimer. It has been proposed that these changes at the dimer interface could be part of a series of sequentially linked conformational changes that result in the activation of F (1). This possibility was supported by the identification of substitutions for I175, E401, and Y526 in the active site of the NDV-L-Kansas HN protein that result in high fusogenicity despite diminished receptor binding. This led to the hypothesis that interface residues may be directly involved in the interaction with F (1).
To begin to explore the role of dimer interface residues in HN function, we have performed a mutational analysis of several residues in a domain at its most membrane-proximal end. We identified substitutions for residues F220, S222, and L224 in this domain which result in a marked reduction in fusogenic activity, resulting from severely diminished hemadsorption activity at 37°C. However, each of these proteins exhibits NA activity at 37°C as well as hemadsorption activity at 4°C in amounts comparable to that of the wild-type protein. Thus, the fusion-deficient phenotype of these mutants is due to a destabilization of the interaction between HN and its receptor(s) rather than to a direct effect on fusion.
MATERIALS AND METHODS
Recombinant plasmids and site-directed mutagenesis.
Construction of the NDV-Australia-Victoria (NDV-AV) HN and F recombinant pBluescript SK(+) (Stratagene Cloning Systems, La Jolla, Calif.) expression vectors has been described previously (15). The HN of the B1-Hitchner NDV strain (NDV-B1) was a gift of Kemal Karaca (Megan Health, Inc., St. Louis, Mo.). It was released from the vector in which it was supplied by XhoI digestion and ligated into the pBSK plasmid.
Site-directed mutagenesis was performed as described previously (3). Briefly, mutagenesis primers, obtained from Bio-Synthesis, Inc. (Lewisville, Tex.), were annealed to single-stranded pBSK HN template previously rescued by R408 helper phage in CJ236 cells. The annealed primer was extended with T4 DNA polymerase and ligated with DNA ligase. This was transformed into MV1190 cells, and transformants were selected by ampicillin resistance. Identification of mutants was facilitated by screening for the presence of unique restriction enzyme sites introduced by the mutagenic primer. The presence of the desired mutation was verified by sequencing of double-stranded DNA with the Sequenase plasmid sequencing kit (United States Biochemicals, Cleveland, Ohio) according to protocols provided by the company. Multiple clones were characterized for each substitution.
Transient expression system and quantitation of cell surface HN.
Wild-type and mutant HN proteins were expressed in BHK-21 cells (American Type Culture Collection, Manassas, Va.), with the vaccinia virus-T7 RNA polymerase expression system (6). All experiments except NA assays were done on 35-mm plates seeded a day earlier at 4 × 105 cells/well. Maintenance of cells, infection with the vaccinia virus recombinant, and transfection were performed as described previously (15), with 1 μg of each plasmid for transfection. Cell surface expression was quantitated by fluorescence-activated cell sorting analysis, with a mixture of monoclonal antibodies to at least five different antigenic sites in the HN globular domain (7, 8, 10, 12).
Hemadsorption assay.
The hemadsorption activity of HN proteins was determined at both 4°C and 37°C by the ability of the expressed protein to adsorb guinea pig erythrocytes (Crane Laboratories, Syracuse, N.Y.). HN-expressing monolayers were incubated for 30 min with a 2% suspension of erythrocytes in phosphate-buffered saline supplemented with 1% CaCl2 and MgCl2. After extensive washing, adsorbed erythrocytes were lysed in 50 mM NH4Cl, and the lysate was clarified by centrifugation. Hemadsorption activity was quantitated by measurement of the absorbance at 540 nm minus the background obtained with cells expressing the vector alone.
Neuraminidase assay.
NA assays were performed on 22.6-mm plates seeded a day earlier at 1.6 × 105 cells/well and transfected with 0.5 μg of DNA. Monolayers were incubated at 37°C for 20 min with 0.5 ml of 625-μg/ml neuraminlactose (Sigma Chemical Co., St. Louis, Mo.) in 0.1 M sodium acetate (pH 6), and NA activity was determined as described previously (15).
Content mixing assay for fusion.
The ability of the mutated HN proteins to complement the F protein in the promotion of fusion was quantitated with a content mixing assay (4). Briefly, monolayers were infected with the vTF7-3 vaccinia virus recombinant as described above and cotransfected with the desired HN and a cleavage site mutant form of the F gene (4). Another set of monolayers was infected with wild-type vaccinia virus (multiplicity of infection of 10) and transfected with 1 μg of plasmid pG1NT7β-gal (19). After 16 h, cells were removed from the wells by trypsinization (0.05% trypsin, 0.53 mM EDTA), which also activates cleavage site mutant F, washed with Dulbecco's modified Eagle's medium, pelleted, and resuspended in 0.8 ml of BHK medium (3).
Equal numbers of the two populations were combined in duplicate wells of a 96-well flat-bottomed plate. After 5 h at 37°C, the cells were lysed by addition to the medium of 10 μl of 10% Nonidet P-40 and incubated at room temperature for 30 min. Then 50 μl of the supernatant from each well was withdrawn and mixed with an equal volume of 16 mM CPRG (chlorophenol red-β-D-galactopyranoside). After incubation for 20 min at room temperature, the extent of fusion was quantitated by determination of the absorbance at 590 nm with a Spectra Max 250 microplate spectrophotometer (Molecular Devices, Sunnyvale, Calif.), with the background obtained with cells expressing vector alone subtracted.
