Abstract
A current view of the emergence of pandemic influenza viruses envisages a gene flow from the aquatic avian reservoir to humans via reassortment in pigs, the hypothetical “mixing vessel.” Understanding arising from recent H5N1 influenza outbreaks in Hong Kong since 1997 and the isolation of avian H9N2 virus from humans raises alternative options for the emergence of a new pandemic virus. Here we report that H9N2 influenza viruses established in terrestrial poultry in southern China are transmitted back to domestic ducks, in which the viruses generate multiple reassortants. These novel H9N2 viruses are double or even triple reassortants that have amino acid signatures in their hemagglutinin, indicating their potential to directly infect humans. Some of them contain gene segments that are closely related to those of A/Hong Kong/156/97 (H5N1/97, H5N1) or A/Quail/Hong Kong/G1/97 (G1-like, H9N2). More importantly, some of their internal genes are closely related to those of novel H5N1 viruses isolated during the outbreak in Hong Kong in 2001. This study reveals a two-way transmission of influenza virus between terrestrial and aquatic birds that facilitates the generation of novel reassortant H9N2 influenza viruses. Such reassortants may directly or indirectly play a role in the emergence of the next pandemic virus.
The introduction and subsequent spread in the human population of influenza A viruses with a novel hemagglutinin (HA) or NA subtype leads to an influenza pandemic. In the last century, four influenza pandemics occurred, resulting in significant mortality and morbidity. Genetic investigations revealed that these pandemic strains were partially or entirely derived from the viruses of avian origin (13, 27, 28) and that most of them first appeared in southern China, a hypothetical influenza epicenter (22).
The accumulated experience of the last decade reminds us that a new influenza pandemic may happen again in the near future since influenza viruses with pandemic potential continue to emerge and reemerge in the southern China region (4, 5, 26). A recent example is the highly pathogenic H5N1 influenza virus which caused an outbreak in the live-poultry markets of Hong Kong (23) and subsequently spread to humans and caused the loss of six human lives in 1997 (26, 29). On this occasion, a pandemic was averted by the depopulation of all live poultry across Hong Kong (24). The continuing outbreaks of H5N1 infections in live-poultry markets of Hong Kong in 2001 (5) and 2002 (authors' unpublished data) highlight the continuing pandemic threat posed by these viruses.
Separate from the H5N1 influenza virus, another subtype of influenza virus, H9N2, has become panzootic in the last decade and has been isolated from different types of terrestrial poultry worldwide (1-3, 9). Two distinct lineages of H9N2 viruses, represented by prototype A/Duck/Hong Kong/Y280/97 (Dk/HK/Y280/97) and A/Quail/Hong Kong/G1/97 (Qa/HK/G1/97) viruses, have become established in terrestrial poultry, predominately in chicken and quail, respectively (7, 9). A Qa/HK/G1/97-like virus is thought to have been involved in the generation of the highly pathogenic H5N1 virus in 1997 (8). H9N2 viruses of each lineage have been isolated from humans (14, 17), and the Dk/HK/Y280/97-like lineage has been isolated from pigs in southern China (16). These H9N2 influenza viruses have acquired receptor-binding specificity to sialic acid with terminal α2-6 Gal linkage found on human cells (10, 15). Thus, H9N2 viruses along with the H5N1 viruses are high on the list of candidates that could potentially cause the next human influenza pandemic.
It has never been possible to be prepared for an influenza pandemic. As a step toward this and to gain a better understanding of the generation of influenza viruses of pandemic potential, live-poultry markets in Shantou, southeastern China, have been investigated for influenza viruses since July 2000. Genetic analysis of representative H9 subtype influenza viruses isolated from ducks revealed that most of the tested duck H9 subtype HAs appear closely related to those of H9 viruses isolated from chickens. However, some of the viruses contain internal genes that are closely related to the genes of the H5N1/97 or Qa/HK/G1-like viruses or to those of the novel H5N1 reassortants isolated in Hong Kong in 2001 (H5N1/01), thus providing evidence that those H5N1/01 viruses might be generated in domestic ducks in the southern region of China. Phylogenetic analyses of each gene segment of these H9N2 influenza viruses from ducks has suggested that they are double or even triple reassortants. Sequence analyses demonstrate that some of these novel reassortant H9N2 viruses in ducks still retain the amino acid signatures in their HA and are therefore compatible with receptor binding preference for humans. This finding raises the possibility of the generation of a pandemic influenza strain directly from aquatic birds in southern China and highlights the need for further study of the influenza ecology in this region as an important component of pandemic preparedness.
MATERIALS AND METHODS
Sampling and virus isolation.
