Abstract
A novel simian immunodeficiency virus (SIV) sequence has been recovered from RNA extracted from the serum of a mona monkey (Cercopithecus mona) wild born in Nigeria. The sequence was obtained by using novel generic (degenerate) PCR primers and spans from two-thirds into the gag gene to the 3′ poly(A) tail of the SIVmonNG1 RNA genome. Analysis of the open reading frames revealed that the SIVmonNG1 genome codes for a Vpu protein, in addition to Gag, Pol, Vif, Vpr, Tat, Rev, Env, and Nef proteins. Previously, only lentiviruses infecting humans (human immunodeficiency virus type 1 [HIV-1]) and chimpanzees (SIVcpz) were known to have a vpu gene; more recently, this has also been found in SIVgsn from Cercopithecus nictitans. Overall, SIVmonNG1 most closely resembles SIVgsn: the env gene sequence groups with HIV-1/SIVcpz env sequences, whereas the pol gene sequence clusters closely with the pol sequence of SIVsyk from Cercopithecus albogaris. By bootscanning and similarity plotting, the first half of pol resembles SIVsyk, whereas the latter part is closer to SIVcol from Colobus guereza. The similarities between the complex mosaic genomes of SIVmonNG1 and SIVgsn are consistent with a shared or common lineage. These data further highlight the intricate nature of the relationships between the SIVs from different primate species and will be helpful for unraveling these associations.
It is becoming increasingly apparent that many African primates are infected with lentiviruses (33). These simian immunodeficiency viruses (SIVs) are of complex evolutionary origin (40, 41). In earlier studies, primate lentiviruses were recovered from chimpanzees (SIVcpz, Pan troglodytes) (13, 21), sooty mangabeys (SIVsm, Cercocebus atys) (20); African green monkeys (SIVagm, Chlorocebus aethiops) (1, 19, 23, 25), Sykes monkeys (SIVsyk, Cercopithecus albogaris) (11, 18), and mandrills (SIVmnd-1, Mandrillus sphinx) (49). More recently, such viruses have been found and characterized in a wide range of primates, including the red-capped mangabey (SIVrcm, Cercocebus torquatus torquatus) (5, 15), guereza colobus monkey (SIVcol, Colobus guereza) (9), L'Hoest's monkey (SIVlhoest, Cercopithecus l'hoesti) (17), sun-tailed monkey (SIVsun, Cercopithecus solatus) (3), talapoin monkey (Myopithecus talapoin) (30), mandrills (SIVmnd-2) (43), and drill monkey (SIVdrl, Mandrillus leucophaeus) (8). Most recently, a novel SIV has been found in the greater spot-nosed monkey (SIVgsn, Cercopithecus nictitans) (10). Some primate lentiviruses are able to cross the species barriers and give rise to new strains: SIVcpz has infected humans more than once to give human immunodeficiency virus type 1 (HIV-1) (13, 21, 22, 34, 50), and SIVsm has infected humans and macaques to yield HIV-2 and SIVmac, respectively (16, 20, 29). SIVagm has been recovered from baboons (Papio spp.) and a patas monkey (Erythrocebus patas), presumably as a result of natural transmissions in the wild (6, 24, 51). Some SIVs have mosaic or recombinant genomes, indicating that they have arisen as a result of infections with more than one distinct SIV. Examples are SIVagmSAB from the Sabaeus monkey (23), SIVrcm (5, 15), SIVdrl (8) (J. P. Clewley, unpublished observations), SIVmnd-2 (43), and SIVgsn (10). Evidence of cross-species transmission and multiple infections with SIVs is consistent with the suggestion that the primate lentiviruses originated and evolved in monkeys of the Cercopithecus genus (3, 17, 25).
The SIVs can be classified into distinct lineages based upon their complete genome sequence relationships (4). To date, six such lineages are recognized: SIVcpz/HIV-1, SIVsm/HIV-2/SIVmac, SIVlhoest/SIVsun/SIVmnd-1, SIVagm, SIVsyk, and SIVcol. New lineages are likely to be defined when the full sequences of several as yet only partially characterized genomes become available (33). The SIVs can also be classified into three groups based on the organization of the accessory genes in their genomes (4). All simian lentiviruses possess vif, vpr, and nef accessory genes and tat and rev regulatory genes but, until recently, only the SIVcpz and HIV-1 lineage viruses were known to have a vpu gene (4, 5, 27, 46). SIVgsn from the greater spot-nosed monkey has now also been shown to have a vpu gene (10). The SIVsm/HIV-2 lineage viruses, including SIVrcm and SIVmnd-2, do not have a vpu but do have a vpx gene (5, 43). The Vpu protein and gene have both been extensively studied in the expectation that they might shed light on the unique pathogenesis of HIV-1. For example, although the Vpu protein is not essential for viral replication, removal of the vpu gene leads to a decrease in pathogenicity (44). The origin of vpu is unclear, but it may have arisen to enhance viral release as a result of increased affinity of the Env protein for the CD4 receptor (45).
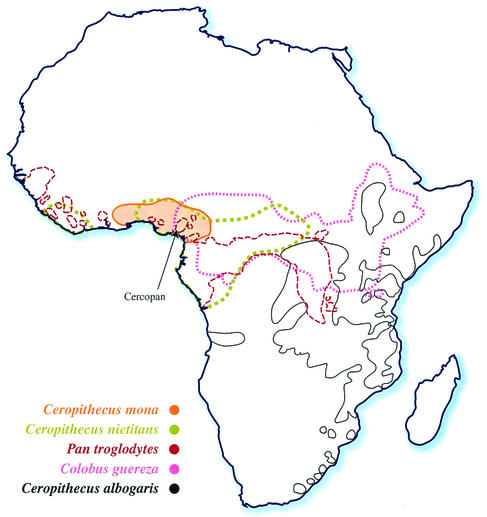
In the present study, we have recovered a genome sequence (SIVmonNG1) from lentivirus RNA extracted from the serum of a mona monkey (Cercopithecus mona). This forest-dwelling guenon has a range from Ghana to Cameroon in western Africa (Fig. 1) (36). There are about 26 species of guenons, of which seven forest species, including the mona, live in southern Nigeria. They are endangered through habitat loss and hunting, and the Cercopan Project, located in southeastern Nigeria, is a nongovernmental organization dedicated to their conservation, rescue, and rehabilitation (http://www.cercopan.org/). A partial pol SIV sequence from a mona monkey from Cameroon has recently been described (33). Here, we show that the SIVmonNG1 genome, like SIVgsn, has a vpu gene, and that its env gene is related to SIVcpz sequences and its pol gene to SIVsyk and SIVcol.
FIG. 1.
Map of Africa showing the natural ranges of the mona monkey, Cercopithecus mona, the greater spot-nosed monkey, Cercopithecus nictitans, the chimpanzee, Pan troglodytes, the Sykes monkey, Cercopithecus albogaris, and the colobus monkey, Colobus guereza. The data were obtained from the study by Rowe (36) and from an online source (http://gorilla.bio.uniroma.it/). The approximate location of the Cercopan Project in Nigeria is arrowed.
MATERIALS AND METHODS
Specimens and serological testing.