RESULTS
Mutation of a domain at the membrane-proximal end of the dimer interface in the HN protein.
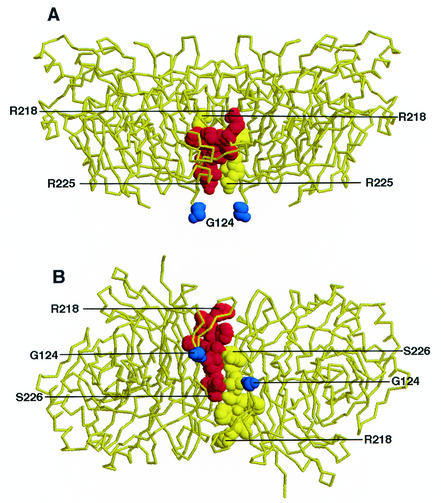
Figure 1 shows the backbone of the pH 6.5 structure of the NDV-L HN dimer (2). The domain targeted in this study is shown in space-filling mode in both a lateral (panel A) and a membrane (panel B) view. The domain constitutes the most membrane-proximal part of the interface between monomers in the HN homodimer and includes residues R218, V219, F220, F221, S222, T223, L224, R225, and S226 in NDV-AV HN.
FIG. 1.
Domain at the interface between monomers in the HN dimer. The domain targeted for mutagenesis in this study is shown in space-filling mode in the backbone of the X-ray crystallographic structure of the pH 6.5 form of the HN dimer of the L-Kansas isolate. The domain defined by residues 218 to 226 is shown from both a lateral (A) and membrane (B) view. The residues on different monomers are shown in space-filling mode in either red or yellow. For reference, residue 124 in each dimer, the most N-terminal residue in the soluble form of the protein presumed to be near the top of the stalk, is also shown in space-filling mode in dark blue. The figure was generated with RasMol.
With the goal of probing the role of this domain in HN function, an alanine substitution was individually introduced at each of the above positions. Based primarily on the properties of this initial panel of mutated proteins, additional substitutions were introduced for several residues in the domain. Thus, to investigate the contribution of the dimer interface residues to HN function, proteins carrying the following mutations were prepared and characterized: R218A and -S; V219A; F220A, -N, and -Y; F221A; S222A, -K, -N, and -T; T223A and -R; L224A, -Q, and -S; R225A; and S226A.
Cell surface expression and antigenic integrity of mutated proteins.
The cell surface expression of each mutated protein was quantitated by fluorescence-activated cell sorting analysis with a cocktail of five different HN-specific monoclonal antibodies. As shown in Fig. 2, all of the mutated proteins were detected at the cell surface in amounts comparable to the wild-type protein. Of the 18 mutated proteins, 15 were expressed at the surface at greater than 90% of the wild-type level, and none of the remaining three mutated proteins was expressed at less than 75% of the wild-type HN level. Thus, all mutated proteins were present at the cell surface in amounts comparable to wild-type HN.
FIG. 2.
Cell surface expression and antigenic integrity of HN interface mutants. The cell surface expression of the HN interface mutants was quantitated by fluorescence-activated cell sorting analysis, with a panel of at least five monoclonal antibodies specific for different antigenic sites on the globular domain of the protein. The results shown represent the mean of at least three determinations. wt, wild type.
Ability of mutated proteins to promote fusion.
The ability of the mutated HN proteins to complement the homologous F protein in the promotion of fusion was quantitated with the content mixing assay (Fig. 3). All substitutions for residue S222, including A, K, N, and T, reduced fusion to less than 10% of the wild-type value.
FIG. 3.
Fusion-promoting activity of HN interface mutants. HN proteins carrying the amino acid substitutions shown were assayed for their ability to complement the F protein in the promotion of fusion. Each value is the mean of a minimum of five determinations. wt, wild type.
Certain F220 mutations also reduced fusion-promoting activity, although the extent depended on the particular substitution. F220N-mutated HN was the weakest in promoting fusion of all mutants tested, inducing only 2.1% of the wild-type level (Fig. 3). F220A-mutated HN also exhibited reduced fusion-promoting activity, although it retained nearly 13% of the wild-type level. On the other hand, a more conservative substitution, F220Y, had no significant effect on fusion. HN carrying this substitution promoted fusion more than 95% as effectively as wild-type HN.
Figure 4 compares the extent of syncytium formation in monolayers in which the poorly fusogenic F220A, F220N, and S222T mutant proteins and the strong fuser F220Y HN were coexpressed with NDV F and stained for fusion. No syncytia were visible in the S222T and F220N HN-expressing cells. Small syncytia were seen in monolayers expressing F220A HN, but the F220Y HN-expressing monolayers were indistinguishable from the wild-type control.
FIG. 4.
Syncytium formation in monolayers coexpressing HN interface mutants and the F protein. The extent of syncytium formation is shown in monolayers coexpressing the following with wild-type F: (A) vector control; (B) wild-type HN; (C) F220A HN; (D) F220N HN; (E) S222T HN; and (F) F220Y HN. At 22 h posttransfection, the monolayers were fixed with methanol and stained with Giemsa stain. Small syncytia are indicated in the F220A-expressing monolayer by arrows (C).