Between July 2000 and November 2001, 498 influenza virus strains were isolated from 2,485 aquatic birds sampled in live-poultry markets in Shantou in the Guangdong Province. Most of these aquatic birds sold in Shantou live-poultry markets, mainly domestic duck, originate from several neighboring provinces such as Jiangxi, Hunan, and Zejiang. Fecal or cloacal swabs were collected from six live-poultry retail markets and one wholesale market. Virus isolates were subtyped by using a panel of reference antisera recommended by the World Health Organization, as described previously (25).
Genetic and phylogenetic analysis.
Viral gene sequencing and analysis were carried out as described previously (5, 6). In brief, viral RNA was extracted from virus-infected allantoic fluids with the RNeasy Mini Kit (Qiagen, Chatsworth, Calif.) and reverse transcribed, and PCR amplification was carried out (primer sequences available upon request). The PCR products were sequenced by using the Big Dye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems) on a Perkin-Elmer model 377 XL DNA sequencer. Sequence data were assembled and edited with the Wisconsin Package Version 10.0 (Genetics Computer Group, Madison, Wis.). Sequence alignment analysis was performed with Genedoc, Version 2.3 (available at http://www.psc.edu/biomed/genedoc/gddl.htm).
For comparison, sequences from H9N2 and H5N1 virus lineages established in southern China since the mid-1990s, including Dk/HK/Y280/97-like, Qa/HK/G1/97-like, Goose/Guangdong/1/96-like (Gs/Gd/96, H5N1), and those newly emerged H5N1 viruses from Hong Kong in 2001 (H5N1/01, genotypes A, B, C, D, and E), were included for phylogenetic analyses (5). In addition, five H9N2 duck viruses isolated in Hong Kong during 1976-1979 were also sequenced (21, 25). In this study, regions used for the phylogenetic analyses are as follows: PB2 1036-2165, PB1 66-1368, PA 1415-2151, HA 43-1047, NP 55-962, NA 41-1393, M 74-889, and NS 39-842.
Nucleotide sequence accession numbers. The nucleotide sequences obtained from this study are available from GenBank under accession numbers AF523372 to AF523519.
RESULTS
In the influenza surveillance studies of Shantou during 2000-2001, 498 influenza virus strains were isolated from aquatic birds, mainly domestic ducks, comprising subtypes H1 to H12 and N1 to N9. Fifty-three of these isolates were of the H9 subtype, an isolation rate around fourfold higher than that found in the 1970s (21, 25). In contrast, chicken and other terrestrial poultry had a more restricted range of subtypes and lineages, largely confined to subtypes H9 and H6 (authors' unpublished data). To understand the relationship between the H9 viruses from ducks and those circulating among terrestrial birds, 14 representative duck H9 viruses were genetically analyzed (Table 1). Twelve of them were identified as H9N2, while two of them were H9N1 viruses.
TABLE 1.
Genotyping of H9 subtype influenza viruses isolated from ducks in southern China (2000-2001)
| Virus | Gene segmenta
|
|||||||
|---|---|---|---|---|---|---|---|---|
| PB2 | PB1 | PA | HA | NP | NA | M | NS | |
| Dk/ST/1796/00 (H9N2) | DK | DK | DK | Y280 | DK | Y280 | Y280 | Y280 |
| Dk/ST/1881/00 (H9N2) | DK | DK | DK | Y280 | DK | Y280 | Y280 | Y280 |
| Dk/ST/2143/00 (H9N2) | DK | DK | DK | Y280 | DK | Y280 | Y280 | Y280 |
| Dk/ST/2144/00 (H9N2) | DK | DK | DK | Y280 | DK | Y280 | Y280 | Y280 |
| Dk/ST/2088/01 (H9N2) | DK | DK | DK | Y280 | DK | Y280 | Y280 | Y280 |
| Dk/ST/830/00 (H9N2) | DK | Y280 | DK | Y280 | DK | Y280 | Y280 | Y280 |
| Ck/ST/1610/01 (H9N2) | DK | Y280 | DK | Y280 | DK | Y280 | Y280 | Y280 |
| Dk/ST/2102/00 (H9N2) | DK | Y280 | DK | Y280 | H5N1/01 E | Y280 | Y280 | Y280 |
| Dk/ST/1042/00 (H9N2) | G1 | G1 | G1 | Y280 | DK | Y280 | G1 | Y280 |
| Dk/ST/1043/00 (H9N2) | G1 | G1 | G1 | Y280 | DK | Y280 | G1 | Y280 |
| Dk/ST/2134/00 (H9N2) | G1 | G1 | DK | Y280 | DK | Y280 | G1 | Y280 |
| WDk/ST/4808/01 (H9N2) | G1 | G1 | (H5N1/01) | Y280 | DK | Y280 | G1 | Y280 |
| Dk/ST/1605/01 (H9N2) | DK | (Gs/Gd) | DK | Y280 | H5N1/01 E | Y280 | Y280 | DK |
| Dk/ST/1588/00 (H9N1) | DK | DK | DK | Korea | DK | (Gs/Gd) | DK | Gs/Gd |
| Dk/ST/2030/00 (H9N1) | DK | DK | DK | Korea | DK | (Gs/Gd) | DK | Gs/Gd |
Genotypes are decided by the phylogenetic relationships in Fig. 1 and 2. Abbreviations: Y280, the lineage of H9N2 viruses established in chicken since 1994; DK, aquatic avian origin; G1, the lineage of H9N2 viruses established in quail and the putative donor of the internal genes for HK156/97-like H5N1 viruses; Gs/Gd, the lineage of Gs/Gd/1/96-like viruses; H5N1/01, the new gene segments detected only in H5N1 viruses from Hong Kong in 2001. Genotypes in parentheses are closely related to prototype gene segments; genotypes without parentheses share the same origin.
