Abstract
Genetic RNA recombination plays an important role in viral evolution, but its molecular mechanism is not well understood. In this work we describe homologous RNA recombination activity that is supported by a subgenomic promoter (sgp) region in the RNA3 segment of brome mosaic bromovirus (BMV), a tripartite plus-strand RNA virus. The crossover frequencies were determined by coinoculations with pairs of BMV RNA3 variants that carried a duplicated sgp region flanked by marker restriction sites. A region composed of the sgp core, a poly(A) tract, and an upstream enhancer supported homologous exchanges in 25% of the analyzed RNA3 progeny. However, mutations in the sgp core stopped both the transcription of the sgp RNA and homologous recombination. These data provide evidence for an association of RNA recombination with transcription.
Genetic recombination is an important process leading to diversity among living organisms. The mechanisms of crossing over (general homologous recombination) and other routes of genetic exchange are relatively well studied in DNA-based organisms. In contrast, although RNA recombination has been studied extensively for several RNA virus groups, including bromoviruses, picornaviruses, coronaviruses, tombusviruses, and bacteriophages (8), its mechanism is not well understood (8, 13). Brome mosaic bromovirus (BMV) is a tripartite RNA virus where RNA components 1 and 2 encode, respectively, the replicase proteins 1a and 2a, while RNA3 encodes the movement (3a) and the coat proteins (CPs) (3). CP is expressed from a subgenomic (sg) RNA4.
The subgenomic promoter (sgp) for RNA4 is located internally on the minus strand of BMV RNA3. It includes a core region that is responsible for binding of the viral RNA polymerase (RdRp) and the initiation of transcription (30), an “enhancing” region, a poly(A) stretch, and a downstream portion (21, 34). Besides BMV, the multielement nature of sgp's has been demonstrated in other RNA viruses (24).
There is only limited information about recombination events in natural populations of viral RNA molecules. Recombination hot spots were observed during the course of infection in poliovirus (16) and in human immunodeficiency virus type 1 (36). It has been proposed that recombination within the sg RNA start sites has led to the formation of genera of luteoviruses (23) or has served as a factor for the modular exchange and rearrangements of the genomes of closteroviruses (5). Also, the recombination hot spots are thought to be associated with RNA replication enhancers, such as those found in turnip crinkle carmovirus (9, 30). RNA structure has been reported to play a role in promoting RNA crossovers in retroviruses (4).
In BMV, the frequency of homologous intersegmental crossovers is approximately 10 times higher than that of nonhomologous crossovers (27). A model of the replicase template switching has been proposed to explain the observed homologous crossovers between BMV RNAs (28, 29). An increased recombination activity has been demonstrated within the intercistronic region during infections with engineered BMV RNAs (7, 11) or cowpea chlorotic mottle virus RNAs, in both nontransgenic and transgenic plants (17). Here we show that a high frequency of RNA3-RNA3 crossovers is supported by the sgp sequence. By introducing mutations at the promoter's core, both the synthesis of the sg RNA and homologous recombination were inhibited. This suggests an association between RNA recombination and subgenomic transcription, and it establishes the sgp as a model recombination hot spot.
MATERIALS AND METHODS
Materials.
Plasmids pB1TP3, pB2TP5, and pB3TP7 (19) were used as templates to synthesize in vitro the infectious capped transcripts of wild-type (wt) BMV RNA1, -2, and -3 by using the MEGAscript T7 kit (Ambion, Austin, Tex.). Plasmids ID1 to -4 (see below) were used to synthesize mutant RNA3 transcripts. The Moloney murine leukemia virus reverse transcriptase and the restriction enzymes were from Promega Corp. (Madison, Wis.).
Generation of BMV RNA3 constructs.
To obtain ID1-1, a region in wt RNA3 between nucleotide (nt) positions 1171 and 1270 was amplified by reverse transcription-PCR (RT-PCR) using primers 1 (5′CGGGATCCCGAATTCTATGTTGTGTGTCTGAGTTAT3′) and 2 (5′CGGGATCCAAGCTTACCAGTTCCTGAAGTCG 3′). Primers 1 and 2 carried, respectively, BamHI, EcoRI, and HindIII marker sites within the overhang (the restriction sites are underlined). Similarly, ID1-2 was generated with primers 3 (5′CGGGATCCCCGGCTATGTTGTGTGTCTGAG3′) and 4 (5′CGGGATCCAACCTTACCAGTTCCTGAAGTCG3′), which did not contain marker restriction sites.
Mutant ID2-1 was obtained as follows. The region between nt 1005 and 1322 of the ID1-2 construct was amplified with primers 9 (5′CGGGGGAGGTTATCCATGTTTGTGGGAATTCTATG3′) and 10 (5′CGACAATTACTGGTTGGACTCGAGCGGTCCAACG3′) (a BstXI restriction site is in bold letters, and EcoRI and XhoI sites are underlined). This introduced A-to-G and T-to-A substitutions at positions 1169 and 1170, respectively (primer 9 EcoRI marker site), as well as A-to-C and G-to-A substitutions at positions 1322 and 1324, respectively (primer 10 XhoI marker site). The resulting PCR product (317 nt) was then used as the 5′ primer in the second PCR, together with the 3′ primer 11 (5′GCTCCTTATTCTTTTCAGAAGAGAGCTCATGGGGC3′), which was complementary to nt 1475 to 1499 (SacI restriction site is underlined). The final PCR product (∼500 nt) was digested with BstXI and SacI and ligated back into ID1-2 between the analogous sites. The resulting ID2-1 mutant carried two marker mutations at nucleotide positions 1170 and 1320 (at INT-1), in addition to the original INT-2 insertion of ID1-2.