Residue L224 may also be important for fusion, although to a lesser degree. L224A- and L224S-mutated HN proteins exhibited reduced fusion-promoting activity, 47.5 and 30.8% of the wild-type level, respectively. However, a substitution of glutamine at this position had no significant effect on fusion. These results strongly suggest that residues F220 and S222 are critical for the fusion-promoting activity of the NDV HN protein and that residue L224 may contribute to a lesser extent. The remaining substitutions at positions 218, 219, 221, 223, 225, and 226 all retained significant fusion-promoting activity, more than 50% of the wild-type HN value.
Hemadsorption activity of mutated proteins at 4°C.
Next, the receptor recognition properties of the mutated proteins were evaluated by assaying their ability to adsorb guinea pig erythrocytes at 4°C (Fig. 5). All, with the lone exception of T223A HN, hemadsorbed at least 60% of the wild-type amount at this temperature. Most important, poorly fusing proteins F220A and F220N hemadsorbed almost as efficiently as the wild-type protein, 82.2 and 91.0%, respectively. The poorly fusogenic proteins carrying substitutions for S222 as well as L224S all hemadsorbed slightly less efficiently, but still 65 to 75% of the wild-type level. Thus, the weakly fusogenic phenotype is not the result of an inherent inability to recognize receptors. Indeed, T223A-mutated HN was the only protein exhibiting a significant reduction in hemadsorption activity at 4°C. It hemadsorbed only about one-third as efficiently as the wild-type protein. However, it fused at 65.0% of the wild-type level.
FIG. 5.
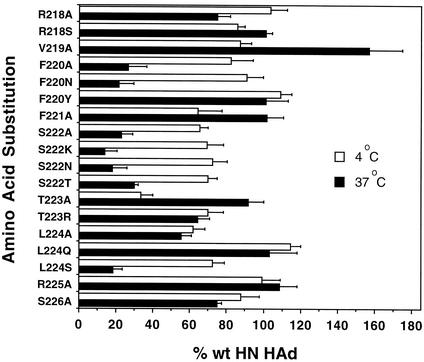
Hemadsorption (HAd) activity of HN interface mutants. The ability of the HN interface mutants to hemadsorb guinea pig erythrocytes was determined by incubation for 30 min at either 4°C or 37°C. The background level obtained with monolayers expressing vector alone was subtracted, and the data are expressed as a percentage of the amount obtained with wild-type (wt) HN-expressing monolayers. The data shown represent the means from at least four determinations.
Diminished fusion-promoting activity correlates with low hemadsorption activity at 37°C.
Hemadsorption assays are routinely performed in the cold to determine receptor binding activity in the absence of NA. However, recognizing that fusion is assayed at 37°C, in order to more accurately determine the relationship between attachment and fusion, we also assayed the hemadsorption activity of the panel of mutated proteins at this temperature.
At 37°C, the hemadsorption activity of several interface mutants was significantly lower than it was at 4°C (Fig. 5). This includes all of the S222-mutated proteins as well as those carrying F220A, F220N, and L224S substitutions. At 37°C, each of these proteins hemadsorbed less than 30% as efficiently as the wild-type protein, whereas they all hemadsorbed greater than 65% of the wild-type value in the cold.
Most of the remaining proteins exhibited hemadsorption activity at the higher temperature that was comparable to that of the wild-type protein. This set included those proteins carrying substitutions of R218S, F220Y, F221A, T223A, L224Q, and R225A. A few mutants, e.g., R218A, T223R, L224A, and S226A, showed slightly reduced hemadsorption at this temperature, 55 to 75% of the wild-type value. V219A HN exhibited an unusual phenotype. It hemadsorbed nearly 60% more than wild-type HN at 37°C (Fig. 5).
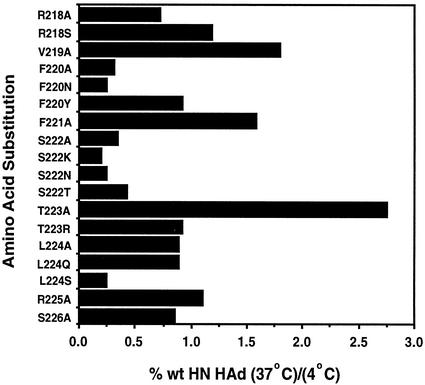
Figure 6 shows the ratio of hemadsorption at 37°C to that at 4°C for each mutant. Presentation of the data in this way groups the poorly fusing proteins (F220A, F220N, S222A, S222K, S222N, S222T, and L224S) together, with ratios that ranged from 0.20 to 0.43. All the other proteins, which retained significant fusogenicity, exhibited 37°C/4°C hemadsorption ratios of at least 0.7. Indeed, most ratios were greater than 0.9. Taken together, these data show a close correlation between weak hemadsorption at 37°C and decreased fusion.
FIG. 6.
Ratio of hemadsorption activity at 37°C to that at 4°C. With the data from Fig. 5, the ratio of the wild-type (wt) HN hemadsorption (HAd) at 37°C to that at 4°C was calculated.
Three substitutions, V219A, F221A, and T223A, resulted in proteins that hemadsorbed much better at 37°C than they did at 4°C, exhibiting ratios of 1.80, 1.58, and 2.76, respectively. This suggests that their hemadsorption activity was more stable at 37°C than that of the wild-type protein.
