Abstract
Latent infection with varicella-zoster virus (VZV) is characterized by restricted virus gene expression and the absence of virus production. Of the ∼70 predicted VZV genes, only five (genes 4, 21, 29, 62, and 63) have been shown by multiple techniques to be transcribed during latency. IE62, the protein product of VZV gene 62, is the major immediate-early (IE) virus-encoded transactivator of viral gene transcription and plays a pivotal role in transactivating viral genes during lytic infection. The protein kinase (66-pk) encoded by VZV gene 66 phosphorylates IE62, resulting in cytoplasmic accumulation of IE62 that mitigates nuclear IE62-induced gene activation. Analysis of latently infected human trigeminal ganglia for 66-pk expression by reverse transcriptase-dependent nested PCR, including DNA sequence analysis, in situ hybridization, and immunohistochemistry, revealed VZV open reading frame 66 to be a previously unrecognized latently expressed virus gene and suggests that prevention of IE62 import to the nucleus by VZV 66-pk phosphorylation is one possible mechanism by which VZV latency is maintained.
After primary infection, varicella-zoster virus (VZV) becomes latent in cranial nerve, dorsal root, and autonomic ganglia along the entire human neuraxis, and no virus is produced. Latent virus DNA assumes an “endless” (episomal) state from which only a few of the approximately 70 predicted virus genes are transcribed (1, 26). cDNA sequences mapping to the 3′ terminus of VZV genes 21, 29, 62, and 63 have been detected in latently infected human trigeminal ganglia (TG). In situ hybridization (ISH) has also identified transcripts mapping to VZV open reading frames (ORFs) 4 and 18 (5, 7, 11, 12, 22), and immunohistochemistry has demonstrated the presence of VZV gene 63 protein in latently infected ganglia (20, 21). There is also one unconfirmed report of proteins corresponding to VZV genes 4, 21, 29, and 62 in latently infected human ganglia (20).
IE62, the essential immediate-early protein product of VZV gene 62, stimulates transcription from all VZV promoters tested (2, 14). IE62 increases the infectivity of transfected VZV DNA (23). IE62 localizes to the nucleus early in virus infection and later accumulates in the cytoplasm (10, 16). Interestingly, this protein is localized predominantly in the cytoplasm during latency (20). Phosphorylation of IE62 by the protein kinase encoded by VZV ORF 66 (66-pk) results in the cytoplasmic accumulation of IE62 (19). While cytoplasmic IE62 is incorporated into enveloped virions (17, 18), the restriction of IE62 from the nucleus also regulates the function of this potent activator of gene transcription (18). Thus, phosphorylation of IE62 by 66-pk appears to be one possible mechanism by which IE62 remains present within a cell without stimulating gene transcription.
We report here the detection of 66-pk expression in latently infected human TG by reverse transcriptase (RT)-PCR, by ISH, and by immunohistochemistry, raising the possibility that a block in nuclear import of IE62 by 66-pk is a crucial determinant of VZV latency.
MATERIALS AND METHODS
Human tissue.
Subjects for this study were from the United States and the United Kingdom. A total of 10 subjects from the Denver, Colo., area and 17 subjects from the Scotland, United Kingdom area were coded for anonymity. In all cases, ganglia were removed within 24 h of death from subjects without cutaneous signs of herpesvirus infection, without clinical evidence of immunosuppression, and who had not received steroids or cytotoxic drugs at least 1 month before death. Ganglia were removed aseptically, and after removal of the nerve roots, ganglia were either flash-frozen in liquid nitrogen for nucleic acid extraction (3) or fixed in formalin or 4% paraformaldehyde in lysine-buffered sodium m-periodate for in situ studies (11, 13).
Nucleic acid extraction, cDNA synthesis, and PCR analysis.