To generate ID3-1 and ID3-2, the region between nt 1151 and 1307 was amplified by RT-PCR using, respectively, the following primers: 5 (5′CGGGATCCCGAATTCTCCATGTTTGTGGATATTCTATGTTGTGTGTCTGAGTTATTATTAAAAAAAAAAAAAAAAGATATAGTTATAATTCAGCTTATTAA3′) and 6 (5′CGGGATCCAGAATTCTGCGAGCGGCAGCACGACGC3′), or 7 (5′CGGGATCCCGGGTTATCCATGTTTGTGGATATTCTATGTTGTGTGTCTGAGTTATTATTAAAAAAAAAAAAAAAAGATATAGTTATAATTCAGCTTATTAA3′) and 8 (5′CGGGATCCAAGCTTCTGCGAGCGGCAGCACGACGC 3′). Primers 5 and 6 carried EcoRI sites while primer 7 carried an SmaI site, and primer 8 had HindIII restriction sites within the overhangs. Additionally, primers 5 and 7 both carried the following single-nucleotide substitutions (mutations within the sgp core region): C to A (nt 1221), T to G (nt 1224), G to T (nt 1225), C to A (nt 1227), and G to T (nt 1238). The mutated nucleotides are in bold letters, while the restriction sites are underlined, except for SmaI which is in bold italic. The resulting PCR products were digested with BamHI restriction enzyme and ligated at the BamHI site of plasmid SF-25 (7). This placed the intercistronic sequence repeat downstream of the CP stop codon (Fig. 1).
FIG. 1.
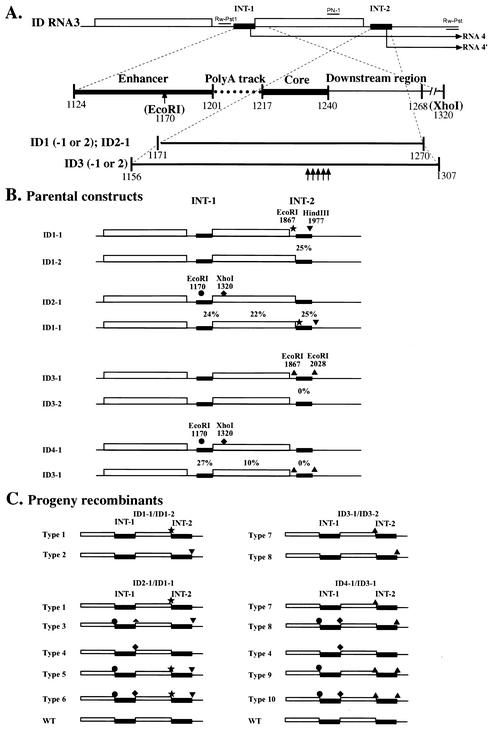
(A) Schematic representation of BMV RNA3 constructs (ID RNA3) carrying repeats of sgp sequences. INT-1 and INT-2 indicate the location of the repeated sg sequence (represented by black bars) and thin lines represent the noncoding regions, while open boxes represent ORFs for 3a and for the CPs. The repeated elements [the enhancer, the poly(A) track, the core, and a downstream hairpin (5-6)] are represented below, as indicated by cut-bars at the bottom for ID1 through ID3. Vertical arrows depict the location of the mutated nucleotides in ID3. The numbers represent nucleotide positions in the wt sequence, as counted from the 5′ end. (B) The location of marker mutations in pairs of coinoculated parental ID1 to ID3 RNAs3. Solid stars, triangles, spades, and circles represent, respectively, EcoRI, HindIII, XhoI, and EcoRI. The numbers on the top show the nucleotide positions of marker mutations. (C) The predicted types of progeny RNA3-RNA3 recombinants (see also Table1) that arose due to crossover events within the INT-1 or INT-2 regions. Solid lines represent INT-1 or INT-2 regions, while open boxes represent the ORFs. The marker restriction sites are depicted, as explained above.
To generate mutant ID4-1, the 151-nt PCR product carrying the mutagenized INT-2 region of ID3-2 was amplified by PCR and was inserted between corresponding BamHI sites of the ID2-1 construct.
RNA analysis.
Extraction and Northern hybridization analysis of total RNA from infected plants were performed as described previously (12). The hybridization signals were quantified on scanned X-ray films using the ImageQuant 5.0 software from Amersham Biosciences.
In vivo recombination assays.
Pairs of RNA3 variants were coinoculated together with wt RNA1 and wt RNA2 on Chenopodium quinoa leaves, as previously described (7). Briefly, a mixture of each transcript in 15 μl of the inoculation buffer solution (10 mM Tris [pH 8.0], 1 mM EDTA, 0.1% Celite) was inoculated on a fully expanded leaf. In each experiment, four separate leaves per plant (two to three plants) were inoculated. Each inoculation experiment was repeated two or three times. The inoculated C. quinoa plants were maintained under greenhouse conditions, as described elsewhere (27-29). Local lesions were counted, collected 10 days postinoculation, and stored at −80°C.
Cloning and analysis of recombinants.