To establish the extent of the temperature sensitivity of the hemadsorption activity, the assay was also performed at the intermediate temperature of 25°C for two of the poorly fusing proteins, F220N and S222N. At this temperature, these proteins still exhibited reduced hemadsorption activity relative to the wild-type protein, 12.5 ± 7.2% for F220N HN and 19.2 ± 3.1% for S222N HN. Thus, even at this intermediate temperature, the mutated proteins were unable to bind red blood cells as tightly as the wild-type protein.
Temperature shift experiments.
To further illustrate the temperature sensitivity of the hemadsorption activity of some of these mutated proteins, a temperature shift experiment was performed. Guinea pig erythrocytes were adsorbed at 4°C to monolayers expressing one of the mutated proteins or wild-type HN. After 30 min in the cold, the plates were shifted to 37°C for 10, 20, or 30 min. The monolayers were then washed, and the amount of bound erythrocytes was compared to that on monolayers not shifted to the higher temperature.
The data are shown for wild-type HN and five of the mutated proteins (Fig. 7). Over the 30-min incubation period at 37°C, the three weakly fusing mutants F220N, S222K, and L224S showed a steady elution of erythrocytes. After 20 min at the higher temperature, at least two-thirds of the originally bound red blood cells had eluted from these monolayers. After 30 min, only 24.7%, 11.1%, and 15.8% of the cells originally bound to the F220N HN-, S222K HN-, and L224S HN-expressing monolayers, respectively, remained attached.
FIG. 7.
Hemadsorption activity of HN interface mutants and wild-type HN after a shift from 4°C to 37°C. Erythrocytes were adsorbed for 30 min in the cold to monolayers expressing either wild-type (wt) HN or F220N, F220Y, S222K, L224Q, or L224S HN. Plates were either washed immediately with cold phosphate-buffered saline with calcium and magnesium or shifted to 37°C for either 10, 20, or 30 min before washing. Hemadsorption (HAd) activity was quantitated spectrophotometrically as described previously. Each point indicates the percentage of hemadsorption remaining from the control plates, which were not shifted to the higher temperature, and represents an average of two determinations.
In contrast, nearly 80% of the erythrocytes originally adsorbed to the wild-type HN-expressing monolayers remained attached at the end of the 30-min incubation at the higher temperature. Similarly, monolayers expressing the fusogenic F220Y HN and L224Q HN proteins still retained 70.2% and 59.4% of the originally bound red blood cells, respectively. Thus, the rate of elution of erythrocytes correlates inversely with the fusogenic activities of the mutated proteins.
NA activity of mutated proteins.
The NA activity of the mutated proteins at the cell surface was determined at 37°C. The data, after correction for differences in cell surface expression, are shown in Fig. 8. Most of the mutated proteins exhibited NA activity that was comparable to that of the wild-type protein. Three of the poor fusers, F220N, S222A, and S222N HN, exhibited marginally significant increases in NA relative to the wild-type protein, 107.5, 115.4, and 113.9% of the wild-type value, respectively. However, the remaining four poorly fusing proteins (F220A, S222K, S222T, and L224S HN) had slightly reduced activity, ranging from 70.5 to 96% of the wild-type level. Thus, the alteration in NA is not consistent and cannot account for the hemadsorption/fusion-deficient phenotype.
FIG. 8.
NA activity of HN interface mutants. The NA activity of each mutant was assayed at 22 h posttransfection by determining the ability of each to cleave sialic acid from neuraminlactose. The data represent the means of at least three determinations, were corrected for differences in cell surface expression (Fig. 2), and are expressed as a percentage of the NA activity of wild-type (wt) HN.
An interesting class of substitutions in the group which included V219A, F221A, and T223A resulted in NA activities that were less than one-third of the wild-type value (Fig. 8). Interestingly, all three exhibited slightly reduced fusion-promoting activity, approximately 55 to 65% of the wild-type level, and more stable hemadsorption activity.
Erythrocytes exposed to monolayers expressing interface mutants can reattach.
Although there is not a consistent change in the NA activity of the mutated proteins, as determined by the ability to release sialic acid from the small substrate neuraminlactose, we cannot strictly rule out the possibility that NA may act differently on receptors than it does on the small substrate used in the NA assays. Thus, we asked whether the fusion-deficient mutants are removing receptors at a faster rate than the wild-type protein. This could account for the hemadsorption defect at the higher temperature at which NA is active.
To address this question, erythrocytes were exposed to independent monolayers expressing three of the fusion-deficient mutants at 37°C for an extended period of time with frequent agitation. Following this treatment, we asked whether the erythrocytes subjected to this treatment could then bind to new monolayers expressing the wild-type HN protein. As shown in Fig. 9, erythrocytes exposed to monolayers expressing either F220N HN, S222A HN, or L224S HN rebound to wild-type HN-expressing cells just as efficiently as erythrocytes that were incubated with wild-type HN-expressing monolayers in the first round. These findings strongly suggest that the hemadsorption deficiency of these mutants at 37°C is not due to the removal of receptors from the erythrocytes that are bound and then released from them, at least not any more than occurs over the same time period with wild-type HN. As a control, erythrocytes treated with Vibrio cholerae NA were unable to bind to HN-expressing monolayers (data not shown).
FIG. 9.