Nucleic acids were extracted as described previously, using DNase treatment of all RNA preparations (3). Reverse transcription of 1 μg of RNA primed with either d(T) or anchor-d(T) (Table 1) was carried out as described elsewhere (6). Oligonucleotide primers for cDNA synthesis and PCR amplification (Table 1) were commercially prepared (IDT, Coralville, Iowa). PCR cycle times and temperatures have been described previously (6). All PCRs include a no-template (water) control, which was uniformly negative. When necessary, PCR products were resolved on 2% agarose gels, transferred to membranes (Zeta probe; Bio-Rad Laboratories, Hercules, Calif.), and probed with 32P-end-labeled DNA oligonucleotides (5). The DNA sequences of PCR products were determined (Macrogene, Seoul, Korea) after ultrafiltration (Micron PCR; Millipore Corp., Bedford, Mass.).
TABLE 1.
Oligonucleotide primers for cDNA synthesis and PCR amplification
| Primer name | 5′ to 3′ sequence | Directiona | Locationb |
|---|---|---|---|
| 66F1 | GTTTGGCAAAACGGTCTTCTCG | Sense | 114,013 |
| 66F2 | CAGGCCATTGTGGACAAATC | Sense | 114,047 |
| 66R2 | GGTCGATGACGTGCGTCAAAC | Antisense | 114,136 |
| 66R1 | GTGGTTAAGCAACACCTCTGC | Antisense | 114,161 |
| ORF 66-P1 | GTGACAGTGAGCGTGAAATTAAAC | Sense | 113,911 |
| ORF 66-P2 | CGACGATCTGGAACTCATCCC | Sense | 113,943 |
| ORF 66-P3 | GGTTTGGCAAAACGGTCTTC | Sense | 114,012 |
| 66-AT-F | CCGTGGACATATGGAGTGCC | Sense | 113,821 |
| 66-AT-R | AAGCAGTGGTAACAACGCAGAGTTATACAATATTTTATTAACAGG | Antisense | 114,251 |
| ORF 66 termc | 114,215 | ||
| ORF 66 PASd | 114,236 | ||
| ORF 40-P1 | TCACACACAATCGGATGTTGC | Sense | 75,280 |
| ORF 40-P2 | ATACGGTGACAGGCTATACAACGGAA | Sense | 75,349 |
| ORF 40-P3 | CCACCACTATGCTCAAGCGAT | Sense | 75,593 |
| 40AT-F | CATGCAATCCTAGAGGTCGAGC | Sense | 75,201 |
| 40AT-R | AAGCAGTGGTAACAACGCAGAGTTTATCGCGGAAGAGGAAGAC | Antisense | 75730 |
| ORF 40 term | 75,728 | ||
| ORF 40 PAS | 75,758 | ||
| 62-AS | GCAAGACGTTTGGTCTTACGAATC | Antisense | 107,836 |
| 62-S | GAGTTTGTTTCGTCTTCATCCTCT | Sense | 108,087 |
| Actin-F1 | GATGCATTGTTACAGGAAGT | Sense | 3,260 |
| Actin-R1 | TCATACATCTCAAGTTGGGG | Antisense | 3,500 |
| Actin term | 3,000 | ||
| Actin PAS | 3,646 | ||
| d(T) | (T)15 | NAe | NA |
| Anchor-d(T) | AAGCAGTGGTAACAACGCAGAGT-(T)15 | NA | NA |
| Anchor | AAGCAGTGGTAACAACGCAGAGT | NA | NA |
ISH and immunohistochemistry.
ISH for VZV RNA using gene-specific oligonucleotides (Table 1) and immunohistochemistry with diluted (1:100) rabbit anti-VZV gene 66 serum were performed as described elsewhere (11).
Antibody production.
The VZV ORF 66 derived from the plasmid PKCMVHA66 (19) was cloned into the pMalCR1 vector (New England Biolabs, Beverly, Mass.), and the maltose binding protein-ORF 66 gene fusion was expressed and purified using amylose affinity chromatography. The purified protein was used to raise antibody to 66-pk in rabbits as described previously (15). The antibody reacts with a 55-kDa protein in Western immunoblotting and stains both the nucleus and cytoplasm of VZV-infected cells (P. R. Kinchington, unpublished data).
RESULTS
VZV ORF 66 transcription in latently infected human TG.