Serum samples were obtained from two mona monkeys during quarantine testing at Cercopan. These monkeys were obtained via the bush meat trade and thus were wild born; they were either confiscated by appropriate wildlife officials and subsequently donated to Cercopan or donated by their owners who had purchased them from hunters. In the majority of cases the monkeys at Cercopan become captive at a very young age (6 months or less) and arrived at the sanctuary soon after. The sera were tested at the Central Public Health Laboratory for antibodies cross-reacting with HIV antigens by Innogenetics and Organon Teknika enzyme immunoassays (EIAs). A positive result in these tests was confirmed by using the Genelabs Western blot test.
RNA extraction, PCR amplification, cloning, and sequencing.
RNA was extracted from serum by a modified guanidinium isothiocyanate-silica method (7) and converted to cDNA by reverse transcription by using Expand reverse transcriptase (RT; Roche Diagnostics, Lewes, United Kingdom). This was PCR amplified by using Expand High Fidelity or Long Template PCR reagents (Roche). Specific primers were used at a concentration of 300 nM, and degenerate primers were used at a concentration of 1,000 nM. Thermocycling conditions were modified appropriately for the target size: typically the first-round amplifications were 94°C for 2 min, followed by 10 cycles of 94°C for 15 s, 50°C for 30 s, and 68°C for 4.5 to 7 min, followed by a further 25 cycles, with an additional 5-s extension per cycle and a 4°C hold at the end. A total of 1.5 μl of the primary PCR was added to a secondary PCR mixture and typically cycled at 94°C for 2 min, followed by 4 to 8 cycles of 94°C for 15 s, 42 to 50°C for 30 s, and 68°C (or 72°C for Expand High Fidelity) for 2 to 7 min, followed by a further 27 to 31 cycles, with an additional 5-s extension per cycle and a 4°C hold. On occasion, AccuPrime Taq polymerase (Invitrogen, Paisley, United Kingdom) was used for nested-PCR amplifications with previously described touchdown conditions (8). A 3′ RACE (rapid amplification of cDNA ends) kit (Invitrogen) was used to recover sequences from the poly(A) tail to the nef gene. PCR products were cloned with TOPO XL kits (Invitrogen) and plasmids purified with Qiagen kits (Qiagen, Crawley, United Kingdom). Sequences were generated by using a capillary sequencer (CEQ 2000XL DNA analysis system; Beckman Coulter, High Wycombe, United Kingdom) or by commercial providers (J. Bartley, Natural History Museum, London, United Kingdom; MWG Biotech, Milton Keynes, United Kingdom; Cytomix, Cambridge, United Kingdom).
Primers.
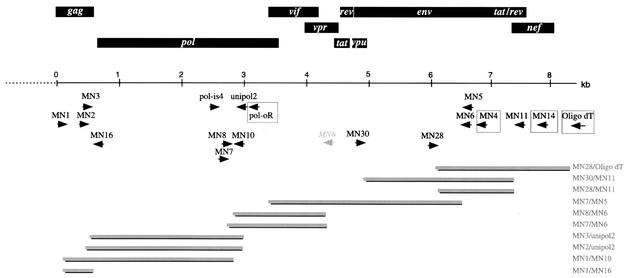
Initially, previously described primers were used to amplify a small region of the pol gene (9, 28). PolOR was used for cDNA synthesis and first-round PCR with Polis4; Polis4/Unipol2 were then used for second-round PCR. New primers were designed in conserved regions of the gag (MN1, MN2, and MN3), env (MN4, MN5, and MN6) and nef (MN11 and MN14) genes based on multiple alignments of previously published SIV and HIV sequences. The sequences of these primers and their locations relative to SIVrcmNG411 (accession no. AF349680) are as follows (Fig. 2): MN1 (GARGAAGSDGCMSANTGGGA; 826 to 847), MN2 (ATNCARAATGCHAAYCCDGAHTG; 1184 to 1206), MN3 (GCHTGYCARGGVGTHGGDGG; 1262 to 1281), MN4 (TKGAGRTTYTTNRYWCCCCA; 7666 to 7685), MN5 (CARVTKYTKCTGYTGCTGCAVTATCCC; 7594 to 7620), MN6 (CCCATTGCAGHDCCHGC; 7531 to 7547), MN11 (CCTTCCAGTCCHCCCTTTBHTTTTA; 8683 to 8709), and MN14 (DBNGRDTYVAABBDCCA; 8954 to 8970).
FIG. 2.
Genome organization and PCR cloning strategy for SIVmonNG1 RNA. The identified ORFs are indicated by rectangular boxes above the scale bar. The alternative binding site of MN6 is shown in italics. The primers used for PCR are shown below the scale bar; primers used for cDNA synthesis are boxed. The position of the clones is shown by solid bars, and the primers used to obtain them are indicated on the right.
In addition to the predicted site in env, MN6 also primed between pol and env. In subsequent work, MN14, MN4, and PolOR were used for cDNA synthesis and then as first-round PCR primers together with MN1, Polis4, or sequence-specific primers designed in the small pol region between Polis4 and Unipol2. Second-round PCR primers included MN2, MN3, MN5, MN6, MN11, Unipol2, and sequence-specific primers designed from sequence obtained from the PCR walk. These included the following: MN7 (CATTGAACCAGCCCGAGA), MN8 (TCCTAGCCAAAGAGATAGTCA), MN10 (CGGCTACTAGGATGATACTTC), MN28 (CTACAGCAGCCCAAAGGAGA), and MN30 (GATAATCCGGGAGTCAAGT).
Sequence and phylogenetic analysis.
Sequences were analyzed by using programs in the Lasergene suite (DNAStar, Madison, Wis.). CLUSTAL alignments (47) were edited by eye in MacClade 4.0.2 (D. R. Maddison and W. P. Maddison, Sinauer Associates, Sunderland, Mass.), exported into PAUP* 4.0 1 (D. L. Swofford, Sinauer Associates) for gap stripping and first- and second-codon position alignments. Concatenated gag, pol, vif, env, and nef nucleotide and corresponding amino acid alignments were also made. Phylogenetic reconstructions were carried out by using PAUP*, and the most appropriate evolutionary models and parameters were chosen by using Modeltest (35). Tree searches were carried out by using a heuristic search strategy and branch swapping. Neighbor joining-derived trees were bootstrapped 1,000 times. Protdist (J. Felsenstein, Department of Genetics, University of Washington, Seattle [http://evolution.genetics.washington.edu/phylip/software.html]) was used to estimate the amino acid similarity of aligned predicted protein sequences. Trees were displayed with Treeview (31). RIP (42) and SimPlot (S. Ray, Division of Infectious Diseases, Department of Medicine, Johns Hopkins University School of Medicine [http://sray.med.som.jhmi.edu/RaySoft/SimPlot/]) were used for similarity and bootscanning analyses (26). Oligo (MedProbe, Oslo, Norway) was used to design SIVmonNG1 sequence-specific primers.
Nucleotide sequence accession numbers.
Nucleotide sequences have been submitted to the EMBL database under accession numbers AJ549283 and AJ549756.
RESULTS
Identification of HIV antibody-positive serum.