Erythrocytes exposed to monolayers expressing interface mutants can reattach. A 2% suspension of erythrocytes was exposed to monolayers expressing either wild-type (wt) HN or the F220N, S222A, or L224S mutant protein for 1 h at 37°C with frequent agitation. The erythrocytes were removed and adjusted to a concentration of 2.5 × 107 cells/ml. One milliliter of this suspension was added to new monolayers expressing the wild-type HN protein, and hemadsorption (HAd) was determined after incubation at 4°C for 30 min. The data are expressed as a percentage of the hemadsorption obtained with erythrocytes exposed to wild-type HN-expressing monolayers in the first round and represent the means from at least four determinations.
Mutated proteins are recognized by conformation-dependent monoclonal antibodies to several different sites on HN at both 4°C and 37°C.
The fluorescence-activated cell sorting data in Fig. 2 strongly suggest that the introduced mutations did not drastically alter the conformation of the protein. However, since the assay is performed with a mixture of monoclonal antibodies, localized alterations in conformation, such as those predicted to take place at the dimer interface, may go undetected. Also, the possibility exists that there may be conformational differences in one or more mutants at 4°C and 37°C that can be detected by individual monoclonal antibodies.
To approach these questions, we individually immunoprecipitated the F220N and S222K mutant proteins from radiolabeled transfected cells with monoclonal antibodies specific for each of the seven antigenic sites previously identified in the globular domain of the protein. All monoclonal antibodies recognized both mutated proteins as efficiently as either wild-type HN or the fusion-competent, F220Y-mutated positive control at both 4°C and 37°C (data not shown). This established that the conformation of the epitopes recognized by these monoclonal antibodies was not significantly altered by either of these mutations or by incubation at the higher temperature at which the attachment defect was evident.
F220A- and S222N-mutated NDV-B1 HN also exhibits diminished fusion and hemadsorption at 37°C.
Next, we were interested in determining whether the effects of substitutions at the interface of the HN dimer would also be manifested in other isolates of NDV. To address this issue, we independently introduced the F220A and S222K substitutions into the non-disulfide-linked HN of the B1-Hitchner isolate. The properties of these mutated proteins are summarized in Table 1. They were assayed for their ability to complement F in fusion with F derived from the AV isolate. Similar to the corresponding mutations in the AV isolate, these two substitutions drastically reduced the fusion-promoting activity of NDV-B1's HN, in both cases to less than 10% of that of the wild-type protein. They also exhibited reduced hemadsorption activity at 37°C compared to 4°C, although the ratio of the two was not as low as it was with NDV-AV. This is most likely because the activity at 4°C itself was low compared with that of wild-type B1 HN. Thus, the diminished fusion due to mutations at the interface of the HN dimer can also be demonstrated with HN from another isolate of the virus.
TABLE 1.
Properties of NDV-B1 HN interface mutantsa
| HN function | % of control
|
|
|---|---|---|
| F220A | S222K | |
| Fusion | 9.6 ± 5.6 | 9.1 ± 2.7 |
| Cell surface expression | 71.4 ± 4.0 | 64.3 ± 3.6 |
| Hemadsorption | ||
| 4°C | 64.6 ± 16.3 | 47.0 ± 11.7 |
| 37°C | 29.6 ± 6.4 | 29.9 ± 11.0 |
| NAb | 58.5 ± 4.8 | 61.7 ± 6.2 |
All values are expressed as the percentage of wild-type NDV-B1 HN activity.
Corrected for differences in cell surface expression.
DISCUSSION
Substitutions for residues F220 and S222, in a domain at the most membrane-proximal part of the NDV HN dimer interface, resulted in a greater than 90% reduction in fusion-promoting activity. However, this phenotype was not due to a direct effect on fusion. Rather, it resulted from an inability of the mutated proteins to bind receptors at 37°C. Since all of the fusion-deficient mutants adsorbed erythrocytes efficiently at 4°C, it is clear that they do not possess an inherent, sequence-based defect in receptor recognition activity. They are, however, unable to maintain binding to receptors at 37°C, the temperature at which fusion-promoting activity is determined.
The simplest explanation for the properties of these mutants is an elevation in NA activity. If this were the case, the NA activity of the protein, which is inactive in the cold, would become active at the higher temperature and destroy receptors, leading to elution of erythrocytes from the mutated HN proteins. However, we found no correlation between hemadsorption/fusion deficiency and an alteration in NA activity. In fact, none of the mutants exhibited NA activity elevated enough to account for this phenotype, the highest being only 15% greater than the wild-type level (Fig. 8). Moreover, mutant S222T, with approximately 30% less NA activity than wild-type HN, was also hemadsorption and fusion deficient. This strongly suggests that the phenotype of these mutants is unrelated to their specific NA activities, at least as assayed with a small substrate.
However, we could potentially argue that NA activity assayed on neuraminlactose may not be a true indicator of its activity on receptors and that the NA activity of the mutated proteins could be destroying receptors at a faster rate than the wild-type protein. It has recently been shown that approximately one-half of bound erythrocytes are released from wild-type human parainfluenza virus type 3 HN-expressing cells in 1 h at 37°C (18). Thus, we compared the ability of erythrocytes to rebind after exposure to monolayers expressing either wild-type HN or the mutants and showed that they rebind with similar efficiencies (Fig. 9). This strongly suggests that the mutants do not destroy receptors any faster than the wild-type protein.