VZV ORF 66 is a contiguous 1,179-bp segment located entirely within the unique short region of the virus genome and encodes a Ser/Thr-specific protein kinase (14). Sequence analysis predicts a poly(A) addition signal 26 bp downstream from the termination codon (8). Initially, six TG were processed separately for DNA and RT-PCR analysis. Serial log10 dilutions of VZV DNA were used to demonstrate the sensitivity of ORF 66 detection by PCR (data not shown). Primary PCR using 66F1 and -R1 primers detected 106 copies, and nesting with primers 66F2 and -R2 increased the sensitivity to 104 copies of purified VZV DNA. The sensitivity of nested PCR increased following probing with 32P-labeled, internally located oligonucleotide; however, since our ultimate goal was to obtain DNA sequence, Southern blot analysis was not performed. DNA extracted from all samples contained both actin- and VZV-specific sequences. Analysis of total RNA extracted from each TG for the presence of residual DNA indicated that amplification of the 240-bp fragment using the actin-F1 and -R1 primers was dependent upon reverse transcription in all RNA samples. VZV gene 66 transcripts were detected in two of six individual TG samples, and this amplification was also RT dependent. These results strongly indicate the presence of ORF 66-specific transcripts in latently infected TG.
Verification of latent VZV ORF 66 transcription by DNA sequence analysis.
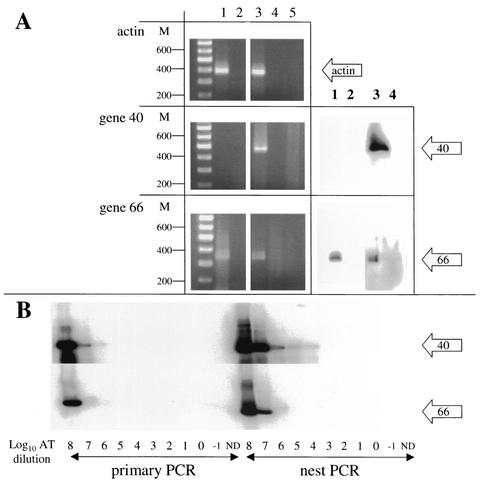
To identify the 3′-terminal sequences of VZV gene 66 transcripts in latently infected human TG, cDNA was synthesized from RNA extracted from the pooled left and right TG of a single subject and analyzed by PCR. Priming cDNA synthesis with the anchor-d(T) primer resulted in modified cDNA, allowing subsequent PCR amplification across the 3′ terminus, including the poly(A) tail. In preliminary experiments, PCR amplification of cDNA synthesized from VZV-infected MeWo cells produced DNA fragments migrating in ethidium bromide-stained agarose gels to the approximate size position expected with primers actin-F1 and anchor (386 bp), ORF 40-P2 and anchor (447 bp), and ORF 66-P2 and anchor (336 bp) (Fig. 1A, left panels, lane 3) and was verified by hybridization with the internally located 32P-end-labeled oligonucleotides (Fig. 1A, right panels, lane 3). In separate experiments, actin and VZV gene 66 transcripts were amplified from latently infected human TG cDNA (Fig. 1A, lane 1). In both instances, the amplification products depended on reverse transcription (Fig. 1A, lane 2). No amplification product was detected when VZV gene 40-specific primers were used (Fig. 1A, lane 1). VZV gene 40 encodes the major capsid protein of the virus, and the lack of gene 40 transcription suggests that VZV had not reactivated in the human ganglia removed at autopsy (5).
FIG. 1.