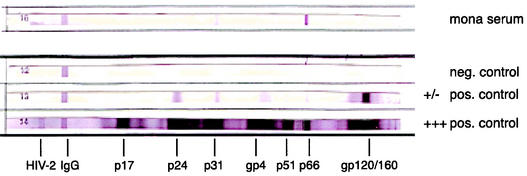
Serum samples were obtained from two mona monkeys at Cercopan in Nigeria. One was found to be reactive in both EIA tests, while the other was unreactive. In the Western blot, the EIA-positive sample was strongly reactive with the HIV-1 p66 antigen (the RT) and had weaker reactivities with gp160 (envelope), p31 (the integrase), and p24 (core) antigens of HIV-1, as well as the HIV-2 p36 transmembrane antigen (Fig. 3). This indicated that the mona monkey was infected with a primate lentivirus.
FIG. 3.
Antibody profile of serum from the mona monkey revealed on a Genelabs Western blot. At the top is the serum from the mona monkey; below are a negative human serum control, a weak anti-HIV-1-positive human serum control, and a strong anti-HIV-1-positive human serum control.
Amplification of SIVmonNG1 RNA.
RNA was extracted from the serum and subjected to nested reverse transcription-PCR with conserved primers in the integrase gene: Polis4/PolOR and then Polis4/Unipol2 (9, 28). The 598 nucleotides of sequence that were derived from this amplicon were found to be phylogenetically related to other primate lentivirus genome sequences, thus confirming that the mona monkey was infected with a novel SIV. Degenerate primers were then designed based on multiple alignments of either gag, env, or nef SIV and HIV sequences in order to be able to extend beyond the pol sequences obtained by using Polis4/Unipol2. These primers (MN1, MN2, and MN3 in gag; MN4, MN5, and MN6 in env; MN11 and MN14 in nef) were used in RT nested-PCR protocols (Fig. 2). For example, MN4 was used as an RT primer and then MN1 and MN4 or MN5 as first-round primers. Alternatively, PolOR or MN14 were used to prime cDNA synthesis. Various combinations of second-round primers were then used, including MN2/Unipol2 and MN1, MN6, and MN11 with specific primers designed on the basis of sequence information derived from amplicons produced with Polis4/Unipol2 (MN7, MN8, and MN10) or with MN7/MN6 (MN28 and MN30). The PCR enzymes and conditions were critical to the successful use of these primers; we mostly used an enzyme mixture with a proofreading polymerase, together with a low annealing temperature for the first 10 cycles of the PCR. By these methods we were able to amplify and clone regions of the genome from about two-thirds into the gag gene to the 3′ poly(A) tail (Fig. 2).
Genomic organization of SIVmonNG1: presence of a vpu gene.
Open reading frames (ORFs) in the sequences were identified by BLAST searches of the Los Alamos HIV sequence database (http://hiv-web.lanl.gov) and by nucleotide and derived amino acid phylogenetic comparisons with other SIV sequences. Potential Gag, Pol, Vif, Tat, Vpr, Rev, and Nef proteins were identified. In addition, an ORF in a similar position to the vpu gene of HIV-1 and SIVcpz was identified (Fig. 2). Therefore, SIVmonNG1 has a genomic organization more similar to HIV-1, SIVcpz, and SIVgsn than to the HIV-2/SIVsm lineage viruses or to the SIVagm, SIVlhoest, SIVsyk, or SIVcol lineages (5).
Putative Vpu amino acid sequence.
The SIVmonNG1 putative Vpu amino acid sequence was not closely related to other Vpu sequences, either HIV-1, SIVcpz, or SIVgsn. The amino acid similarity of SIVmonNG1 Vpu to other Vpu proteins was estimated by using the PHYLIP program Protdist with a manually adjusted CLUSTAL W amino acid alignment and ranged from 15.2% for SIVcpzUS to 17.9% for HIV-1 to 23.4% for SIVgsn to 29% for SIVcpzANT (Table 1). Like all Vpu proteins, the N-terminal half of the sequence (residues 6 to 33) has many hydrophobic residues and is characteristic of a transmembrane domain. The C-terminal half of the sequence has many acidic residues but not the EDSGNESXG(E/D) domain that is highly conserved among HIV-1 and SIVcpz strains, nor does it have a similar sequence to SIVgsn in this region (10). It does, however, have the run of glutamic and aspartic acid residues present in all Vpu proteins, and it also has a DNP motif near the C terminus, in common with other HIV-1 and SIVcpz Vpu sequences.
TABLE 1.
Amino acid sequence identity between SIVmonNG1 and other SIV and HIV-1 genomes in Vpu
| Sequence | Amino acid similarity (%)
|
||||||
|---|---|---|---|---|---|---|---|
| gsn71 | cpzUS | cpzANT | cpzGAB | M | N | O | |
| SIVmonNG1 | 23.4 | 15.2 | 29.0 | 20.9 | 17.9 | 18.5 | 23.9 |
| SIVgsn71 | 100 | 20.3 | 23.9 | 16.9 | 15.9 | 20.0 | 21.7 |
| SIVcpzUS | 100 | 24.6 | 32.9 | 35.6 | 44.3 | 30.4 | |
| SIVcpzANT | 100 | 24.6 | 20.0 | 29.2 | 28.2 | ||
| SIVcpzGAB | 100 | 32.9 | 28.2 | 31.3 | |||
| HIV-1 M Z2Z6 | 100 | 42.9 | 28.4 | ||||
| HIV-1 N YBF | 100 | 27.6 | |||||
| HIV-1 O ANT70 | 100 | ||||||
Genomic relatedness of SIVmonNG1 to other SIVs.
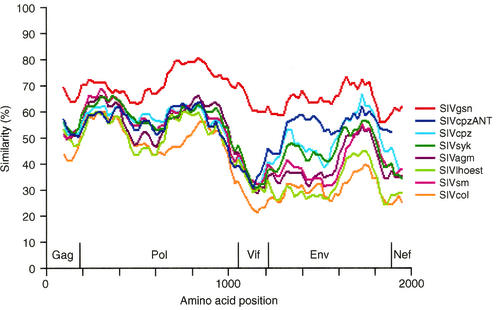
The relationship of SIVmonNG1 to other primate lentiviruses was investigated by comparing the sequence obtained to representatives of the six other SIV lineages and to SIVgsn. First, diversity plots were performed by using a multiple alignment of concatenated predicted Gag, Pol, Vif, Env, and Nef gene products. Ambiguous sites, as well as sites that contained a gap, were removed from the alignment. The sequence similarity was calculated for a window of 200 amino acids moved along the alignment (Fig. 4). The SIVmonNG1 sequence showed the greatest similarity to SIVgsn across the whole genome, and the most diverse protein was Vif.
FIG. 4.
Similarity plotting of SIVmonNG1 against the six major SIV lineages (SIVsyk, SIVlhoest, SIVcol, SIVagm, SIVcpz, and SIVsm) and SIVcpzANT and SIVgsn. Alignments of the nonoverlapping regions of gag, pol, vif, env, and nef were concatenated together; gaps were removed from the amino acid alignment; a window of 200 residues was used. The relative position of the genome proteins is shown. Sequences for comparison were obtained from the Los Alamos HIV Database/GenBank.