Another NA-related caveat is whether the interface mutant hemadsorption/fusion-deficient phenotype is specific for the AV isolate of NDV. This isolate is the only one shown to have cooperative NA activity, loss of which results in enhanced fusogenic activity in a variant virus (14). Thus, it seems possible that the diminished fusogenic activity could be related to an alteration in the relationship between the NA active sites on the same spike. However, we have been unable to demonstrate cooperativity with transiently expressed NDV-AV HN. As an alternative approach, F220A and S222K substitutions were introduced in the HN of the B1-Hitchner isolate of the virus, which has noncooperative NA activity (14). Analogous to the results with NDV-AV HN, these substitutions resulted in decreased fusion due to a temperature-dependent decrease in hemadsorption. This indicates that the fusion-deficient phenotype of the NDV-AV HN interface mutants is not related to cooperative NA activity. Thus, we have ruled out a role for the NA activities of the phenotype of the interface mutants.
In order to try to understand the basis for the hemadsorption/fusion-deficient phenotype, we examined the structure of the interface domain in more detail. Figure 10 shows the antiparallel alignment of the two domains from the pair of monomers in a dimer, along with the major hydrogen bonds in the region. Table 2 summarizes all the hydrogen bonds as well as the side chain van der Waals contacts involving critical residues in the domain. While there are several important intermolecular hydrogen bonds, the vast majority of side chain van der Waals interactions are intramolecular.
FIG. 10.
Membrane-proximal region of the interface between monomers in the HN dimer. The domain targeted for mutagenesis in this study is shown for both monomers in a dimer. The membrane view shows the antiparallel orientation of the two domains. Hydrogen bonds are indicated by dotted lines. The figure was generated from the pH 6.5 structure of the HN of the L-Kansas NDV isolate with Swiss PDB-Viewer (Glaxo Wellcome Experimental Research). Note that residue 219, an isoleucine in NDV-L HN, is a valine in NDV-AV HN.
TABLE 2.
Interactions involving HN residues 216 to 228a
| Residue | Side chain hydrogen bondsb | Side chain van der Waals contacts |
|---|---|---|
| F220 | S228cbackbone amide N | R212, T213, T214,dI227 |
| S222 | S226 backbone C = O | V210, R212, F220, S226 |
| L224 backbone C = Oe | ||
| L224 | — | P126, L208, V210, R212, R225 |
| S226 | Y205 side chain | L160, S222, T223, T223 |
| F221 backbone C = Oe |
Based on the crystallographic structure of the pH 6.5 form of the HN from the L-Kansas isolate. Interactions were predicted by the UCSF Midas Plus Program.
All hydrogen bond lengths were less than 3.0 Å. Intermonomeric interactions are shown in bold-faced.
HN residue 228 is an asparagine in HN of the AV isolate.
HN residue 214 is a serine in HN of the AV isolate.
Hydrogen bond mediated via an H2O bridge.
The side chain of S222, for which none of the substitutions was tolerated, is involved in hydrogen bonds with the backbones of both residues S224 (via a water bridge) and S226. Of particular importance are the intermonomeric hydrogen bonds between the hydroxyls on the S222 side groups and the carbonyl groups of the S226 residues. The serine side group at position 222 is a requirement for this hydrogen bond. Neither alanine, lysine, nor asparagine was tolerated at this position. This stresses the importance of the side chain hydroxyl for formation of the hydrogen bond. The fact that threonine, with only an additional methyl group in its side chain, was also not tolerated points to the tight fit for the serine side chain at this position.
The electron cloud around the aromatic side chain of F220 is involved in an intermonomeric hydrogen bond with the backbone of residue 228, which is a serine in the L isolate but an asparagine in the AV isolate. This explains why a tyrosine but not an alanine or asparagine was tolerated at position 220.
The rationale for the importance of residue L224 seems to be based on its van der Waals interactions. An L224Q substitution was tolerated at this position, but L224S and, to a lesser extent, L224A were not, suggesting that only the glutamine side chain has the proper steric properties to maintain the necessary van der Waals contacts. The S226A mutation did not alter HN function, even though it is involved in a hydrogen bond with the backbone of residue F221 on the opposite monomer. Perhaps this is related to the fact that a water molecule mediates the hydrogen bond.
Substitutions for other residues, such as I219 (a valine in the AV isolate), F221, and T223 resulted in proteins with greater hemadsorption activity at 37°C than at 4°C (Fig. 6). Each of these mutants had very low NA activity, less than one-third of the wild-type value (Fig. 8). Presumably, the low NA activity is responsible for the increased stability of the HN-receptor interaction at the higher temperature.
Based on the importance of residues 220, 222, and 224 in subunit interactions at the interface, we propose that the hemadsorption/fusion-deficient phenotype of mutants carrying substitutions at these positions is related to a disruption of these interactions as a result of a subtle change in conformation that destabilizes the structure of the dimer. Such a change in structure would not result in an inability to detect the dimer form of the protein because the HN dimer in the AV isolate is disulfide linked via a cysteine at position 123 in the stalk (16, 22). Given this, as one might expect, the gel migration of each mutant is identical to that of the wild type (data not shown), consistent with disulfide-linked dimer formation.