PCR amplification of VZV cDNA. (A) Total TG RNA extracted was reverse transcribed with (lane 1) or without (lane 2) RT. In a second reaction, VZV-infected MeWo cell RNA was reverse transcribed with (lane 3) or without (lane 4) RT. Each cDNA product was amplified with primers designed to yield products spanning the poly(A) tract. Amplified products were resolved by agarose gel electrophoresis and stained with ethidium bromide (left). VZV-specific amplification products were transferred to nylon membranes and probed with 32P-end-labeled, internally located oligonucleotides (right). Both TG and VZV-infected MeWo cell cDNA contained actin transcripts (top gels); VZV gene 40 transcripts were detected only in virus-infected MeWo cell cDNA (middle gels); and VZV gene 66 transcripts were detected in TG and VZV-infected MeWo cell cDNA (bottom gels). In all instances, amplification was RT dependent (lanes 1 and 3). No product was seen when RT (lanes 2 and 4) or template DNA (lane 5) was omitted from the PCR. M, double-stranded DNA size markers. (B) Primer efficiencies were determined by PCR amplification of serial log10 dilutions of VZV ORF 40 and ORF 66 artificial transcripts (AT). In the primary PCR, both ORF primer sets gave strong signals with 108 copies of AT. In nested PCR, the ORF 40 primer set detected 104 to 105 copies of AT, while ORF 66 primers detected 106 to 107 of added AT. ND, no AT template DNA added to the PCR.
PCR primer efficiency was determined by amplification of known numbers of artificial transcripts. Artificial transcripts were constructed by amplification of VZV DNA with 66AT-F and -R and 40AT-F and -R. These fragments extend ∼85 nucleotides (nt) upstream from the location of the primers used to detect the transcripts (ORF 40- and 66-P1) and extended to the respective ORF termination codon. The artificial transcripts were also modified by incorporating the anchor primer site at the 3′ end of the DNA fragments. Following PCR amplification, the artificial transcripts were cleaned and quantitated, and serial log10 dilutions were PCR amplified using the same primer sets as were employed to generate those shown in Fig. 1A. The amplification products were transferred to nylon-based membranes and probed with 32P-end-labeled ORF 40- or 66-P3. The results demonstrate that ORF 40 primer sets were ∼1 order of magnitude more efficient than the ORF 66 primer sets.
In separate experiments, the 3′-terminal cDNA sequence was determined for gene 66 transcripts from VZV-infected MeWo cells and latently infected human TG (Table 2). Both lytic and latent gene 66 transcripts terminate at nearly identical sites: 114,238 nt (lytic) and 114,236 nt (latent). This very slight difference in poly(A) addition sites has been seen in latent gene 21 and 63 transcription (5) and most likely reflects inherent inaccuracies in poly(A) addition or slight virus strain differences, as opposed to a mechanism of latent gene transcriptional control. However, both lytic and latent gene 66 termination sites are located within the poly(A) stretch, which suggests that alternative signal sites to those of the proposed polyadenylation signal (8) are employed. Two such potential alternative poly(A) addition signaling sites are located within 30 bp of the 3′ end of the transcript (Table 2).
TABLE 2.
3′ cDNA sequence of ORF 66 transcripts
| Nucleotides and source | Sequence |
|---|---|
| nta 114, 153 | |
| TGb | CTT AAC CAC TCT GTT TTC CAA ACT CTT CCC GAT CCA TAT |
| cDNAc | CTT AAC CAC TCT GTT TTC CAA ACT CTT CCC GAT CCA TAT |
| Genomicd | CTT AAC CAC TCT GTT TTC CAA ACT CTT CCC GAT CCA TAT |
| nt 114,192 | |
| TG | CCA AAT CCA ATG GAA GTT GGA GAT TAA |
| cDNA | CCA AAT CCA ATG GAA GTT GGA GAT TAA |
| Genomic | CCA AAT CCA ATG GAA GTT GGA GAeT TAA |
| nt 114,216 | |
| TG | AATTC ATTAA GCCTGTT AAAAAAAAAAAAAAAAAAA |
| cDNA | AATTC ATTAA GCCTGTT AAT AAAAAAAAAAAAAAAAA |
| Genomic | AATTC ATTAA GCCTGTT AfATAAAATATTGTATAAATTGTG |
Nucleotide number relative to the VZV genomic sequence (8).
cDNA sequence obtained from latently infected human TG.
cDNA sequence obtained from VZV-infected MeWo cells.
Genomic VZV DNA sequence (8).