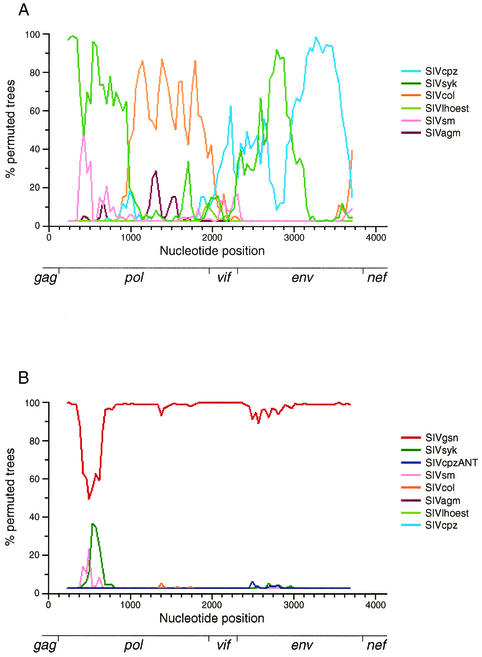
The second method used for examining the relationship of SIVmonNG1 to other SIV genomes was bootscanning (Fig. 5). For this, as for subsequent phylogenetic analyses, third-codon bases were removed from the alignment since they are saturated with nucleotide substitutions and have been shown to be phylogenetically uninformative (10). Bootscanning against single sequences from the six major SIV lineages showed that the SIVmonNG1 genome is a complex mosaic, with the 5′ region related to SIVsyk and SIVcol and the 3′ region related to SIVcpz and SIVsyk sequences (Fig. 5A). When SIVgsn and SIVcpzANT sequences were included in the bootscanning analysis, the SIVmonNG1 sequence was seen to be most closely related to SIVgsn, apart from a small region in pol resembling SIVsyk (Fig. 5B). This substantiates the conclusion that SIVmonNG1 and SIVgsn form a shared lineage.
FIG. 5.
(A) DNA bootscanning comparisons of SIVmonNG1 against single sequences from the major SIV lineages (SIVsyk, SIVlhoest, SIVcol, SIVagmGRI, SIVcpz, and SIVsm). (B) Similar analysis including SIVgsn and SIVcpzANT. Alignments of the nonoverlapping regions of gag, pol, vif, env, and nef were concatenated together, and third-codon positions and gaps were excluded. A sliding window of 450 bases in 40 base steps and 1,000 bootstrap replicates was used for bootscanning with the Kimura 2 parameter model and a transition/transversion ratio of 0.75. The relative positions of the genes are shown at the bottom of the figures.
Comparing the predicted protein sequences of SIVmonNG1 with representatives of other primate lentivirus lineages, SIVmonNG1 was most closely related to SIVgsn (Table 2). For example, for Pol, the similarity score ranged from 34.9% with SIVcpzGAB to 68.3% with SIVgsn. For Env, the similarity ranged from 30.1% with SIVcol to 56.2% for SIVcpzANT to 61.9% for SIVgsn. The SIVmonNG1 Env protein V3 loop is predicted to be 35 amino acids in length and has a QIGAGMTFYS central motif identical to that of SIVgsn; overall, these two V3 loops have 94% identity. The SIVmonNG1 V3 loop is also closely related to SIVcpzANT (71% identity) and similar to other SIVcpz V3 sequences.
TABLE 2.
Amino acid sequence identity between SIVmonNG1 and other SIV isolates
| Sequence | Amino acid similarity (%)a
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gag | Pol | Vif | Vpr | Tat | Rev | Vpu | Env | Nef | |
| SIVgsn71 | 65.4 | 68.3 | 64.0 | 67.7 | 46.8 | 19.4 | 23.4 | 61.9 | 71.4 |
| SIVcpzANT | 52.1 | 36.2 | 29.9 | 28.6 | 53.0 | 18.9 | 29.0 | 56.2 | NA |
| SIVcpzGAB | 53.6 | 34.9 | 26.2 | 27.8 | 46.3 | 19.1 | 20.9 | 49.9 | 33.7 |
| SIVsyk | 53.3 | 48.9 | 29.4 | 24.2 | 42.4 | 17.6 | NA | 45.7 | 32.2 |
| SIVsm | 45.0 | 37.1 | 28.1 | 26.1 | 30.0 | 20.2 | NA | 40.5 | 39.4 |
| SIVagmGRI | 45.3 | 40.9 | 23.7 | 28.9 | 36.8 | 21.2 | NA | 39.8 | 36.7 |
| SIVlhoest | 44.8 | 54.6 | 24.0 | 19.8 | 29.4 | 9.4 | NA | 31.8 | 33.8 |
| SIVcol | 39.9 | 48.5 | 26.4 | 24.4 | 38.9 | 17.2 | NA | 30.1 | 24.7 |
NA, not applicable or available. Values in boldface indicate highest sequence identity scores.
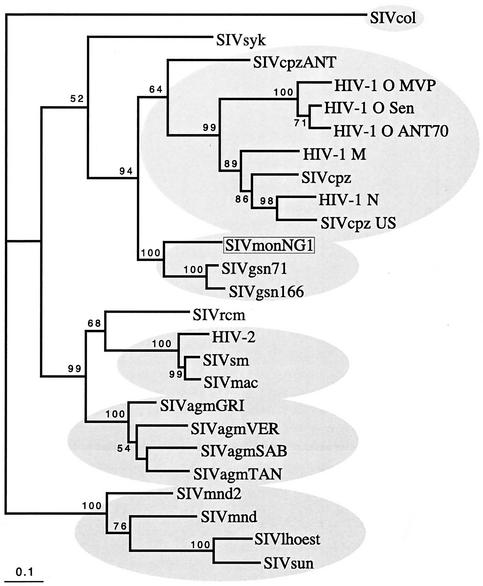
The predicted mosaicism of the genome of SIVmonNG1 observed by bootscanning was further analyzed by phylogenetic analysis of selected genomic regions. Several gene regions were investigated, but only the env and pol gave trees with significant bootstrap support for the branching of the SIVmonNG1 and SIVgsn sequences. Analysis of the env gene by both standard alignments and gap-stripped alignments, and by first- and second-codon positions only, all gave rise to phylogenetic trees in which SIVmonNG1 env clustered with SIVgsn sequences. For example, Fig. 6 shows a phylogenetic tree derived by maximum likelihood searching, in which the best-fitting model of evolution was evaluated by using Modeltest and PAUP* (35). Support for the branching order was found by bootstrapping (1,000 times) a congruent neighbor-joining tree derived with the same model of evolution. Further, the SIVmon and SIVgsn env sequences also formed a clade with HIV-1/SIVcpz env sequences, supported by a 94 bootstrap proportion, further strengthening the idea that these envelope genes are related.
FIG. 6.
Phylogenetic comparison of env SIV and HIV sequences. An in-frame nucleotide alignment of env sequences was derived from the corresponding amino acid alignment, and gaps and codon third-position bases were excluded. The best-fitting model of evolution found by using Modeltest (35) with PAUP* was the general time reversible with the following parameters. The rates were as follows: A-C = 1.3738, A-G = 0.9821, A-T = 0.3369, C-G = 1.5349, C-T = 1.2473, and G-T = 1.0. The base frequencies were as follows: A = 0.3135, C = 0.1323, G = 0.2273, and T = 0.3269. The gamma distribution shape parameter (alpha) was 4.6652, and the proportion of invariable sites was 0.0093. The branch lengths of the tree shown were derived by a maximum-likelihood search with the above parameters. The bootstrap values (of 1,000) were derived from a congruent distance/neighbor-joining tree found with the same model of evolution. The scale bar indicates the number of nucleotide substitutions per site. Sequences for comparison were obtained from the Los Alamos HIV Database/GenBank.