The recombination frequency was calculated as the fraction of recombinants in the total number of cDNA clones that were amplified by RT-PCR from a total RNA extract of the combined local lesion tissue (25). For pairs ID1-1-ID1-2 and ID3-1-ID3-2, the region of recombination (INT-2) was amplified by RT-PCR using the primers RW-Pst (5′AAAACTGCAGCCAATGGTCTCTTTTAGAGATTTACAGTG3′), complementary to 3′-terminal nt 2113 to 2089 of wt RNA3, and primer PN1 (5′CTGAAGCAGTGCCTGCTAAGGCGGTC3′), representing nt 1731 to 1756 of wt RNA3. For pairs ID2-1-ID1-1, ID3-1-ID3-2, and ID4-1-ID3-1, the corresponding primers were RW-Pst and RW-PstI (5′AAAACTGCAGCCAACTTACCTTACAACGGCGTGTTGAG3′), the latter annealing to nt 1112 to 1134 of wt RNA3. The PCR products were purified with the QIAquick PCR purification kit (QIAGEN Inc.) and ligated into the 3′-U overhangs of the cloning vector pDrive (Qiagen PCR cloning kit; catalog no. 231122).
Two kinds of control RT-PCRs were performed. First, the transcribed BMV RNA3 mutants were mixed pairwise (in the same order as during coinoculation experiments), 100 ng each, and used directly as templates for RT-PCR amplification with corresponding primers (RW-Pst, PN-1, and RW-PstI). The resulting PCR fragments were cloned and analyzed, as described above. In the second control, each counterpart of the RNA3 mutant pair was inoculated separately on C. quinoa plants, and the progeny RNA was isolated from separate infections, combined together, and subjected to the RT-PCRs, followed by cloning and analysis, as for the first control.
RESULTS
Generation, infectivity, and in vivo accumulation of the ID RNA3 constructs.
Figure 1 shows RNA3 constructs (named ID RNA3s) that carry duplications of sgp sequences just downstream to the CP termination codon (at the INT-2 locus). This allowed for modifications of the sgp sequences without losing the infectivity. Also, it allowed for probing the sgp activity at INT-2 by measuring the concentration of a smaller, extra sg RNA (called RNA4′) synthesized from the sgp repeat (Fig. 1A). ID1 carried the sgp repeat of nt 1171 to 1270, which included part of the enhancer sequence, the poly(A) tract, the core region, and a downstream region. The truncated sgp was duplicated to avoid potential instability of the insert during infection. Two versions of ID-1 were designed (ID1-1 and ID1-2). ID1-1 carried the EcoRI and HindIII marker sites at flanking positions of the INT-2 insert, while ID1-2 did not.
ID2-1 was similar to ID1-2, except that it carried flanking marker mutations at INT-1:EcoRI at position 1170 and XhoI at position 1320. Two ID3 variants carried a slightly longer sgp insertion at INT-2 (nt 1156 to 1307), encompassing the whole enhancer sequence and a longer downstream sequence, in addition to the elements carried by ID1 and ID2 mutants. ID3-1 carried EcoRI markers at flanking positions of INT-2, while ID3-2 did not. Also, ID3-1 and ID3-2 carried the following five nucleotide substitutions within the sgp core of the INT-2 insert: C-1221 to A, U-1224 to G, G-1225 to U, C-1227 to A, and G-1238 to U (1, 32). These mutations aimed at inactivating the sgp by destabilizing the putative stem-loop structure, and substituting for the initiation guanylate, at the essential nucleotide positions (18).
Construct ID4-1 was comparable to construct ID3-2 except that it carried EcoRI and XhoI marker mutations at INT-1. Thus, coinoculation of ID4-1 and ID3-1 allowed for testing of the recombination activity at both INT-1 and INT-2 loci.
To test the infectivity, equal amounts of transcribed ID RNA3s plus wt RNA1 and RNA2 (for brevity, the wt components of RNA1 and RNA2 will not be mentioned throughout the rest of the text) were inoculated on leaves of C. quinoa, and the number of local lesions was estimated 10 days postinfection. This demonstrated that all the constructs generated a comparable number of local lesions and, thus, they were equally infectious. Based on sequencing of five cDNA clones per construct, there were no size alterations observed in the RNA progeny within the region of interest (data not shown). This confirmed the stability of ID RNAs during infection.
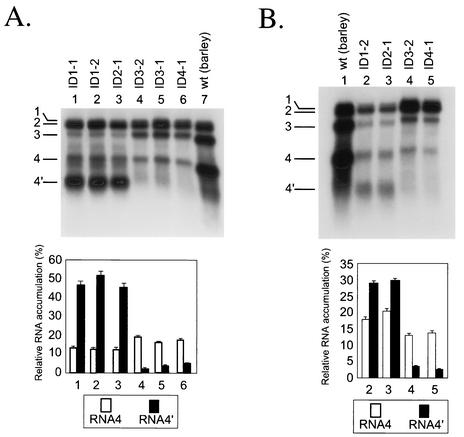
To determine the accumulation of BMV RNA in vivo, total RNA preparations were extracted from an equal weight of the combined local lesion tissue (100 mg) and analyzed by Northern blotting, as described in Materials and Methods. Figure 2A shows that ID1-1 (lane 1), ID1-2 (lane 2), ID2-1 (lane 3), ID3-2 (lane 4), ID3-1 (lane 5), and ID4-1 (lane 6) subgenomic RNA4s accumulated to comparable levels, reflecting similar transcriptional activity from the INT-1 sgp. Also, ID1-1, ID1-2, and ID2-1 generated the sg RNA4′ (lanes 1 to 3), while ID3-1, ID3-2, and ID4-1 did not (lanes 4 to 6). This proved that the core region nucleotide substitutions inhibited the transcriptional activity of the INT-2 sgp, verifying the in vitro results of Siegel et al. (32). The concentration of subgenomic RNA4′ was higher than that of RNA4 (lanes 1 to 3; from 45.8 to 52.3% of the total amount of transcripts for RNA4′, and from 12.8 to 13.3% for RNA4), which verified earlier observations in barley protoplasts reported by French and Ahlquist (15). The accumulation of RNA3 was higher for the ID constructs with inactive INT-2 than for those with active INT-2 (results not quantified). This could be due to the competition for encapsidation with RNA4′ (10) or competition for viral replicase. Both RNA3 and RNA4 accumulated to lower levels in C. quinoa (lanes 1 to 6) than in barley (lane 7), which has been observed before for the wt virus (14, 26).