Moreover, a panel of monoclonal antibodies specific for seven different antigenic sites, six of which are conformation dependent, did not detect a difference between the mutated proteins at 4°C and at 37°C. Although these sites form a continuum over the surface of the globular domain, the one site that is most informative about the local integrity of the mutated proteins is antigenic site 23. This conformation-dependent site overlaps the NA active site (11) and maps close to the interface region (2, 9), selecting variants with substitutions between residues 193 and 203 (11, 12, 14). A monoclonal antibody to this site recognized the mutant and wild-type proteins to similar extents, again at both 4°C and 37°C, suggesting that there is not an alteration in the conformation of the protein in this region.
If there is a conformational change in the hemadsorption/fusion-deficient proteins, it is likely to be a very subtle one. The antigenic structure of the monomers appears to be intact in the mutants. Thus, we propose that the interface mutations described here act by disrupting the association between the monomers. The disruption of the intermolecular hydrogen bonds results in a separation of the monomers. This was not detected by the monoclonal antibodies because the epitopes that they recognize remained antigenically intact. It may be that hemadsorption or attachment is dependent on bivalent binding of HN to receptors, i.e., binding by the receptor recognition site on both monomers in a dimer. The interface mutations could destabilize the dimer at 37°C relative to its structure at 4°C, enough to minimize multivalent binding.
Recently, Takimoto et al. (23) performed a mutational analysis of surface residues in the HN dimer of the L-Kansas strain of NDV. Similar to our findings in the membrane-proximal region, they showed that mutations of residues in widely separated parts of the hydrophobic interface but including F220 and L224 almost completely abolished fusion. These findings led them to propose that the hydrophobic interface of the molecule undergoes a conformational change that, either directly or indirectly, triggers the activation of the F protein. They proposed three alternative models for the role of these residues in the HN-F interaction. However, the hemadsorption activity of these mutants was determined only at 4°C. Even at this temperature, six of the eight fusion-deficient mutants exhibited significantly reduced hemadsorption activity, as little as 20% of the wild-type value. Hemadsorption activity at 37°C was not reported. It will be informative to determine whether the phenotype that we have identified will apply to fusion-blocking mutations in other regions of the interface.
Furthermore, it would appear that the possibility that interface residues can directly mediate the virus-specific interaction with F can be ruled out based on the fusion specificity of HN chimeras composed of regions from the HN proteins of NDV and human parainfluenza virus type 3 (5) and the lack of conservation of the domain among paramyxovirus HN proteins (17). Specificity for the homologous F protein is determined primarily by the stalk region of the HN spike. An HN protein composed of 135 N-terminal amino acids from human parainfluenza virus type 3 HN will fuse only with human parainfluenza virus type 3 F despite having an entire globular domain, including the interface region, derived from NDV HN (5). In light of this earlier result, and given the complete lack of homology between the interface regions of different paramyxovirus HN proteins, it is not clear how the interface domain could mediate a virus-specific interaction with a complementary domain on F.
In summary, site-directed mutational analysis of a short domain at the membrane-proximal end of the interface between monomers in the HN dimer has identified residues that decrease the strength of the interaction between HN and receptors, leading to a fusion-deficient phenotype. These results indicate that subunit interactions in this region are critical for high-avidity binding of HN to its receptors.
Acknowledgments
We gratefully acknowledge Paul Mahon for helpful discussions and help with preparation of the manuscript. We thank him, Jianrong Li, and Nikhat Parveen for critical reading of the manuscript. We acknowledge the advice of Celia Schiffer in analyzing the subunit interactions at the dimer interface. We also thank Bernard Moss for the recombinant vaccinia virus, Walter Demkowicz for wild-type vaccinia virus, Richard A. Morgan for the pGINT7β-gal plasmid, and Kemal Karaca for B1-Hitchner HN.
This work was made possible by grant AI-49268 from the National Institutes of Health.
REFERENCES
- 1.Connaris, H., T. Takimoto, R. Russell, S. Crennell, I. Moustafa, A. Portner, and G. Taylor. 2002. Probing the sialic acid binding site of the hemagglutinin-neuraminidase of Newcastle disease virus: identification of key amino acids involved in cell binding, catalysis, and fusion. J. Virol. 76:1816-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crennell, S., T. Takimoto, A. Portner, and G. Taylor. 2000. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat. Struct. Biol. 7:1068-1074. [DOI] [PubMed] [Google Scholar]
- 3.Deng, R., Z. Wang, R. L. Glickman, and R. M. Iorio. 1994. Glycosylation within an antigenic site on the HN glycoprotein of Newcastle disease virus interferes with its role in the promotion of membrane fusion. Virology 204:17-26. [DOI] [PubMed] [Google Scholar]
- 4.Deng, R., Z. Wang, P. J. Mahon, M. Marinello, A. M. Mirza, and R. M. Iorio. 1999. Mutations in the NDV HN protein that interfere with its ability to interact with the homologous F protein in the promotion of fusion. Virology 253:43-54. [DOI] [PubMed] [Google Scholar]