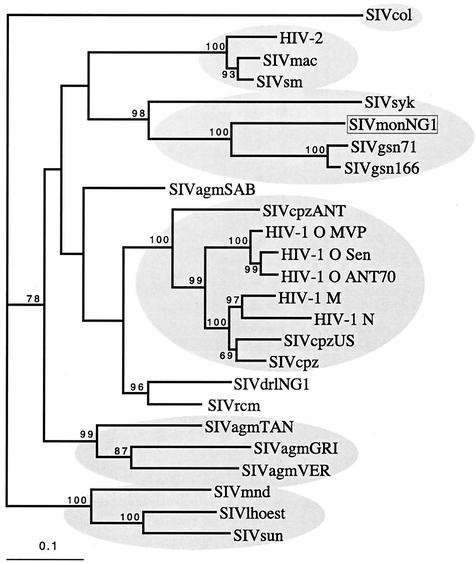
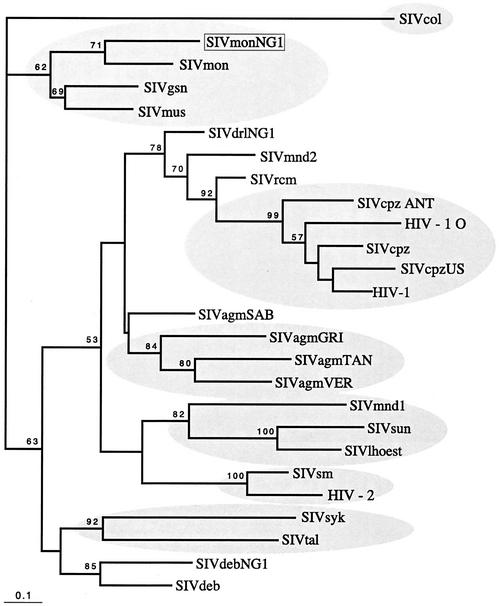
In contrast to the env gene, when the pol gene was examined by similar phylogenetic methods the SIVmonNG1 sequence formed a clade with SIVgsn and SIVsyk, separate from SIVcpz/HIV-1 sequences (Fig. 7). Using bootstrapping with distance methods a high (98 bootstrap proportion) support was found for this grouping. Recently, partial pol sequences (522 nucleotides) have been determined for several previously uncharacterized SIVs from African primates in Cameroon (33), including mona monkeys and the mustached monkey (Cercopithecus cephus). Comparing the equivalent region from SIVmonNG1 indicated that it was closest to another mona monkey sequence, SIVmon-99CM-CML1 (Fig. 8). These sequences formed a separate cluster with the Cercopithecus nicitans (SIVgsn-99CM-CN71) and Cercopithecus cephus (SIVmus-01CM-S1239) sequences, away from other SIV sequences, including SIVsyk, which grouped with SIVtal (Fig. 8). Additional sequence data from SIVmus and SIVtal, and any as-yet-uncharacterized SIVs, will enable greater understanding of the relationships between these SIVs.
FIG. 7.
Phylogenetic comparison of pol SIV and HIV sequences. An in-frame nucleotide alignment of pol sequences was derived from the corresponding amino acid alignment, and gaps and codon third-position bases were excluded. The best-fitting model of evolution found by using Modeltest (35) with PAUP* was the general time reversible. The rates were as follows: A-C = 2.9306, A-G = 3.2906, A-T = 1.9048, C-G = 3.1502, C-T = 5.2621, and G-T = 1.0. The base frequencies were as follows: A = 0.3709, C = 0.2021, G = 0.2353, and T = 0.1917. The gamma distribution shape parameter was alpha = 1.2972. The proportion of invariable sites was 0.2594. The branch lengths of the tree shown were derived by a maximum-likelihood search with the above parameters. The bootstrap values (of 1,000) were derived from a congruent distance/neighbor-joining tree found with the same model of evolution. The scale bar indicates the number of nucleotide substitutions per site. Apart from SIVmonNG1, sequences were obtained from the Los Alamos HIV Database/GenBank.
FIG. 8.
Maximum-likelihood tree of a 520-bp pol sequence, with significant bootstrap values (of 100) indicated. The sequences represent the amplicon derived by using the Polis4/Unipol2 primers (9, 28). The best-fitting model of evolution found by using Modeltest (35) with PAUP* was the general time reversible. The rates were as follows: A-C = 2.4379, A-G = 5.8145, A-T = 2.2893, C-G = 3.5281, C-T = 12.703, and G-T = 1.0. The base frequencies were as follows: A = 0.4341, C = 0.1827, G = 0.2021, and T = 0.1811. The gamma distribution shape parameter was alpha = 0.9683. The proportion of invariable sites was 0.2127. Both the branch lengths and the bootstrap proportions (of 100) were derived by maximum-likelihood searching. The scale bar indicates the number of nucleotide substitutions per site. SIVmonNG1 (AJ549283), SIVdrlNG1 (AJ310481), and SIVdebNG1 (AJ549756) were sequenced as part of the present study; SIVmon (99CM-CML1), SIVgsn (99CM-CN71), SIVmus (01CM-S1239), SIVmnd2, SIVtal, and SIVdeb are from Peeters et al. (33): these latter sequences and others were obtained from the Los Alamos HIV Database/GenBank.
DISCUSSION
The sequence of more than 8,200 bases of the RNA genome [partial gag to 3′ poly(A)] of an SIV from a mona monkey (Cercopithecus mona) was derived by a PCR amplification strategy by using novel degenerate primers located in conserved regions of SIV/HIV genomes. Attempts to recover the 5′ end by RACE and amplification with a primer based on the presumptive tRNA-binding site were unsuccessful, and insufficient sample remained for further work. Further characterization of the SIVmonNG1 genome will, therefore, require the collection of more serum. The SIVmonNG1 sequence was obtained from serum collected from a young wild-born mona monkey in the Cercopan Conservation Centre in Nigeria. As with any primate lentivirus that can jump from one species to another, it has to be asked whether the mona monkey is the primary host for this virus or whether it could have come from another monkey. The young age of the monkey (<2 months old) is consistent with it being the primary host. The availability of a small pol region sequence independently obtained from a mona monkey from the Cameroon (33) allowed a comparison with the sequence we obtained (Fig. 8). The two sequences were clearly related (82% identity over 522 nucleotides), a finding consistent with them being the same SIV from animals at extremes of the species range, which is from Ghana to the Cameroon (Fig. 1) (36). We do not have any evidence that SIVmonNG1 was associated with any specific disease.