FIG. 2.
Northern blot analysis of the accumulation of BMV ID RNA3 constructs in C. quinoa (A) or in barley (B). The leaves were coinoculated mechanically with the in vitro-transcribed wt RNA1 and RNA2 and with ID RNA3 mutants (as indicated on the top). The progeny BMV RNA was extracted from the infected tissue, separated electrophoretically in a 1% agarose-formamide-formaldehyde gel, blotted to a nylon membrane (HybondN+; Amersham), and probed with a 32P-labeled RNA complementary to the 3′ 200 nt of the plus strand. Lanes 1 to 6 (A) or 2 to 5 (B) show the RNAs extracted from the local lesion tissue of C. quinoa or of barley, respectively. The control lanes 1 and 7 show wt BMV RNA from barley. The positions of individual BMV RNA segments are indicated on the left. The histograms below show the quantification of the accumulation of sg RNA4 and RNA4′. The Northern blot autoradiograms were scanned and subjected to computer-based densitometric analysis by using the program ImageQuant 5.0 of Amersham Biosciences. The bars show the fraction (percent) by which the band intensity for RNAs 4 and 4′ contributed to the total intensity of all BMV RNA components.
To determine the level of BMV accumulation after a large number of replication cycles, individual ID RNA3 constructs were inoculated on barley seedlings, a systemic host for BMV, and the RNA was extracted 10 days after infection. As shown in Fig. 2B, the concentration of BMV RNA reached similar levels among the constructs ID1-2, ID2-1, ID3-2, and ID4-1 (lanes 2 to 5) and, more importantly, RNA4′ accumulated only for those constructs that carried the unmodified INT-2 sgp sequence (lanes 2 and 3). This confirmed that the nucleotide substitutions within the sgp core effectively arrested transcription at INT-2.
Coinfections with mutants carrying a transcriptionally active sgp at INT-2.
To establish whether the sgp sequence could support homologous crossovers, C. quinoa plants were coinoculated with mutants ID1-1 and ID1-2 (Fig. 1). The INT-2 region of the progeny RNA3 was amplified by RT-PCR, and the cDNA products were cloned. Of 91 clones analyzed (Table 1), 68 represented the parental constructs, 8 were type 1 recombinants (ID1-1 to ID1-2 [Fig. 1]), and 15 were type 2 recombinants (ID1-2 to ID1-1). Thus, the recombination frequency was 25%, demonstrating that the sgp subset at INT-2 was capable of supporting crossover events. The ratio of parental ID1 RNA3 mutants in the RNA progeny was almost even (33 clones represented ID1-1 and 35 clones represented ID1-2), demonstrating the lack of selective advantage among the mutants and, thus, equal availability of both parental RNAs for recombination.
TABLE 1.
Restriction analysis of the parental and recombinant cDNA clones in the progeny BMV RNA3
| Mutant RNA3 paira | No. of parents
|
No. of INT-2 recombinants
|
No. of INT-1 recombinants
|
No. of CP recombinants (either direction) | |||
|---|---|---|---|---|---|---|---|
| P1 | P2 | P1 to P2 | P2 to P1 | P1 to P2 | P2 to P1 | ||
| ID1-1 (P1) and ID1-2 (P2) | 33 | 35 | 8 | 15 | |||
| ID2-1 (P1) and ID1-1 (P2) | 9 | 11 | 10 | 8 | 10 | 7 | 16 |
| ID3-1 (P1) and ID3-2 (P2) | 34 | 35 | 0 | 0 | |||
| ID4-1 (P1) and ID3-1 (P2) | 10 | 9 | 0 | 0 | 3 | 5 | 3 |
Plants were coinoculated with different combinations of BMV ID RNA3 variants (as specified; see also Fig. 1). RT-PCR products were obtained from RNAs extracted from local lesions and analyzed as described in Materials and Methods. P1 and P2 stand for parental ID RNA3 variants 1 and 2, respectively.