Proposed poly(A) addition signal (single underlined).
Predicted poly(A) addition signal (double underlined) as identified previously (8).
Detection of VZV transcription by ISH.
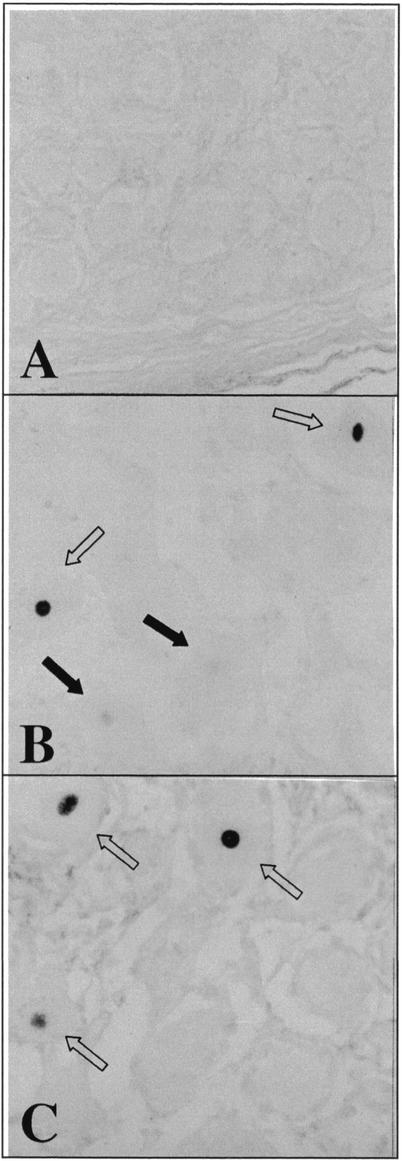
A total of 27 ganglia (26 TG and 1 dorsal root ganglion) from 22 individuals were analyzed by ISH for transcripts corresponding to VZV genes 66 and 62. In serial sections from five ganglia (four TG and one dorsal root ganglion), both viral gene transcripts were detected exclusively in the nuclei of neurons. In one instance, RNA for VZV gene 62 was detected in the absence of gene 66 transcripts. Figure 2 shows the presence of both VZV gene 66 and 62 transcripts in TG neurons. Note the difference in signal strengths obtained within the same section using the same probe. This may indicate (i) each neuron has different amounts of latent VZV, (ii) some neurons are showing early reactivation, or (iii) a technical variation indicating unequal probe access. The coded samples were harvested within 24 h postmortem; however, the precise interval between death and autopsy is unknown. Therefore, correlations between ISH signal strength and harvest time could not be obtained.
FIG. 2.
Detection of VZV transcripts by ISH. Adjacent sections of a latently infected human TG were hybridized to digoxigenin-labeled VZV-specific probes (66F1, 66R1, and 62-AS; Table 1). (A and B) No signal was detected after hybridization with a sense VZV gene 66 probe (A), whereas a strong signal was seen in the nuclei of two neurons (open arrows) and a weaker signal was seen in two neurons (dark arrows) after hybridization with an antisense VZV gene 66 probe (B). (C) A strong signal in three neurons (open arrows) was also detected after hybridization with an antisense probe to VZV gene 62. Magnification, ×450.
VZV gene 66 protein expression.
Immunohistochemical analysis of 11 latently infected TG from seven subjects revealed gene 66 protein in 3 TG exclusively in the cytoplasm of neurons. Figure 3A demonstrates VZV gene 66 protein in the cytoplasm of two TG neurons; no VZV gene 66 protein-specific staining was detected in the TG of another subject (Fig. 3B). Analysis of TG from both of these subjects revealed VZV gene 66 transcripts only in the former.
FIG. 3.