The molecular characterization of this genome provides new insights into the phylogeny and evolution of the primate lentiviruses. Significantly, the genome has a putative vpu gene, which has previously only been found in HIV-1 and SIVcpz genomes and, more recently, in SIVgsn (10). The conceptual translation of this gene yields a sequence that is consistent with it being related to the Vpu proteins of HIV-1, SIVcpz, and SIVgsn although, like these proteins, it is clearly divergent with an average similarity of 21% (Table 1). HIV-1 Vpu has two domains: an N-terminal transmembrane anchor and a C-terminal cytoplasmic domain (38). The transmembrane domain enhances virus release from infected cells, whereas the cytoplasmic domain is involved in the degradation of CD4 (32, 37, 48). When the derived protein sequence of SIVmonNG1 Vpu was analyzed by molecular modeling (data not shown), the amino terminus (residues 6 to 33) was found, as expected, to be characteristic of a transmembrane domain. The C-terminal (cytoplasmic) half, however, was predicted to have a single alpha-helix, located before two candidate phosphoserine residues. The SIVgsn Vpu was also predicted to have only one alpha-helix. In comparison, by using the same prediction algorithms, HIV-1 and SIVcpz Vpu proteins had two alpha-helices, one on either side of the phosphoserines, a finding consistent with the structural data (12, 52, 53). This could be interpreted as indicating a difference in biological activity of the cytoplasmic domain of the SIVmonNG1 Vpu compared to the equivalent region of the HIV-1/SIVcpz protein. Of course, this would need to be established experimentally.
Sequence comparisons of the SIVmonNG1 genome further illustrate the complexity of the relationships between the primate lentiviruses. The SIVmonNG1 sequence is most closely related to SIVgsn, as can be seen from the amino acid similarity plot (Fig. 4) and, in common with that genome, it can be interpreted as having a complex mosaic structure, as shown by bootscanning (Fig. 5A). Aside from SIVgsn, the 5′ half of the SIVmonNG1 genome shows similarity to SIVsyk and SIVcol by bootscanning. Again, apart from SIVgsn (Fig. 5B), the 3′ half of the genome resembles SIVcpzANT by similarity plotting and SIVsyk and SIVcpz by bootscanning. The vif gene is the most diverse region of the SIVmonNG1 genome (Fig. 4).
More definitive analyses of the genetic relationships of SIVmonNG1 were attempted by phylogenetic comparisons of selected genome regions. Trees derived from alignments of small genome regions suggested by the bootscans have did not high bootstrap support, as was also observed for SIVgsn (10). Trees made from the pol and env regions were, however, unambiguous. The env region of SIVmonNG1 and SIVgsn formed a clade with SIVcpz/HIV-1 with high bootstrap support (94 BP) (Fig. 6). It is perhaps not surprising that env sequences resembling those of the HIV-1/SIVcpz clade are found in another SIV that has a vpu gene, since the env and vpu genes overlap and are translated from a bicistronic mRNA (39). Thus, the env gene similarity and the presence of a vpu gene could be interpreted as indicating that the vpu/env gene regions of SIVmonNG1, SIVgsn, and SIVcpz have a common origin or ancestor. Analysis of the pol region showed that SIVmonNG1, SIVsyk, and SIVgsn formed a clade with high bootstrap support (98 BP) (Fig. 7), indicating that the pol gene region of SIVmonNG1 has a different origin from the env gene region. Thus, from the similarity plots, bootscans, and phylogenetic trees, it is clear that SIVmonNG1, like SIVgsn, has a complex mosaic genome, related to the SIVs of other Cercopithecinae monkeys. The present-day ranges of the mona monkey and those of the four other species (chimpanzee, greater spot-nosed monkey, Sykes monkey, and colobus monkey), whose SIVs show the most similarity to SIVmonNG1, are shown in Fig. 1. The ranges of the mona monkey, chimpanzee, greater spot-nosed monkey, and colobus monkey overlap. The Sykes monkey range is adjacent to that of the greater spot-nosed monkey and overlaps with the colobus monkey. This geographical distribution, as well as the similarity of the SIVmonNG1 and SIVgsn genomes from distinct species, is consistent with the idea that the same lineage has infected both species or that SIVgsn passed from the greater spot-nosed to the mona monkey.
The similarity of SIVmonNG1 and SIVgsn to SIVcpz in env raises the question of whether they or the SIVcpz lineage are recombinant. Previously characterized recombinants include SIVagmSAB from the sabaeus African green monkey (23), SIVrcm from red-capped mangabeys (5, 15), SIVmnd-2 from the mandrill (43), and SIVdrl from the drill monkey (8) (Clewley, unpublished). As Souquière et al. have pointed out, the Cercopithecinae monkeys, the mandrills, and drill monkeys are close relatives (43), and the existence of recombinant forms in these species supports the suggestion that the SIVs from them are ancestral and that recombinants play a major role in primate lentivirus evolution (3, 17, 25). The present study provides evidence consistent with an SIVcpz ancestral or progenitor virus being present in a Cercopithecinae monkey, suggesting that the SIVcpz group is mosaic. As more SIV genomes are characterized, it is likely to become increasingly difficult to recognize pure lineages.
The PCR primers we used to recover sequence information from SIVmonNG1 RNA in serum may help the characterization of previously unknown SIVs. Many recent SIV studies have used proviral DNA from cultured infected cells as the starting point for the recovery of whole-genome sequence information, either by PCR amplification of unintegrated circular DNA or by screening a genomic library (2, 3, 5, 9, 10, 15, 17-19, 43). These experiments have often relied on degenerate primers to obtain starting sequence information or for inverse PCR of circular DNA (5, 9, 10, 15, 17, 43). Several sets of degenerate primers have been designed and used by different investigators to obtain, for example, small regions of sequence information in the conserved pol gene (8, 9, 14, 28). Perhaps the most comprehensive use of degenerate primers has been Peeters et al. study of SIV sequences recovered from bushmeat in Cameroon (33).
In conclusion, we have identified a novel SIV RNA sequence from Cercopithecus mona that has a mosaic (recombinant) genome and which has a vpu-like gene, potentially expressing a Vpu protein. Collection of further samples will be necessary for characterization of the 5′ end of the genome and growth of the virus.
Acknowledgments
We thank John Lewis and Zena Tooze for supplying the serum, without which this work could not have been done, and the Cercopan Project (www.cercopan.org) for their conservation work with the Cercopithecinae. We also thank Grace McCormack for phylogenetic advice, John Parry and the CPHL HIV-1 Reference Laboratory staff for serological tests, Iain Tatt for performing sequencing reactions, and Philip Mortimer and David Brown for encouragement.