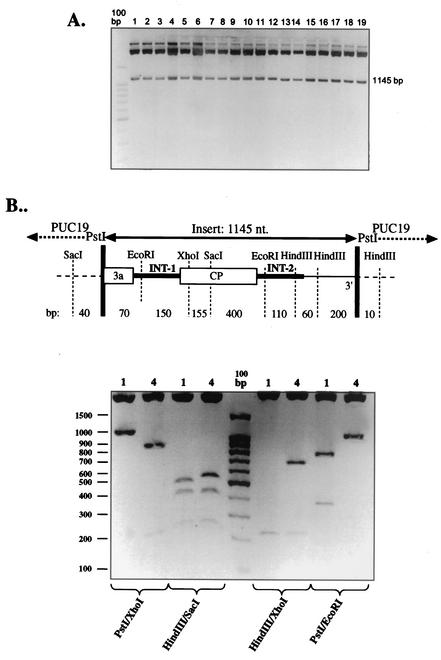
To check whether RT-PCR could generate recombinants, C. quinoa plants were infected separately with either ID1-1 or ID1-2 RNA3s, all at the same time as the coinfections described above. The RNA extracts were harvested 10 days after infection (to secure the same concentration as in the coinoculation experiments) and mixed together at the 1:1 ratio, and the cDNA was amplified by RT-PCR and cloned. All 19 clones analyzed carried the cDNA insert of the same size (Fig. 3A), and a restriction analysis of 19 clones (two exemplary digestions of clones 1 and 4 are shown in Fig. 3B) confirmed only the presence of parental RNA3 sequences, demonstrating that the observed crossovers occurred during coinfection.
FIG. 3.
Electrophoretical analysis of cDNA clones obtained during a control experiment towards testing if RT-PCR generated RNA3 recombinants. (A) The progeny BMV RNAs from separately inoculated plants (with ID2-1 or ID1-1 RNA3s) were mixed together and coamplified with RT-PCR, using the primers RW-Pst and RW-PstI. The DNA was cloned in the pUC19 vector between PstI sites, and the inserts (of 1,145 nt) were released from the resulting plasmids with PstI digestion and separated in a 1.5% agarose gel. No size variability was demonstrated among the 19 analyzed clones. (B) Exemplary restriction analysis of clones 1 and 4 from lanes 1 and 4 in panel A, respectively. Plasmid DNA was digested with the indicated restriction enzymes, and the products were separated in a 1.5% agarose gel against the 100-bp DNA standard (Promega). The size of the individual bands corresponds to the parental fragments, as predicted with the diagram (not to scale) on top.
During the course of the experiment, we did not observe any variability in the length of the obtained PCR products, and none of the clones carried a modified sequence. This confirmed the precision of the observed homologous crossovers (see Discussion).
To determine whether the recombination frequencies at INT-2 reflected the wt recombination activity, we examined the recombination activity at the original sgp locus (INT-1). The ID2-1 and ID1-1 RNA3s were coinoculated on C. quinoa leaves, and equal concentrations of the progeny RNA were analyzed for the recombinants at both loci as described above. Of the 71 cDNA clones analyzed (Table 1), 17 represented the recombinants at INT-1 (types 4 and 5 in Fig. 1B) and 18 represented recombinants at INT-2 (types 1 or 3), whereas 16 crossed within the control CP open reading frame (ORF) (type 6). The corresponding recombination frequencies for these regions were 24, 22, and 25%, respectively. Similarly to ID1-1 × ID1-2 infection, there was an even distribution of parental RNA3 variants in the RNA3 progeny (9 clones for ID2-1 and 11 clones for ID1-1), reflecting an equal availability of both templates for crossovers. These results demonstrated that the truncated sgp at INT-2 supported a recombination frequency similar to that for an intact sgp sequence at INT-1, with the latter being embraced by the markers only at slightly wider locations. In contrast, an almost 4 times larger distance between the markers flanking the CP region made the actual frequency at the CP ORF almost 4 times lower.
To find out if RT-PCR could generate the observed recombinants, parallel amplification reactions were performed using equal concentrations of total RNA preparations that were obtained from plants inoculated separately with either ID2-1 or ID1-1 RNAs and harvested at the same time, as described above for ID1-1 × ID1-2 infections. Similar analyses to those described for Fig. 3 confirmed the lack of RNA3 recombinants among 30 analyzed cDNA clones (data not shown).
Coinfections with mutants carrying a transcriptionally inactive sgp at INT-2.
To test whether the transcriptional activity of the sgp was required for recombination, mutants ID3-1 and ID3-2 were coinoculated on C. quinoa hosts. The INT-2 region of the progeny RNA3 was amplified by RT-PCR, and the cDNA products were cloned. The analysis of 69 clones (Table 1) revealed the accumulation of only parental RNA3 constructs, almost equally distributed: 34 clones for ID3-1 and 35 clones for ID3-2. This demonstrated that, although both parental RNAs could accumulate to equal levels during infection, they did not recombine with each other within INT-2. Thus, the transcriptionally active sgp was essential for homologous crossovers to occur at INT-2.
To compare the recombination frequency at the active loci (INT-1) with that at the inactive loci (INT-2), another coinfection experiment involved mutants ID4-1 and ID3-1 (Fig. 1). The analysis of 30 clones demonstrated the presence of five recombinants in one direction (type 9) and three recombinants in another direction (type 4) at INT1, but no recombinants at INT-2 (Table 1). This gave a 27% recombination frequency at INT-1, comparable to the frequency determined at INT-1 for the ID2-1 × ID1-1 infection (see above). The parental ID4-1 and ID3-1 RNAs accumulated to similar levels, yielding 10 and 9 clones, respectively (Table 1), which reflected the lack of selection among the recombining molecules. These data confirmed that recombination could occur at the transcriptionally active sgp but not at the inactive sgp. A significantly lower recombination frequency (10%) was again observed within the larger region of the CP ORF (two type 10 recombinants and one wt RNA3).