Phylogenetic analysis.
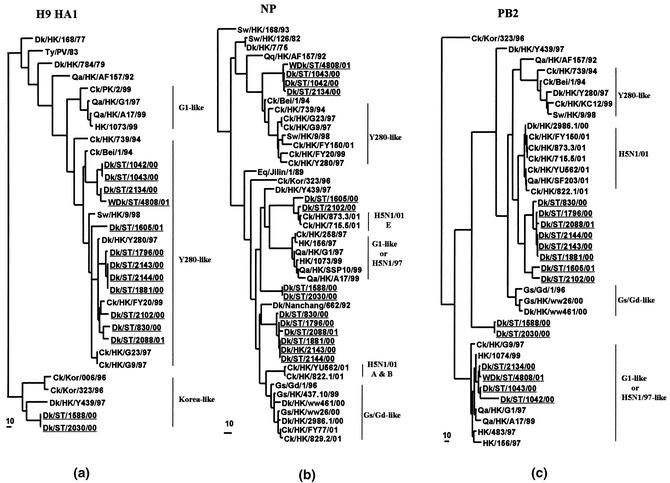
Phylogenetic analysis of the H9 HA1 section shows that all of the H9N2 viruses are incorporated into the lineage represented by Ck/HK/G9/97 or Dk/HK/Y280/97 (Fig. 1a). All of these viruses were derived from CK/BEI/94 or Ck/HK/739/94-like viruses. Topologically, the HA1 genes of four viruses (Dk/ST/2134/00, Dk/ST/1042/00, Dk/ST/1043/00, and WDk/ST/4808/01) are more closely related to Ck/Bei/1/94, which caused the first outbreak in chickens in Guangdong Province in 1994, while another five viruses (Dk/ST/1796/00, Dk/ST/2143/00, Dk/ST/2144/00, Dk/ST/1881/00, and Dk/ST/1605/00) seem to form an intermediate group more closely related to the 1997 isolates, i.e., Dk/HK/Y280/97. There are an additional three viruses (Dk/ST/2088/01, Dk/ST/830/00, and Dk/ST/2102/00) that are more closely related to the recent H9N2 viruses isolated in 1999. This suggests that the H9N2 viruses circulating in chickens in southern China may have, at intervals, been transmitted back to domestic ducks. It is noteworthy that the HA1 of both H9N1 viruses joined the lineage, represented by Dk/HK/Y439/97, similar to those of viruses circulating in chickens in southern Korea since 1996 (Korea-like lineage). None of the Qa/HK/G1/97-like HA was detected among these H9 viruses tested.
FIG. 1.
Phylogenetic trees for the H9 HA1 (a), NP (b), and PB2 (c) genes of influenza A viruses. The nucleotide sequences were analyzed by PAUP using a maximum-parsimony algorithm. The H9 HA1 phylogenetic tree is rooted to Duck/Alberta/60/76 (H12N5). The NP and PB2 gene trees were rooted to A/Equine/Prague/1/57 (H7N7). The lengths of the horizontal lines are proportional to the minimum number of nucleotide differences required to join the nodes. Vertical lines are used for spacing branches and labels. All viruses sequenced in the present studies are underlined, and the remaining sequences can be found in GenBank. Dk, duck; Gs, goose; Ty, turkey; Qa, quail; Ck, chicken; Pg, pigeon; SCk, silky chicken; WDk, wild duck; Sw, swine; Eq, equine; AqB, aquatic bird; HK, Hong Kong; ST, Shantou; Bei, Beijing; Kor, Korea; Ger, Germany; Gd, Guangdong.