REFERENCES
- 1.Allan, J. S., M. Short, M. E. Taylor, S. Su, V. M. Hirsch, P. R. Johnson, G. M. Shaw, and B. H. Hahn. 1991. Species-specific diversity among simian immunodeficiency viruses from African green monkeys. J. Virol. 65:2816-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beer, B. E., E. Bailes, G. Dapolito, B. J. Campbell, R. M. Goeken, M. K. Axthelm, P. D. Markham, J. Bernard, D. Zagury, G. Franchini, P. M. Sharp, and V. M. Hirsch. 2000. Patterns of genomic sequence diversity among their simian immunodeficiency viruses suggest that L'Hoest monkeys (Cercopithecus lhoesti) are a natural lentivirus reservoir. J. Virol. 74:3892-3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beer, B. E., E. Bailes, R. Goeken, G. Dapolito, C. Coulibaly, S. G. Norley, R. Kurth, J. P. Gautier, A. Gautier-Hion, D. Vallet, P. M. Sharp, and V. M. Hirsch. 1999. Simian immunodeficiency virus (SIV) from sun-tailed monkeys (Cercopithecus solatus): evidence for host-dependent evolution of SIV within the C. lhoesti superspecies. J. Virol. 73:7734-7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beer, B. E., E. Bailes, P. M. Sharp, and V. M. Hirsch. 1999. Diversity and evolution of primate lentiviruses, p. 460-474. In C. L. Kuiken, B. Foley, B. Hahn, P. A. Marx, F. McCutchan, J. W. Mellors, J. I. Mullins, S. Wolinsky, and B. Korber (ed.), Human retroviruses and AIDS. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 5.Beer, B. E., B. T. Foley, C. L. Kuiken, Z. Tooze, R. M. Goeken, C. R. Brown, J. Hu, M. S. Claire, B. T. Korber, and V. M. Hirsch. 2001. Characterization of novel simian immunodeficiency viruses from red-capped mangabeys from Nigeria (SIVrcmNG409 and -NG411). J. Virol. 75:12014-12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bibollet-Ruche, F., A. Galat-Luong, G. Cuny, P. Sarni-Manchado, G. Galat, J. P. Durand, X. Pourrut, and F. Veas. 1996. Simian immunodeficiency virus infection in a patas monkey (Erythrocebus patas): evidence for cross-species transmission from African green monkeys (Cercopithecus aethiops sabaeus) in the wild. J. Gen. Virol. 77:773-781. [DOI] [PubMed] [Google Scholar]
- 7.Boom, R., C. J. A. Sol, M. M. M. Salimans, C. L. Jansen, P. M. E. Wertheim-van Dillen, and J. van der Noorda. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clewley, J. P., J. C. M. Lewis, D. W. G. Brown, and E. L. Gadsby. 1998. A novel simian immunodeficiency virus (SIVdrl) pol sequence from the drill monkey, Mandrillus leucophaeus. J. Virol. 72:10305-10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Courgnaud, V., X. Pourrut, F. Bibollet-Ruche, E. Mpoudi-Ngole, A. Bourgeois, E. Delaporte, and M. Peeters. 2001. Characterization of a novel simian immunodeficiency virus from guereza colobus monkeys (Colobus guereza) in Cameroon: a new lineage in the nonhuman primate lentivirus family. J. Virol. 75:857-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Courgnaud, V., M. Salemi, X. Pourrut, E. Mpoudi-Ngole, B. Abela, P. Auzel, F. Bibollet-Ruche, B. Hahn, A.-M. Vandamme, E. Delaporte, and M. Peeters. 2002. Characterization of a novel simian immunodeficiency virus with a vpu gene from greater spot-nosed monkeys (Cercopithecus nictitans) provides new insights into simian/human immunodeficiency virus phylogeny. J. Virol. 76:8298-8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emau, P., H. M. McClure, M. Isahakai, J. G. Else, and P. N. Fultz. 1991. Isolation from African Sykes' monkeys (Cercopithecus mitis) of a lentivirus related to human and simian immunodeficiency viruses. J. Virol. 65:2135-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Federau, T., U. Schubert, J. Flossdorf, P. Henklein, D. Schomburg, and V. Wray. 1996. Solution structure of the cytoplasmic domain of the human immunodeficiency virus type 1 encoded virus protein U (Vpu). Int. J. Peptide Protein Res. 47:297-310. [DOI] [PubMed] [Google Scholar]
- 13.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 14.Gelman, I. H., J. Zhang, E. Hailman, H. Hanafusa, and S. S. Morse. 1992. Identification and evaluation of new primer sets for the detection of lentivirus proviral DNA. AIDS Res. Hum. Retrovir. 8:1981-1989. [DOI] [PubMed] [Google Scholar]
- 15.Georges-Courbot, M. C., C. Y. Lu, M. Makuwa, P. Telfer, R. Onanga, G. Dubreuil, Z. Chen, S. M. Smith, A. Georges, F. Gao, B. H. Hahn, and P. A. Marx. 1998. Natural infection of a household pet red capped mangabey (Cercocebus torquatus torquatus) with a new simian immunodeficiency virus. J. Virol. 72:600-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henderson, L. E., R. E. Benveniste, R. Sowder, T. D. Copeland, A. M. Schultz, and S. Oroszlan. 1988. Molecular characterization of Gag proteins from simian immunodeficiency virus (SIVmne). J. Virol. 62:2587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirsch, V. M., B. J. Campbell, and E. Bailes. 1999. Characterization of a novel simian immunodeficiency virus (SIV) from L'Hoest monkeys (Cercopithecus l'hoesti): implications for the origins of SIVmnd and other primate lentiviruses. J. Virol. 73:1036-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirsch, V. M., G. Dapolito, S. Goldstein, H. M. McClure, P. Emau, P. N. Fultz, M. Isahakai, R. Neroot, G. Myers, and P. R. Johnson. 1993. A distinct African lentivirus from Sykes' monkeys. J. Virol. 67:1517-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirsch, V. M., C. McGann, G. Dapolito, S. Goldstein, A. Ogen-Odoi, B. Biryawaho, T. Lakwo, and P. R. Johnson. 1993. Identification of a new subgroup of SIVagm in tantalus monkeys. Virology 197:426-430. [DOI] [PubMed] [Google Scholar]
- 20.Hirsch, V. M., R. A. Olmsted, M. Murphy-Corb, R. H. Purcell, and P. R. Johnson. 1989. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature 339:389-392. [DOI] [PubMed] [Google Scholar]
- 21.Huet, T., R. Cheynier, A. Meyerhans, G. Roelants, and S. Wain-Hobson. 1990. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature 345:356-359. [DOI] [PubMed] [Google Scholar]
- 22.Janssens, W., K. Fransen, M. Peeters, L. Heyndrickx, J. Motte, L. Bedjabaga, E. Delaporte, P. Piot, and G. van der Groen. 1994. Phylogenetic analysis of a new chimpanzee lentivirus SIVcpz-gab2 from a wild-captured chimpanzee from Gabon. AIDS Res. Hum. Retrovir. 10:1191-1192. [DOI] [PubMed] [Google Scholar]
- 23.Jin, M. J., H. Hui, D. L. Robertson, M. C. Muller, F. Barre-Sinoussi, V. M. Hirsch, J. S. Allan, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1994. Mosaic genome structure of simian immunodeficiency virus from west African green monkeys. EMBO J. 13:2935-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin, M. J., J. Rogers, J. E. Phillips-Conroy, J. S. Allan, R. C. Desrosiers, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1994. Infection of a yellow baboon with simian immunodeficiency virus from African green monkeys: evidence for cross-species transmission in the wild. J. Virol. 68:8454-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson, P. R., A. Fomsgaard, J. Allan, M. Gravell, W. T. London, R. A. Olmsted, and V. M. Hirsch. 1990. Simian immunodeficiency viruses from African green monkeys display unusual genetic diversity. J. Virol. 64:1086-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkami, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCormick-Davis, C., S. Dalton, D. Singh, and E. Stephens. 2000. Comparison of Vpu sequences from diverse geographical isolates of HIV type 1 identifies the presence of highly variable domains, additional invariant amino acids, and a signature sequence motif common to subtype C isolates. AIDS Res. Hum. Retrovir. 16:1089-1095. [DOI] [PubMed] [Google Scholar]
- 28.Miura, T., J. Sakuragi, M. Kawamura, M. Fukasawa, E. N. Moriyama, T. Gojobori, K. Ishikawa, J. A. Mingle, V. B. Nettey, H. Akari, M. Enami, H. Tsujimoto, and M. Hayami. 1990. Establishment of a phylogenetic survey system for AIDS-related lentiviruses and demonstration of a new HIV-2 subgroup. AIDS 4:1257-1261. [DOI] [PubMed] [Google Scholar]
- 29.Novembre, F. J., V. M. Hirsch, H. M. McClure, P. N. Fultz, and P. R. Johnson. 1992. SIV from stump-tailed macaques: molecular characterization of a highly transmissible primate lentivirus. Virology 186:783-787. [DOI] [PubMed] [Google Scholar]
- 30.Osterhaus, A. D., N. Pedersen, G. van Amerongen, M. T. Frankenhuis, M. Marthas, E. Reay, T. M. Rose, J. Pamungkas, and M. L. Bosch. 1999. Isolation and partial characterization of a lentivirus from talapoin monkey (Myopithecus talapoin). Virology 260:116-124. [DOI] [PubMed] [Google Scholar]
- 31.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 32.Paul, M., S. Mazumder, N. Raja, and M. A. Jabbar. 1998. Mutational analysis of the human immunodeficiency virus type 1 Vpu transmembrane domain that promotes the enhanced release of virus-like particles from the plasma membrane of mammalian cells. J. Virol. 72:1270-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peeters, M., V. Courgnaud, B. Abela, P. Auzel, X. Pourrut, F. Bibollet-Ruche, S. Loul, F. Liegeois, C. Butel, D. Koulagna, E. Mpoudi-Ngole, G. M. Shaw, B. H. Hahn, and E. Delaporte. 2002. Risk to human health from a plethora of simian immunodeficiency viruses in primate bushmeat. Emerg. Infect. Dis. 8:451-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peeters, M., C. Honore, T. Huet, L. Bedjabaga, S. Ossari, P. Bussi, R. W. Cooper, and E. Delaporte. 1989. Isolation and partial characterization of an HIV-related virus occurring naturally in chimpanzees in Gabon. AIDS 3:625-630. [DOI] [PubMed] [Google Scholar]
- 35.Posada, D., and K. A. Crandall. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 36.Rowe, N. 1996. The pictorial guide to the living primates. Pogonias Press, East Hampton, N.Y.