Immunohistochemical detection of VZV ORF 66 protein. TG sections from two subjects were incubated with antibody to VZV gene 66 protein as described in Materials and Methods. (A and B) Strong labeling in the cytoplasm of one neuron (open arrow) and weaker labeling in another neuron (closed arrow) from the first subject was detected (A), whereas no signal was detected in the second subject (B). TG sections from the first subject but not the second subject were positive for VZV gene 66 transcripts by ISH (data not shown). (C and D) VZV gene 66 protein was present in VZV-infected MeWo cells (C), but not in uninfected MeWo cells (D). Images were captured at a magnification of ×427.5; panel B was printed at a magnification of ×541.5.
DISCUSSION
Three separate methods were used to study VZV gene 66 expression in latently infected human TG. RT-dependent PCR and DNA sequence analysis revealed the 3′ terminus of latently transcribed gene 66, including its poly(A) tail, ∼20 nt distal to the termination codon; ISH identified 66-pk transcripts in the nuclei of latently infected neurons; and immunohistochemistry detected 66-pk protein exclusively in the cytoplasm of latently infected neurons. The sequence analysis and cellular localization of 66-pk within human TG parallels previous analyses of VZV transcripts in latently infected human ganglia (5, 11, 12). Together, the results indicate that VZV gene 66 is both transcribed and translated in latently infected human ganglia.
Given the presence of at least 70 VZV ORFs, the extent of virus transcription in latently infected human ganglia is obviously restricted. The presence of gene 62 transcripts (5, 22) and, by one account, VZV gene 62-encoded protein (20) during latency coupled with the function of IE62 in transcriptional activation (2, 4, 23) suggest that the functional activity of IE62 during latency is modulated. We recently showed that 21p and 29p, proteins encoded by VZV genes 21 and 29, do not affect IE62-induced transactivation of VZV gene 20, 21, 28, or 29 promoters in transfection studies (4). Since activation of VZV gene transcription by IE62 most likely occurs in the nucleus, nuclear import of IE62 may be a critical event in gene transactivation, and this can clearly be regulated. Kinchington et al. (16, 18, 19) showed that phosphorylation of IE62 by 66-pk leads to cytoplasmic accumulation of IE62. While the phosphorylation of IE62 late in VZV lytic infection appears to be a mechanism by which IE62 is incorporated into progeny virions (18), the cytoplasmic location of IE62 during latency that results from 66-pk phosphorylation might also modulate IE62-dependent gene transactivation.
IE62 is an essential, immediate-early protein pivotal to the production of infectious virus (10, 25). Thus, it is surprising that IE62 transcripts are detected during latency. In this report, VZV gene 66 transcripts were detected by independent means in two separate laboratories using autopsy samples collected from separate population pools. The known effect of 66-pk on the cellular location of IE62 and the detection of VZV gene 66 transcription during latent infection suggest that latent IE62 is functionally restricted through 66-pk phosphorylation.
Although our experiments were not designed to address the prevalence of ORF 66 transcription in the human population or the abundance of ORF 66 transcripts within individual ganglia, we did find that 2 of 6 (33%) individual TG were positive for VZV gene 66 transcripts by RT-PCR and that 5 of 27 (18%) individual ganglia were positive for VZV gene 66 transcription by ISH. The apparent twofold increase in the frequency of gene 66 detection by RT-PCR compared to ISH most likely reflects the different sensitivities of each technique. For example, RT-PCR uses RNA extracted from the entire powdered TG and samples from >3 × 1010 mRNA molecules per reaction mixture, while ISH samples individual sections of each TG; thus, only a portion of the entire TG is sampled in each ISH assay. Further, the focal and patchy distribution of ISH signal in TG sections makes it difficult to give a precise number for the percentage of neurons which are 66 and/or 62 positive. However, we estimate ∼2 to 5% of neurons are positive for gene 66 or gene 62 during latent infection. No obvious difference in the positivity rate between genes 66 and 62 was discerned. It was also difficult to determine if the same neuronal cell is positive for both gene 66 and 62 transcripts; however, our analysis of multiple serial sections within the same ganglion has indicated that both gene transcripts are present in the same ganglion. Also, we could not detect TG neurons in which VZV gene 66 protein was expressed in both the nucleus and the cytoplasm.