Phylogenetic analysis of the N2 gene shows that all of the N2 genes from these duck H9N2 viruses tested, like their HA genes, join and are closely related to the Dk/HK/Y280/97 lineage. These findings further confirm their terrestrial origins. Also, none of the Qa/HK/G1/97-like N2 was detected (data not shown). However, phylogenetic analysis revealed that the N1 genes of two H9N1 viruses of ducks from Shantou, together with Aq/HK/M603/98, clustered into the Gs/Gd/96 virus lineage that differentiates from the HK/97-like lineage (Table 1).
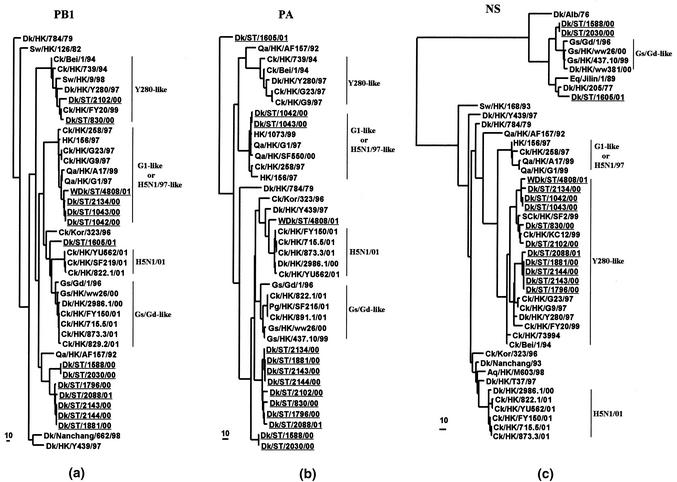
Phylogenetic analyses of the six internal genes revealed that many were of aquatic avian origin. The NP genes of H9N2 viruses form four groups, all of aquatic avian origin (Fig. 1b). It was noted that the NP genes of two H9N2 viruses from ducks in Shantou (Dk/ST/1605/01 and Dk/ST/2102/00) share the same clade with Ck/HK/715.5/01 (H5N1, genotype E). The polymerase genes (PB2, PB1, and PA) are mainly of aquatic avian origin. Of these polymerase genes, four PB2, four PB1, and two PA genes are closely related to the H9N2 viruses of the Qa/HK/G1/97-like lineage (Fig. 1c and 2a and b; Table 1). Like the NP gene tree, the PB2 genes of eight H9N2 duck virus strains from Shantou also joined the same clade with the PB2 of those novel H5N1 viruses associated with the outbreak in poultry markets of Hong Kong in 2001 (Fig. 1c). It is also noted that the PB1 gene of Dk/ST/1605/01 and the PA gene of WDk/ST/4808/01 are also closely related to those of H5N1/01 viruses (Fig. 2a and b).
FIG. 2.
Phylogenetic trees for the PB1 (a), PA (b), and NS (c) genes of influenza A viruses. Evolutionary analyses of nucleotide sequences of PB1, PA, and NS genes were performed as described in the legend to Fig. 1. The sequence ranges were listed in Materials and Methods. The PB1 and PA trees were rooted to B/Lee/40 and B/Singapore/222/79, respectively. The NS gene tree was rooted to A/Equine/Prague/1/56 (H7N7). Virus names and abbreviations can be found in the legend of Fig. 1.
Phylogenetic analysis shows that the NS gene of most of the H9 influenza virus isolated in the gene pool in 2000-2001 is located in the Ck/Bei/1/94 virus lineage (A allele), while two H9N1 (Dk/ST/1588/00 and Dk/ST/2030/00) and one H9N2 (Dk/ST/1605/01) virus lineages belong to the B allele. It is interesting that the NS genes of the H9N1 viruses show the same evolutionary pathway as the Gs/Gd/1/96 virus lineage (Fig. 2c), suggesting that Gs/Gd/1/96-like viruses may be directly derived from the aquatic gene pool in southern China. In contrast to the NS gene tree, the M genes of four H9N2 viruses are closely related to the Qa/HK/G1/97 lineage, while eight viruses are closely related to the Dk/HK/Y280/97 lineage (Table 1). Only the two H9N1 viruses are incorporated into other aquatic avian influenza virus lineages (Table 1).