To rule out the possibility of recombinant RT-PCR artifacts, the control RT-PCR amplifications were performed on mixtures of RNA preparations that were extracted from singly infected plants (with either ID4-1 or ID3-1). Similar analyses to those presented in Fig. 3 did not reveal recombinant RNA3 sequences among the 18 cDNA clones (data not shown).
DISCUSSION
Many diverse virus groups utilize subgenomic RNA promoters for expression of their genetic information. Although sg RNA synthesis has been studied in several virus groups (24) and the BMV sgp sequences have been mapped (21, 22, 32, 34), additional functions of sgp's are poorly understood. It has been suggested previously that recombination has taken place preferentially at sg RNA promoters (5, 23), and it has been proposed that viral RdRps could select the recombination sites on the basis of resemblance to RNA replication promoters or enhancers (30). In this work we report a novel activity for the sgp on BMV RNA3, i.e., that of homologous recombination. We demonstrate for the first time that a functional sg RNA promoter was required for recombination to occur.
We used two pairs of RNA3 constructs (ID1 and ID2) that carried the sgp repeat at an additional, functionally neutral locus (called INT-2), just downstream to the CP stop codon. One pair (ID1-1-ID1-2) had marker mutations only at INT-2 flanking positions, while another pair (ID2-1-ID1-1) carried flanking mutations at both INT-1 (the wt sgp site) and INT-2. The coinfections revealed comparable recombination frequencies (24 to 27%) at both loci. The third pair (ID3-1-ID3-2), which carried a transcriptionally silent INT-2 sgp sequence, did not exhibit the recombination activity at INT-2. However, the use of a fourth pair (ID4-1-ID3-1) revealed a regular level of recombination (27%) at the transcriptionally active INT-1 site, while the crossovers at the silent INT-2 remained undetectable (Fig. 1 and Table 1).
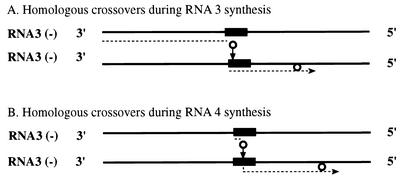
To explain these findings, a model is presented in Fig. 4. Earlier studies have demonstrated that BMV RdRp could initiate on minus strands of RNA3 either at the 3′ end (to copy full-length positive RNA3 strands [3]) or internally (to synthesize subgenomic RNA4 [1, 22, 32]). That BMV RdRp can switch between templates has been demonstrated in vitro by Kim and Kao (20) and by Dzianott et al. (12). Figure 4A predicts that, after initiation at the 3′ end, the RdRp pauses and detaches at the sgp region and reinitiates at the sgp region on another RNA3 minus-strand template. It is known, for instance, that secondary structures or AU-rich regions can destabilize polymerase ternary complexes (33, 35), or they can facilitate the RdRp slippage (6). In addition, the RdRp complexes (or other proteins) that are bound to the sgp sequence may hinder the trafficking of other RdRp molecules. The reinitiation could occur by priming with the detached 3′ portion of the nascent strand, which rehybridizes to another RNA3 template.
FIG. 4.
Diagrammatic representation of a putative mechanism of crossovers at the sgp recombination hot spot. Thick lines represent minus strands of BMV RNA3, while dotted lines represent progeny plus strands. Arrows show the direction of migration of the RdRp complex (represented by a solid circle). (A) RNA-RNA crossovers during copying of plus strands of RNA3. The replicase initiates the synthesis of plus strands at the 3′ end, pauses at the sgp, and switches onto another RNA3 molecule. (B) De novo initiation of RNA4 transcription.
De novo initiation of RNA4 transcription is shown in Fig. 4B. Theoretically, the crossovers could occur at this stage as well. Caused by similar factors to those mentioned above, the RdRp could pause or detach within the sgp region after internal initiation of RNA4 synthesis, and the aborted short RNA products could prime on another minus-sense RNA3 template. A stepwise mechanism of the initiation of RNA4 has been described based on the in vitro accumulation of short oligonucleotide products (2). However, it is difficult to detect such RNA4 recombinants by RT-PCR amplification, because the upper-side oligonucleotide primers would need to cover short 5′ sequences in RNA4. The recombination model with abortive RNAs as primers has been proposed by others before (30, 31).
It is possible that the events of transcription and recombination do not share a straight cause-and-effect relationship. One mechanism (predicted by the model in Fig. 4) might be that both transcription and recombination require the same sgp stem-loop structure for the RdRp binding and reattachment, respectively. Indeed, mutations at known RdRp contact sites (1, 18, 32) inhibited the synthesis of RNA4′ and recombination at INT-2. However, transcription and recombination may use different mechanisms for initiation, i.e., unprimed (de novo) and primed, respectively. The role of priming by the nascent strand was demonstrated for recombination in turnip crinkle carmovirus RdRp (30), and the capability of both primer extension and de novo initiation were shown in vitro for BMV RdRp (31). Also, the destabilization of the stem-loop structure may prevent stalling of the RdRp, so the limiting factor might be the dissociation from the donor strand, rather than the reinitiation on the acceptor strand.