- 5.Deng, R., Z. Wang, A. M. Mirza, and R. M. Iorio. 1995. Localization of a domain on the paramyxovirus attachment protein required for the promotion of cellular fusion by its homologous fusion protein spike. Virology 209:457-469. [DOI] [PubMed] [Google Scholar]
- 6.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eucaryotic transient expression system based on recombinant vaccinia virus virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iorio, R. M., J. B. Borgman, R. L. Glickman, and M. A. Bratt. 1986. Genetic variation within a neutralizing domain on the haemagglutinin-neuraminidase glycoprotein of Newcastle disease virus. J. Gen. Virol. 67:1393-1403. [DOI] [PubMed] [Google Scholar]
- 8.Iorio, R. M., and M. A. Bratt. 1983. Monoclonal antibodies to Newcastle disease virus: delineation of four epitopes on the HN glycoprotein. J. Virol. 48:440-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iorio, R. M., G. M. Field, J. M. Sauvron, A. M. Mirza, R. Deng, P. J. Mahon, and J. Langedijk. 2001. Structural and functional relationship between the receptor recognition and neuraminidase activities of the Newcastle disease virus hemagglutinin-neuraminidase protein: receptor recognition is dependent on neuraminidase activity. J. Virol. 75:1918-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iorio, R. M., R. L. Glickman, A. M. Riel, J. P. Sheehan, and M. A. Bratt. 1989. Functional and neutralization profile of seven overlapping antigenic sites on the HN glycoprotein of Newcastle disease virus: monoclonal antibodies to some sites prevent viral attachment. Virus Res. 13:245-262. [DOI] [PubMed] [Google Scholar]
- 11.Iorio, R. M., R. J. Syddall, R. L. Glickman, A. M. Riel, J. P. Sheehan, and M. A. Bratt. 1989. Identification of amino acid residues important to the neuraminidase activity of the HN glycoprotein of Newcastle disease virus. Virology 173:196-204. [DOI] [PubMed] [Google Scholar]
- 12.Iorio, R. M., R. J. Syddall, J. P. Sheehan, M. A. Bratt, R. L. Glickman, and A. M. Riel. 1991. Neutralization map of the hemagglutinin-neuraminidase glycoprotein of Newcastle disease virus: domains recognized by monoclonal antibodies that prevent receptor recognition. J. Virol. 65:4999-5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamb, R. A., and D. Kolakofsky. 1996. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Field's virology, 4th ed. Lippincott-Raven, Philadelphia, Pa.
- 14.Mahon, P. J., R. Deng, A. M. Mirza, and R. M. Iorio. 1995. Cooperative neuraminidase activity in a paramyxovirus. Virology 213:441-444. [DOI] [PubMed] [Google Scholar]
- 15.Mirza, A. M., R. Deng, and R. M. Iorio. 1994. Site-directed mutagenesis of a conserved hexapeptide in the paramyxovirus hemagglutinin-neuraminidase glycoprotein: effects on antigenic structure and function. J. Virol. 68:5093-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mirza, A. M., J. P. Sheehan, L. W. Hardy, R. L. Glickman, and R. M. Iorio. 1993. Structure and function of a membrane anchorless form of the hemagglutinin-neuraminidase glycoprotein of Newcastle disease virus. J. Biol. Chem. 268:21425-21431. [PubMed] [Google Scholar]
- 17.Morrison, T. G., and A. Portner. 1991. Structure, function and intracellular processing of the glycoproteins of Paramyxoviridae, p. 347-382. In D. W. Kingsbury (ed.), The paramyxoviruses. Plenum, New York, N.Y.
- 18.Murrell, M., M. Porotto, T. Weber, O. Greengard, and A. Moscona. 2003. Mutations in human parainfluenza virus type 3 hemagglutinin-neuraminidase causing increased receptor binding activity and resistance to the transition state sialic acid analog 4-GU-DANA (zanamivir). J. Virol. 77:309-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nussbaum, O., C. C. Broder, and E. A. Berger. 1994. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus virus-based assay quantitating cell fusion-dependent reporter gene activation. J. Virol. 68:5411-5422. [DOI] [PMC free article] [PubMed]
- 20.Scheid, A., and P. W. Choppin. 1973. Isolation and purification of the envelope proteins of Newcastle disease virus. J. Virol. 11:263-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheid, A., and P. W. Choppin. 1974. Identification and biological activities of paramyxovirus glycoproteins. Activation of cell fusion, hemolysis and infectivity by proteolytic cleavage of an inactive precursor protein of Sendai virus. Virology 57:475-490. [DOI] [PubMed] [Google Scholar]
- 22.Sheehan, J. P., R. M. Iorio, R. J. Syddall, R. L. Glickman, and M. A. Bratt. 1987. Reducing agent-sensitive dimerization of the hemagglutinin-neuraminidase glycoprotein of Newcastle disease virus correlates with the presence of cysteine at residue 123. Virology 161:603-606. [DOI] [PubMed] [Google Scholar]
- 23.Takimoto, T., G. L. Taylor, H. C. Connaris, S. J. Crennell, and A. Portner. 2002. Role of the hemagglutinin-neuraminidase protein in the mechanism of paramyxovirus-cell membrane fusion. J. Virol. 76:13028-13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanabayashi, K., and R. W. Compans. 1996. Functional interaction of paramyxovirus glycoproteins: identification of a domain in HN that promotes cell fusion. J. Virol. 70:6112-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson, S. D., W. G. Laver, K. G. Murti, and A. Portner. 1988. Isolation of a biologically active soluble form of the hemagglutinin-neuraminidase protein of Sendai virus. J. Virol. 62:4653-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsurodome, M., M. Kawano, T. Yuasa, N. Tabata, M. Nishio, H. Komada, and Y. Ito. 1995. Identification of regions on the hemagglutinin-neuraminidase protein of human parainfluenza virus type 2 important for promoting cell fusion. Virology 213:190-203. [DOI] [PubMed] [Google Scholar]