Taken together, the phylogenetic analyses of all eight gene segments of these H9 influenza viruses isolated from ducks in Shantou revealed that, except for two H9N1 viruses, all of the H9N2 viruses are double or even triple reassortants (Table 1). Among these H9N2 influenza viruses, all of their surface genes (HA and NA) were derived from the Y280-like virus lineage, but most of their RNP gene complexes are of aquatic avian origin. Some of these genes are closely related to those of the newly emerged H5N1/HK/01 viruses (e.g., NP). However, the NS and M genes of these H9N2 duck viruses are mainly related to either Y280-like or G1-like virus lineages (Table 1). Overall, the available information indicates that the H9N2 influenza viruses isolated from ducks in Shantou from 2000 to 2001 arose through interspecies transmission from the terrestrial gene pool in this region. After entering their aquatic hosts, these viruses further reassorted with those influenza viruses originally residing there.
Alignment studies.
The deduced amino acid sequences of the duck H9 HA genes were aligned and compared with those of representative H9 viruses. All H9 duck isolates, except for two H9N1 and one wild-duck H9N2 (WDk/ST/4808) virus lineages, had amino acid residue leucine (L) at position 226 (H3 numbering) and glycine (G) at position 228 at the receptor-binding site (Table 2). Amino acid L at position 226 is found in H9N2 viruses of the Dk/HK/Y280/97 and Ck/HK/G9/97 lineages isolated in Hong Kong and their more recent descendants isolated from chicken (15). However, the H9 subtype viruses isolated from ducks in the 1970s as well as the earliest viruses of the Dk/HK/Y280/97 lineage such as Ck/Bei/1/94 and Ck/HK/739/94 have glutamine (Q) at position 226 (Table 2). The two recent H9N1 viruses (Dk/ST/1588/00 and Dk/ST/2030/00) also have Q at amino acid residue 226. It is also noted that only those H9 viruses established in land-based poultry and the recent H9N2 duck isolates from Shantou maintain the Arg-Ser-Ser-Arg (R-S-S-R) motif at the connecting peptide of their HA. In contrast, the H9 virus isolates obtained from ducks in the 1970s as well as recent duck H9N1 viruses have the X-S-X-R motif (Table 2) (where X represents any amino acid other than Arg, Ser, or Lys). Thus, the genetic and phylogenetic analyses of the H9N2 influenza viruses isolated from domestic ducks in southern China during 2000-2001 provide convincing evidence that the H9N2 influenza virus lineages established since the mid-1990s in chicken and quail (7, 8) have been transmitted back to domestic ducks, thereby generating double or triple reassortants with influenza viruses resident in ducks.
TABLE 2.
Comparison of the amino acid sequences of the HA of the H9 subtype of representative viruses isolated in southern China (1976 to 2001)
| Virusa | RBSb
|
(←HA1) connecting peptide
|
||||
|---|---|---|---|---|---|---|
| 226 | 228 | −4 | −3 | −2 | −1 | |
| Ck/Bei/1/94 | Q | G | R | S | S | R |
| Ck/HK/G9/97 | L | G | R | S | S | R |
| Ck/HK/G1/97 | L | G | R | S | S | R |
| Dk/ST/830/00 | L | G | R | S | S | R |
| WDk/ST/4808/00 | Q | G | R | S | S | R |
| Dk/HK/Y439/97 | Q | G | A | S | N | R |
| Dk/ST/1588/00 | Q | G | A | S | D | R |
| Dk/HK/86//76 | Q | G | A | S | G | R |
| Dk/HK/289/78 | Q | G | A | S | N | R |
| DK/HK/366/78 | Q | G | V | S | N | R |
| Dk/HK/610/79 | Q | G | A | S | G | R |
| Dk/HK/552/79 | Q | G | V | S | N | R |
Only representative viruses are chosen. Ck/HK/739/94 is similar to Ck/Bei/1/94; Dk/HK/Y280/97 is similar to Ck/HK/G9/97; Dk/ST/830 represents the other 11 duck H9N2 viruses isolated in Shantou; Qa/HK/G1/97 and Ck/HK/G9/97 represent two major lineages of H9N2 viruses established in terrestrial poultry in southern China. Dk/ST/1588/00 represents the H9N1 viruses Dk/ST/2030/00.
RBS, receptor binding site.
DISCUSSION
Aquatic birds are recognized as natural gene reservoirs of influenza A viruses around the world (1, 18, 28). Interspecies transmission usually happens from aquatic birds to terrestrial birds or to mammals such as pigs, horses, and even humans. These interspecies transmissions establish a very complex ecological system in nature. Since most of these interspecies transmission events were identified and associated with zoonotic outbreaks in new hosts, influenza gene flow was usually considered to occur from aquatic birds to other animal species. This idea inhibited our understanding of the whole influenza ecology system and the emergence of new influenza viruses with pandemic propensity.