Correlation between sg RNA synthesis at INT-1 and INT-2 (RNA4 versus RNA4′) (Fig. 2) and recombination was poor. The sg RNA concentration may depend upon such postevent factors as RNA stability, encapsidation, RNA transport, positional effects favoring synthesis of shorter sg RNAs, or other, yet unknown, regulation mechanisms (15). It is therefore unclear whether the original rate of transcription parallels the original rate of recombination. On the other hand, the effects of RNA fitness on recombination frequency and on the profiles of recombinants seem to be insignificant, for no substantial differences in the accumulation levels were observed among the ID RNA3 constructs (Fig. 1). Alternatively, mutations in ID3 constructs could inhibit transcription and recombination independently. For instance, transcription could be affected directly by mutations, whereas recombination could be affected by a changed sequence context. Our laboratory has shown previously that the sequence context was important for homologous recombination (28, 29). Otherwise, the flanking marker nucleotides have been at functionally neutral positions.
Theoretically, reciprocal inter- and/or intramolecular crossovers could take place between homologous sequences of INT-1 and INT-2. The products of such crossovers would, however, carry a deleted or duplicated CP gene and, thus, be out-competed by selection pressure. Regardless, INT-2 differs from INT-1 by the presence of specific marker sites, which would have been redistributed in case of reciprocal recombinants.
Our data reveal only precise homologous RNA3-RNA3 recombinants within the INT-1 or INT-2 sgp sequences. In contrast, for recombination at the 3′ noncoding region, a significant fraction of imprecise homologous recombinants has been observed (27-29). This might reflect differences in the mechanism of recombination at the 3′ versus sgp regions. For instance, 3′ recombination occurred upstream to the minus-strand promoter (27), whereas sgp-mediated recombination occurred within the transcription promoter. Also, 3′ recombination was between different BMV RNA segments (RNA2 and RNA3), while sgp recombination was among the same (RNA3) segment. One may speculate that the promoter-driven crossovers (such as sgp crossovers) would tend to occur more precisely than those at other regions because the polymerase binds to the predetermined signal sequences.
The observed recombination frequency at the control CP ORF (for pairs ID2-1-ID1-1 and ID4-1-ID3-1) must be considered lower than at INT-1 and INT-2 sites, due to larger distances between marker mutations. A computer analysis did not reveal obvious homologies of the CP ORF to the sgp sequence (data not shown). A more detailed mapping is required to find out whether the CP region carries a distinct recombination hot spot, thus adding to the broader inventory of sequences that support homologous RNA-RNA crossovers in BMV.
Acknowledgments
We thank Barbara Ball for excellent assistance with the figures.
This work was supported by a grant from the National Science Foundation (MCB-9983033), by the Polish Government through a grant (6 P04C 046 19) from the State Committee for Scientific Studies, awarded to J.J.B., and by the Plant Molecular Biology Center at Northern Illinois University.
REFERENCES
- 1.Adkins, S., and C. C. Kao. 1998. Subgenomic RNA promoters dictate the mode of recognition by bromoviral RNA-dependent RNA polymerases. Virology 252:1-8. [DOI] [PubMed] [Google Scholar]
- 2.Adkins, S., S. S. Stawicki, G. Faurote, R. W. Siegel, and C. C. Kao. 1998. Mechanistic analysis of RNA synthesis by RNA-dependent RNA polymerase from two promoters reveals similarities to DNA-dependent RNA polymerase. RNA 4:455-470. [PMC free article] [PubMed] [Google Scholar]
- 3.Ahlquist, P. 1992. Bromovirus RNA replication and transcription. Curr. Opin. Genet. Dev. 2:71-76. [DOI] [PubMed] [Google Scholar]
- 4.Balakrishnan, M., P. J. Fay, and R. A. Bambara. 2001. The kissing hairpin sequence promotes recombination within the HIV-I 5′ leader region. J. Biol. Chem. 276:36482-36492. [DOI] [PubMed] [Google Scholar]
- 5.Bar-Joseph, M., G. Yang, R. Gafny, and M. Mawass. 1997. Subgenomic RNAs: the possible building blocks for modular recombination of Closteroviridae genomes. Semin. Virol. 8:113-119. [Google Scholar]
- 6.Barr, J. N., and G. W. Wertz. 2001. Polymerase slippage at vesicular stomatitis virus gene junctions to generate poly(A) is regulated by the upstream 3′-AUAC-5′ tetranucleotide: implications for the mechanism of transcription termination. J. Virol. 75:6901-6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruyere, A., M. Wantroba, S. Flasinski, A. Dzianott, and J. J. Bujarski. 2000. Frequent homologous recombination events between molecules of one RNA component in a multipartite RNA virus. J. Virol. 74:4214-4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bujarski, J. J. 1996. Introduction: experimental systems of genetic recombination and defective RNA formation in RNA viruses. Semin. Virol. 7:361-362.