Finally, the identification of VZV gene 66 RNA and protein in latently infected human ganglia suggests a possible mechanism by which the gene-activating function of latently expressed IE62 is regulated. An interesting alternative interpretation is that 66-pk expression in neuronal TG is a means of damping reactivation of latent VZV. Feldman et al. (9) have documented rare occurrences of spontaneous subclinical herpes simplex virus type 1 reactivation in a small subset of latently infected mouse TG neuronal cells. If a population of VZV-latently infected neuronal cells undergoes reactivation, one would predict IE62 to be among the first virus genes expressed. Thus, induction of 66-pk transcription would limit virus reactivation. In this case, gene 66 transcription could be seen as a “smoking gun,” indicating those neuronal cells which have undergone recent (and now abortive) reactivation. Although this hypothesis is difficult to test in humans, it is testable in developing animal models of VZV latency and reactivation (27).
Acknowledgments
This work was supported in part by Public Health Service grants AG 06127 (D.H.G.), NS 32623 (D.H.G. and R.J.C.), and EY 09397 and EY 08098 (P.R.K.), all from the National Institutes of Health, by the Wellcome Trust, and by the Chief Scientist's Office of Scotland (P.G.E.K.).
Expert technical assistance was provided by John Smith, Carrie Essman, Jeannie Welsher, Stephanie E. Turse, and Michael Hurley. We also thank Marina Hoffman for editorial help and Cathy Allen for manuscript preparation.
REFERENCES
- 1.Clarke, P., T. Beer, R. Cohrs, and D. H. Gilden. 1995. Configuration of latent varicella-zoster virus DNA. J. Virol. 69:8151-8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen, J. I., and S. E. Straus. 2001. Varicella-zoster virus and its replication, p. 2707-2730. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, New York, N.Y.
- 3.Cohrs, R., R. Mahalingam, A. N. Dueland, W. Wolf, M. Wellish, and D. H. Gilden. 1992. Restricted transcription of varicella-zoster virus in latently infected human trigeminal and thoracic ganglia. J. Infect. Dis. 166:S24-S29. [DOI] [PubMed] [Google Scholar]
- 4.Cohrs, R. J., J. Wischer, C. Essman, and D. H. Gilden. 2002. Characterization of varicella-zoster virus gene 21 and 29 proteins in infected cells. J. Virol. 76:7228-7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohrs, R. J., M. Barbour, and D. H. Gilden. 1996. Varicella-zoster virus (VZV) transcription during latency in human ganglia: detection of transcripts mapping to genes 21, 29, 62, and 63 in a cDNA library enriched for VZV RNA. J. Virol. 70:2789-2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohrs, R. J., M. B. Barbour, R. Mahalingam, M. Wellish, and D. H. Gilden. 1995. Varicella-zoster virus (VZV) transcription during latency in human ganglia: prevalence of VZV gene 21 transcripts in latently infected human ganglia. J. Virol. 69:2674-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Croen, K. D., J. M. Ostrove, L. J. Dragovic, and S. E. Straus. 1988. Patterns of gene expression and sites of latency in human nerve ganglia are different for varicella-zoster and herpes simplex viruses. Proc. Natl. Acad. Sci. USA 85:9773-9777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1759-1816. [DOI] [PubMed] [Google Scholar]
- 9.Feldman, L. T., A. R. Ellison, C. C. Voytek, L. Yang, P. Krause, and T. P. Margolis. 2002. Spontaneous molecular reactivation of herpes simplex virus type 1 latency in mice. Proc. Natl. Acad. Sci. USA 99:978-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forghani, B., R. Mahalingam, A. Vafai, J. W. Hurst, and K. W. Dupuis. 1990. Monoclonal antibody to immediate early protein encoded by varicella-zoster virus gene 62. Virus Res. 16:195-210. [DOI] [PubMed] [Google Scholar]
- 11.Kennedy, P. G. E., E. Grinfeld, and J. E. Bell. 2000. Varicella-zoster virus gene expression in latently infected and explanted ganglia. J. Virol. 74:11893-11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kennedy, P. G. E., E. Grinfeld, and J. W. Gow. 1999. Latent varicella-zoster virus in human dorsal root ganglia. Virology 258:451-454. [DOI] [PubMed] [Google Scholar]
- 13.Kennedy, P. G. E., E. Grinfeld, and J. W. Gow. 1998. Latent varicella-zoster virus is located predominantly in human trigeminal ganglia. Proc. Natl. Acad. Sci. USA 95:4658-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kinchington, P. R., and J. I. Cohen. 2002. Viral proteins, p. 74-104. In A. Arvin and A. Gershon (ed.), Varicella zoster virus. Cambridge University Press, Cambridge, United Kingdom.