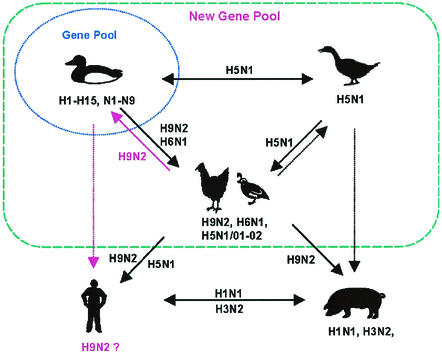
Antigenic and genetic analyses of the H9N2 influenza viruses isolated from domestic ducks in southern China during 2000-2001 provide convincing evidence that the H9N2 influenza virus lineages established since the mid-1990s in chicken and quail (8) have been transmitted back to domestic ducks, generating double or triple reassortants with influenza viruses already resident in ducks. Rather than indicating a one-way flow of H9N2 viruses and their genes from aquatic to terrestrial poultry as previously recognized (7, 9), these findings indicated reverse transmission back to duck, i.e., there is a two-way transmission between terrestrial and aquatic birds (Fig. 3). This resulted in the generation of multiple genotypes of H9N2 viruses containing internal genes of aquatic avian origin. Further interspecies transmission of these reassortants to other hosts may subsequently occur. This parallels the reemergence of H5N1 influenza viruses in Hong Kong in 2001 (5) and 2002 (authors' unpublished data). The lessons learned from these H5N1 incidents are that when an established virus lineage such as the A/Goose/Guangdong/1/96-like virus is reintroduced into the duck (6), it leads to the generation of multiple reassortants with new aquatic gene segments (5). Novel reassortants are biologically unstable (12) and may transmit further to other hosts—in this case terrestrial poultry, lower mammals, and possibly humans (see below). This may be one method by which the next pandemic virus will emerge.
FIG. 3.
Current understanding of the ecology of influenza virus in southern China that favors the emergence of H9N2 as a possible new pandemic influenza virus. The concept of the classic gene pool and the interplay between aquatic birds and influenza viruses may be explained by frequent interspecies transmission between terrestrial and aquatic poultry in the southern region of China.
Phylogenetic analyses of the internal genes of the H9N2 virus showed that some of them are closely related to the internal gene complex found in H5N1/01 viruses. This information suggests that the precursors of H5N1/01 viruses are still circulating in the gene pool in this region and are reassorting promiscuously. This finding also provides an example of the interaction between those established lineages in terrestrial or semiaquatic poultry and the domestic duck. In the long term, this kind of interaction will increase virus diversity in the ecosystem in southern China.
The findings of the present investigation suggest that these H9N2 reassortant viruses may have pandemic potential since they contain a receptor-binding profile (10, 15) that favors infection of humans and that some (e.g., the Qa/HK/G1/97-like lineage) have gene segments previously associated with human disease (14, 17). As H9N2 influenza viruses are not highly pathogenic for poultry, it makes them more, rather than less, likely to be of pandemic relevance. In fact, viruses that are less pathogenic for poultry have a greater opportunity to become widespread since they do not raise concern and permit their hosts to survive unhindered. Thus, they are free to continue to reassort and are more likely to have the opportunity to find the best gene constellation (19, 28) that permits infection of humans and facilitates further person-to-person transmission. The connecting peptides of the HA of the 1968 H3N2 and 1957 H2N2 pandemic viruses indicate that they are unlikely to have been highly pathogenic for poultry. Two-way transmissions between different types of poultry in southern China increase the opportunity to generate influenza viruses with pandemic potential. Our findings demonstrate that such viruses may be directly generated from ducks.
It was noteworthy that the NS genes of two H9N1 viruses are closely related to the counterparts in the Gs/Gd/1/96 virus. This fact suggests that Gs/Gd/1/96-like H5N1 influenza virus might also be generated from ducks in this region. As more sequence data have been accumulated, precursors of important emerging influenza viruses can be identified. Southern China is considered to be an influenza epicenter mainly because the region was the site of two previous pandemics (Asian and Hong Kong influenza) (22). The current status of the influenza ecosystem in this region makes it very likely that the next pandemic virus will also be generated in this region (Fig. 3).