- 9.Cascone, P. J., T. F. Haydar, and A. E. Simon. 1993. Sequences and structures required for recombination between virus-associated RNAs. Science 260:801-805. [DOI] [PubMed] [Google Scholar]
- 10.Choi, Y. G., T. W. Dreher, and A. L. Rao. 2002. tRNA elements mediate the assembly of an icosahedral RNA virus. Proc. Natl. Acad. Sci. USA 99:655-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dzianott, A., S. Flasinski, S. Pratt, and J. J. Bujarski. 1995. Foreign complementary sequences facilitate genetic recombination in brome mosaic virus. Virology 208:370-375. [DOI] [PubMed] [Google Scholar]
- 12.Dzianott, A., N. Rauffer-Bruyere, and J. J. Bujarski. 2001. Studies on functional interaction between brome mosaic virus replicase proteins during RNA recombination, using combined mutants in vivo and in vitro. Virology 289:137-149. [DOI] [PubMed] [Google Scholar]
- 13.Figlerowicz, M., and J. J. Bujarski. 1998. RNA recombination in brome mosaic virus, a model plus strand RNA virus. Acta Biochim. Pol. 45:847-868. [PubMed] [Google Scholar]
- 14.Figlerowicz, M., P. D. Nagy, and J. J. Bujarski. 1997. A mutation in the putative RNA polymerase gene inhibits nonhomologous, but not homologous, genetic recombination in an RNA virus. Proc. Natl. Acad. Sci. USA 94:2073-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.French, R., and P. Ahlquist. 1988. Characterization and engineering of sequences controlling in vivo synthesis of brome mosaic virus subgenomic RNA. J. Virol. 62:2411-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gmyl, A. P., E. V. Belousov, S. V. Maslova, E. V. Khitrina, A. B. Chetverin, and V. I. Agol. 1999. Nonreplicative RNA recombination in poliovirus. J. Virol. 73:8958-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greene, A. E., and R. F. Allison. 1994. Recombination between viral RNA and transgenic plant transcripts. Science 263:1423-1425. [DOI] [PubMed] [Google Scholar]
- 18.Haasnoot, P. C., F. T. Brederodem, R. C. Olsthoorn, and J. F. Bol. 2000. A conserved hairpin structure in Alfamovirus and Bromovirus subgenomic promoters is required for efficient RNA synthesis in vitro. RNA 6:708-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janda, M., R. French, and P. Ahlquist. 1987. High efficiency T7 polymerase synthesis of infectious RNA from cloned brome mosaic virus cDNA and effects of 5′ extensions of transcript infectivity. Virology 158:259-262. [DOI] [PubMed] [Google Scholar]
- 20.Kim, M. J., and C. C. Kao. 2001. Factors regulating template switch in vitro by viral RNA-dependent RNA polymerases: implications for RNA-RNA recombination. Proc. Natl. Acad. Sci. USA 98:4972-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marsh, L. E., T. W. Dreher, and T. C. Hall. 1988. Mutational analysis of the core and modulator sequences of the BMV RNA3 subgenomic promoter. Nucleic Acids Res. 16:981-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, W. A., T. W. Dreher, and T. C. Hall. 1985. Synthesis of brome mosaic virus subgenomic RNA in vitro by internal initiation on (−)-sense genomic RNA. Nature 313:68-70. [DOI] [PubMed] [Google Scholar]
- 23.Miller, W. A., S. P. Dinesh-Kumar, and C. P. Paul. 1995. Luteovirus gene expression. Crit. Rev. Plant Sci. 13:179-211. [Google Scholar]
- 24.Miller, W. A., and G. Koev. 2000. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology 273:1-8. [DOI] [PubMed] [Google Scholar]
- 25.Nagy, P. D., and J. J. Bujarski. 1992. Genetic recombination in brome mosaic virus: effect of sequence and replication of RNA on accumulation of recombinants. J. Virol. 66:6824-6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagy, P. D., and J. J. Bujarski. 1993. Targeting the site of RNA-RNA recombination in brome mosaic virus with antisense sequences. Proc. Natl. Acad. Sci. USA 90:6390-6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagy, P. D., and J. J. Bujarski. 1995. Efficient system of homologous RNA recombination in brome mosaic virus: sequence and structure requirements and accuracy of crossovers. J. Virol. 69:131-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagy, P. D., and J. J. Bujarski. 1997. Engineering of homologous recombination hotspots with AU-rich sequences in brome mosaic virus. J. Virol. 71:3799-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagy, P. D., C. Ogiela, and J. J. Bujarski. 1999. Mapping sequences active in homologous RNA recombination in brome mosaic virus: prediction of recombination hot spots. Virology 254:92-104. [DOI] [PubMed] [Google Scholar]
- 30.Nagy, P. D., J. Pogany, and A. E. Simon. 1999. RNA elements required for RNA recombination function as replication enhancers in vitro and in vivo in a plus-strand RNA virus. EMBO J. 18:5653-5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ranjith-Kumar, C. T., X. Zhang, and C. C. 2003. Enhancer-like activity of a brome mosaic virus RNA promoter. J. Virol. 77:1830-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siegel, R. W., S. Adkins, and C. C. Kao. 1997. Sequence-specific recognition of a subgenomic RNA promoter by a viral RNA polymerase. Proc. Natl. Acad. Sci. USA 94:11238-11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steitz, T. 1998. A mechanism for all polymerases. Nature 391:231-232. [DOI] [PubMed] [Google Scholar]
- 34.Sullivan, M. L., and P. Ahlquist. 1999. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J. Virol. 73:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Hippel, P. H. 1998. An integrated model of the transcription complex in elongation, termination, and editing. Science 281:660-665. [DOI] [PubMed] [Google Scholar]
- 36.Zhuang, J., A. E. Jetzt, G. Sun, H. Yu, G. Klarmann, Y. Ron, B. D. Preston, and J. P. Dougherty. 2002. Human immunodeficiency virus type 1 recombination: rate, fidelity, and putative hot spots. J. Virol. 76:11273-11282. [DOI] [PMC free article] [PubMed] [Google Scholar]