- 37.Schubert, U., L. C. Anton, J. H. Cox, S. Bour, J. R. Bennink, M. Orlowski, K. Strebel, and J. W. Yewdell. 1998. CD4 glycoprotein degradation induced by human immunodeficiency tirus type 1 Vpu protein requires the function of proteasomes and the ubiquitin-conjugating pathway. J. Virol. 72:2280-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schubert, U., S. Bour, A. Ferrer-Montiel, M. Montal, F. Maldarell, and K. Strebel. 1996. The two biological activities of human immunodeficiency virus type 1 Vpu protein involve two separable structural domains. J. Virol. 70:809-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwartz, S., B. K. Felber, E. M. Fenyo, and G. N. Pavlakis. 1990. Env and Vpu proteins of human immunodeficiency virus type 1 are produced from multiple bicistronic mRNAs. J. Virol. 64:5448-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharp, P. M., E. Bailes, F. Gao, B. E. Beer, V. M. Hirsch, and B. H. Hahn. 2000. Origins and evolution of AIDS viruses: estimating the time scale. Biochem. Soc. Trans. 28:275-282. [DOI] [PubMed] [Google Scholar]
- 41.Sharp, P. M., E. Bailes, D. L. Robertson, F. Gao, and B. H. Hahn. 1999. Origins and evolution of AIDS viruses. Biol. Bull. 196:338-342. [DOI] [PubMed] [Google Scholar]
- 42.Siepel, A. C., A. L. Halpern, C. Macken, and B. T. M. Korber. 1995. A computer program designed to screen rapidly for HIV type 1 intersubtype recombinant sequences. AIDS Res. Hum. Retrovir. 11:1413-1416. [DOI] [PubMed] [Google Scholar]
- 43.Souquière, S., F. Bibollet-Ruche, D. L. Robertson, M. Makuwa, C. Apetrei, R. Onanga, C. Kornfeld, J. C. Plantier, F. Gao, K. Abernethym, L. J. White, W. Karesh, P. Telfer, E. J. Wickings, P. Mauclère, P. A. Marx, F. Barre-Sinoussi, B. H. Hahn, M. C. Müller-Trutwin, and F. Simon. 2001. Wild Mandrillus sphinx are carriers of two types of lentivirus. J. Virol. 75:7086-7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stephens, E., C. McCormick, E. Pacyniak, D. Griffin, D. Pinson, F. Sun, W. Nothnick, S. Wong, R. Gunderson, N. Berman, and D. Singh. 2002. Deletion of the vpu sequences prior to the env in a simian-human immunodeficiency virus results in enhanced Env precursor synthesis but is less pathogenic for pig-tailed macaques. Virology 293:252-261. [DOI] [PubMed] [Google Scholar]
- 45.Strebel, K. 1996. Structure and function of HIV-1 Vpu, p. III and 19-27. In G. Myers, B. Korber, B. Foley, K.-T. Jeang, J. W. Mellors, and S. Wain-Hobson (ed.), Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 46.Strebel, K., T. Klimkait, and M. A. Martin. 1988. A novel gene of HIV-1, vpu, and its 16-kilodalton product. Science 241:1221-1223. [DOI] [PubMed]
- 47.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tiganos, E., J. Friborg, B. Allain, N. G. Daniel, X. J. Yao, and E. A. Cohen. 1998. Structural and functional analysis of the membrane-spanning domain of the human immunodeficiency virus type 1 Vpu protein. Virology 251:96-107. [DOI] [PubMed] [Google Scholar]
- 49.Tsujimoto, H., A. Hasegawa, N. Maki, M. Fukasawa, T. Miura, S. Speidel, R. W. Cooper, E. N. Moriyama, T. Gojobori, and M. Hayami. 1989. Sequence of a novel simian immunodeficiency virus from a wild-caught African mandrill. Nature 341:539-541. [DOI] [PubMed] [Google Scholar]
- 50.Vanden Haesevelde, M. M., M. Peeters, G. Janssens, W. Janssens, G. van der Groen, P. M. Sharp, and E. Saman. 1996. Sequence analysis of a highly divergent HIV-1-related lentivirus isolated from a wild captured chimpanzee. Virology 221:346-350. [DOI] [PubMed] [Google Scholar]
- 51.van Rensburg, E. J., S. Engelbrecht, J. Mwenda, J. D. Laten, B. A. Robson, T. Stander, and G. K. Chege. 1998. Simian immunodeficiency viruses (SIVs) from eastern and southern Africa: detection of a SIVagm variant from a chacma baboon. J. Gen. Virol. 79:1809-1814. [DOI] [PubMed] [Google Scholar]
- 52.Willbold, D., S. Hoffmann, and P. Rosch. 1997. Secondary structure and tertiary fold of the human immunodeficiency virus protein U (Vpu) cytoplasmic domain in solution. Eur. J. Biochem. 245:581-588. [DOI] [PubMed] [Google Scholar]
- 53.Zheng, S., J. Strzalka, C. Ma, S. J. Opella, B. M. Ocko, and J. K. Blasie. 2001. Structural studies of the HIV-1 accessory protein Vpu in langmuir monolayers: synchrotron X-ray reflectivity. Biophys. J. 80:1837-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]