One remaining question is why the HA and NA genes of Qa/HK/G1/97-like H9N2 viruses have not been recently detected in ducks even though some of their internal gene segments were found incorporated into some Ck/Bei/1/94-like duck viruses. One explanation is that the HA and NA genes of the Qa/HK/G1/97-like virus are no longer adaptable to aquatic birds since the viruses have become more adapted to terrestrial poultry. Another possible reason is that we sampled ducks from specific regions where Qa/HK/G1-like viruses were not locally prevalent. It is believed that these gene segments were acquired in terrestrial birds, since reassortment events are frequently observed between two established H9N2 lineages in quail isolates (authors' unpublished data).
Virus gene movements in both directions between different types of poultry seem to be not uncommon in this region in the recent past. The species barriers between the birds have become much more permeable than previously anticipated. Increasing the heterogeneity of influenza viruses in these hosts results in an enlarged and dynamic influenza gene pool in continuous flux rather than one that is limited to aquatic birds and therefore in evolutionary stasis (Fig. 3). The diversity of genotypes, gene constellations, and host receptor specificities will provide these viruses and their progeny with multiple options of hosts (11, 12, 19, 20). Such a situation would conceivably increase the opportunity for the generation of a new pandemic influenza virus.
Acknowledgments
This study was supported by the World Health Organization (HQ/00/428534), Li Ka-Shing Foundation, Wellcome Trust grant 067072/D/02/Z, Public Health Research grant A195357 from the National Institute of Allergy and Infectious Diseases, and the Research Grant Council of Hong Kong (7334/01 M) from the Hong Kong SAR Government.
We gratefully acknowledge the excellent technical assistance of L. J. Zhang from the Department of Microbiology and of C. Cheung from the Computer Center at the University of Hong Kong.
REFERENCES
- 1.Alexander, D. J. 2001. Ecology of avian influenza in domestic birds, p. 25-33. In B. Dodet and M. Vicari (ed.), Emergence and control of zoonotic ortho- and paramyxovirus diseases. John Libbey Eurotext, Paris, France.
- 2.Banks, J., E. C. Speidel, P. A. Harris, and D. J. Alexander. 2000. Phylogenetic analysis of influenza A viruses of H9 haemagglutinin subtype. Avian Pathol. 29:353-360. [DOI] [PubMed] [Google Scholar]
- 3.Cameron, K. R., V. Gregory, J. Banks, I. H. Brown, D. J. Alexander, and Y. P. Lin. 2000. H9N2 subtype influenza viruses in poultry in Pakistan are closely related to the H9N2 viruses responsible for human infection in Hong Kong. Virology 278:36-41. [DOI] [PubMed] [Google Scholar]
- 4.de Jong, J. C., E. C. Claas, A. D. Osterhaus, R. G. Webster, and W. L. Lim. 1997. A pandemic warning? Nature 389:554.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan, Y., J. S. M. Peiris, A. S. Lipatov, T. M. Ellis, K. C. Dyrtings, S. Krauss, L. J. Zhang, R. G. Webster, and K. F. Shortridge. 2002. Emergence of multiple genotypes of H5N1 avian influenza viruses in Hong Kong SAR. Proc. Natl. Acad. Sci. USA 99:8950-8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guan, Y., J. S. M. Peiris, K. F. Kong, K. C. Dyrting, T. M. Ellis, T. Sit, L. J. Zhang, and K. F. Shortridge. 2002. H5N1 influenza viruses isolated from geese in southeastern China: evidence for genetic reassortment and interspecies transmission to ducks. Virology 292:16-23. [DOI] [PubMed] [Google Scholar]
- 7.Guan, Y., K. F. Shortridge, S. Krauss, P. S. Chin, K. C. Dyrting, T. M. Ellis, R. G. Webster, and M. Peiris. 2000. H9N2 influenza viruses possessing H5N1-like internal genomes continue to circulate in poultry in southeastern China. J. Virol. 74:9372-9380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan, Y., K. F. Shortridge, S. Krauss, and R. G. Webster. 1999. Molecular characterization of H9N2 influenza viruses: were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc. Natl. Acad. Sci. USA 96:9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo, Y., J. Dong, M. Wang, Y. Zhang, J. Guo, and K. Wu. 2001. Characterization of hemagglutinin gene of influenza A virus subtype H9N2. Chin. Med. J. (Engl. Ed.) 114:76-79. [PubMed] [Google Scholar]