- 15.Kinchington, P. R., and S. E. Turse. 1995. Transcriptional mapping of varicella-zoster virus regulation protein. Neurology 45:S13-S14. [DOI] [PubMed] [Google Scholar]
- 16.Kinchington, P. R., and S. E. Turse. 1998. Regulated nuclear localization of the varicella-zoster virus major regulatory protein IE62. J. Infect. Dis. 178:S16-S21. [DOI] [PubMed] [Google Scholar]
- 17.Kinchington, P. R., J. K. Hougland, A. M. Arvin, W. T. Ruyechan, and J. Hay. 1992. The varicella-zoster virus immediate-early protein IE62 is a major component of virus particles. J. Virol. 66:359-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kinchington, P. R., K. Fite, A. Seman, and S. E. Turse. 2001. Virion association of IE62, the varicella-zoster virus (VZV) major transcriptional regulatory protein, requires expression of the VZV open reading frame 66 protein kinase. J. Virol. 75:9106-9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinchington, P. R., K. Fite, and S. E. Turse. 2000. Nuclear accumulation of IE62, the varicella-zoster virus (VZV) major transcriptional regulatory protein, is inhibited by phosphorylation mediated by the VZV open reading frame 66 protein kinase. J. Virol. 74:2265-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lungu, O., C. A. Panagiotidis, P. W. Annunziato, A. A. Gershon, and S. J. Silverstein. 1998. Aberrant intracellular localization of varicella-zoster virus regulatory proteins during latency. Proc. Natl. Acad. Sci. USA 95:7080-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahalingam, R., M. Wellish, R. Cohrs, S. Debrus, J. Piette, B. Rentier, and D. H. Gilden. 1996. Expression of protein encoded by varicella-zoster virus open reading frame 62 in latently infected human ganglionic neurons. Proc. Natl. Acad. Sci. USA 93:2122-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meier, J. L., R. P. Holman, K. D. Croen, J. E. Smialek, and S. E. Straus. 1993. Varicella-zoster virus transcription in human trigeminal ganglia. Virology 193:193-200. [DOI] [PubMed] [Google Scholar]
- 23.Moriuchi, M., H. Moriuchi, S. E. Straus, and J. I. Cohen. 1994. Varicella-zoster virus (VZV) virion-associated transactivator open reading frame 62 protein enhances the infectivity of VZV DNA. Virology 200:297-300. [DOI] [PubMed] [Google Scholar]
- 24.Nakajima-Iijima, S., H. Hamada, P. Reddy, and T. Kakunaga. 1985. Molecular structure of the human cytoplasmic beta-actin gene: interspecies homology of sequences in the introns. Proc. Natl. Acad. Sci. USA 82:6133-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shiraki, K., and R. W. Hyman. 1987. The immediate early proteins of varicella-zoster virus. Virology 156:423-426. [DOI] [PubMed] [Google Scholar]
- 26.Silverstein, S. J., and S. E. Straus. 2000. Pathogenesis of latency and reactivation, p. 123-141. In A. M. Arvin and A. A. Gershon (ed.), Varicella zoster virus. Cambridge University Press, Cambridge, United Kingdom.
- 27.White, T. M., D. H. Gilden, and R. Mahalingam. 2001. An animal model of varicella virus infection. Brain Pathol. 11:475-479. [DOI] [PMC free article] [PubMed] [Google Scholar]