- 10.Ha, Y., D. J. Stevens, J. J. Skehel, and D. C. Wiley. 2001. X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs. Proc. Natl. Acad. Sci. USA 98:11181-11186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatta, M., P. Gao, P. Halfmann, and Y. Kawaoka. 2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293:1840-1842. [DOI] [PubMed] [Google Scholar]
- 12.Holland, J. J., J. C. de la Torre, D. K. Clarke, and E. Duarte. 1991. Quantitation of relative fitness and great adaptability of clonal populations of RNA viruses. J. Virol. 65:2960-2967. [DOI] [PMC free article] [PubMed]
- 13.Kawaoka, Y., S. Krauss, and R. G. Webster. 1989. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J. Virol. 63:4603-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin, Y. P., M. Shaw, V. Gregory, K. Cameron, W. Lim, A. Klimov, K. Subbarao, Y. Guan, S. Krauss, K. Shortridge, R. Webster, N. Cox, and A. Hay. 2000. Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc. Natl. Acad. Sci. USA 97:9654-9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matrosovich, M. N., S. Krauss, and R. G. Webster. 2001. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology 281:156-162. [DOI] [PubMed] [Google Scholar]
- 16.Peiris, J. S. M., Y. Guan, D. Markwell, P. Ghose, R. G. Webster, and K. F. Shortridge. 2001. Cocirculation of avian H9N2 and contemporary “human” H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J. Virol. 75:9679-9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peiris, M., K. Y. Yuen, C. W. Leung, K. H. Chan, P. L. S. Ip, R. W. M. Lai, W. K. Orr, and K. F. Shortridge. 1999. Human infection with influenza H9N2. Lancet 354:916-917. [DOI] [PubMed] [Google Scholar]
- 18.Rohm, C., N. Zhou, J. Suss, J. Mackenzie, and R. G. Webster. 1996. Characterization of a novel influenza hemagglutinin, H15: criteria for determination of influenza A subtypes. Virology 217:508-516. [DOI] [PubMed] [Google Scholar]
- 19.Scholtissek, C., H. Burger, O. Kistner, and K. F. Shortridge. 1985. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology 147:287-294. [DOI] [PubMed] [Google Scholar]
- 20.Scholtissek, C., S. Ludwig, and W. M. Fitch. 1993. Analysis of influenza A virus nucleoproteins for the assessment of molecular genetic mechanisms leading to new phylogenetic virus lineages. Arch. Virol. 131:237-250. [DOI] [PubMed] [Google Scholar]
- 21.Shortridge, K. F. 1992. Pandemic influenza—a zoonosis? Sem. Resp. Infect. 7:11-25. [PubMed] [Google Scholar]
- 22.Shortridge, K. F., and C. H. Stuart-Harris. 1982. An influenza epicentre? Lancet ii:812-813. [DOI] [PubMed]
- 23.Shortridge, K. F., N. N. Zhou, Y. Guan, P. Gao, T. Ito, Y. Kawaoka, S. Kodihalli, S. Krauss, D. Markwell, K. G. Murti, M. Norwood, D. Senne, L. Sims, A. Takada, and R. G. Webster. 1998. Characterization of avian H5N1 influenza viruses from poultry in Hong Kong. Virology 252:331-342. [DOI] [PubMed] [Google Scholar]
- 24.Shortridge, K. F., M. Peiris, Y. Guan, K. Dyrting, T. Ellis, and L. Sims. 2001. H5N1 virus: beaten, but is it vanquished? p. 91-97. In B. Dodet and M. Vicari (ed.), Emergence and control of zoonotic ortho- and paramyxovirus diseases. John Libbey Eurotext, Paris, France.
- 25.Shortridge, K. F., W. K. Butterfield, R. G. Webster, and C. H. Campbell. 1977. Isolation and characterization of influenza A viruses from avian species in Hong Kong. Bull. W. H. O. 55:15-19. [PMC free article] [PubMed] [Google Scholar]
- 26.Subbarao, K., A. Klimov, J. Katz, H. Regenery, W. Lim, H. Hall, M. Perdue, D. Swayne, C. Bender, J. Huang, M. Hemphill, T. Rowe, M. Shaw, X. Xu, K. Fukuda, and N. Cox. 1998. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 279:393-396. [DOI] [PubMed] [Google Scholar]
- 27.Taubenberger, J. K., A. H. Reid, A. E. Krafft, K. E. Bijwaard, and T. G. Fanning. 1997. Initial genetic characterization of the 1918 “Spanish”influenza virus. Science 275:1793-1796. [DOI] [PubMed] [Google Scholar]
- 28.Webster, R. G., W. J. Bean, O. T. Gorman, T. M. Chambers, and Y. Kawaoka. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuen, K. Y., K. S. Chan, M. Peiris, D. N. C. Tsang, T. L. Que, K. F. Shortridge, P. T. Cheung, W. K. To, E. T. F. Ho, R. Sung, A. F. B. Cheng, and members of the H5N1 Study Group. 1998. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet 351:467-471. [DOI] [PubMed] [Google Scholar]