Abstract
Trigger factor, a ribosome-associated chaperone and peptidyl-prolyl cis-trans isomerase (PPIase), is essential for the secretion and maturation of the cysteine protease of the pathogenic gram-positive bacterium Streptococcus pyogenes. In the absence of trigger factor, the nascent protease polypeptide is not targeted to the secretory pathway. Some partial-function mutations restore targeting. However, the secreted protease does not efficiently mature into an enzymatically active form, suggesting that trigger factor has an additional role in protease biogenesis. Here, we show that, while not required for targeting, the PPIase activity of trigger factor is essential for maturation of the protease following its secretion from the bacterial cell. Site-specific mutations introduced into ropA, the gene which encodes trigger factor in S. pyogenes, produced mutant proteins deficient in PPIase activity. When these mutant alleles were used to replace the wild-type gene on the streptococcal chromosome, analysis of protease biogenesis revealed that, although the protease was secreted normally, it did not efficiently mature to an active form. Furthermore, mutation of a single proline residue in the protease prodomain suppressed the requirement for PPIase activity, suggesting that this residue is the target of trigger factor. These data support a model in which trigger factor-mediated prolyl isomerization influences the conformation of the prodomain, which in turn directs the protease into one of several alternative folding pathways.
The folding of a protein into its native structure requires the coordinated interaction of a network of accessory proteins known as molecular chaperones. There has been considerable progress in understanding how chaperones function to fold proteins in the cytoplasm (for a review see reference 1); however, the mechanisms by which they contribute to the folding of secreted proteins are poorly understood. An example of a cytoplasmic chaperone is trigger factor, an enigmatic protein that was originally identified in a cell-free secretion system as a factor that was required to present the substrate precursor protein proOmpA to the secretory apparatus in a translocation-competent form (5). However, subsequent studies suggested that trigger factor was neither essential for secretion of any known protein in Escherichia coli or Bacillus subtilis nor essential for cell viability (4, 9, 14, 38). Trigger factor has many remarkable properties: it associates with the ribosome, binds to nascent polypeptides in vivo, and possesses peptidyl-prolyl cis-trans isomerase (PPIase) activity (21, 23). Recent studies have indicated that, while mutants lacking the gene for trigger factor or for the chaperone DnaK are viable under normal growth conditions, simultaneous mutation of both genes is lethal (8, 38). Thus, there is considerable evidence that trigger factor plays an important role in protein synthesis; however, its function is far from understood.
A new model for understanding trigger factor function has come from the observation that trigger factor is required for the secretion and maturation of the cysteine protease of the gram-positive bacterium Streptococcus pyogenes (group A streptococcus) (26). As the causative agent of diseases, which include pharyngitis (strep throat) and necrotizing fasciitis, S. pyogenes is an important human pathogen, and the secreted cysteine protease (also known as SpeB) may contribute to the organism's ability to cause disease (3, 24, 25). Interestingly, trigger factor was shown to be required for two distinct steps in the biogenesis of the active protease. First, while a trigger factor mutant produced the protease polypeptide, the protease was not secreted from the bacterial cell, suggesting that trigger factor is required for targeting the protease to the secretory pathway. Second, an in-frame deletion mutation that excised just the central region of trigger factor resulted in normal levels of secretion of the protease zymogen. However, the secreted protease from this mutant exhibited a multipartite defect in maturation, including a kinetic defect in the autologous processing of the zymogen to the mature form, and the mature protease had a catalytic defect (26). These data indicate that, while this mutant form of trigger factor is competent for targeting the protease, the central domain has activity which influences the maturation of the protease following its translocation across the cellular membrane. Furthermore, this activity is required in response to some specific feature of the protease since streptococcal trigger factor mutants do not exhibit a general defect in protein secretion and function (26).
Multiple roles for trigger factor in the biogenesis of the streptococcal protease are consistent with the modular structure of trigger factor. Of the three distinct domains defined for trigger factor, the N- and C-terminal domains have been shown to interact with each other and are required for binding to substrate polypeptides (41). In addition, the N-terminal 118 amino acids are required for binding to ribosomes (17). The central domain has homology to the family of FK506-binding PPIase proteins (13, 16, 35). This class of proteins can catalyze the isomerization of the peptide bond preceding a prolyl residue, and the central domain of trigger factor can function independently in vitro as an active PPIase (16, 35). Prolyl isomerization can be a rate-limiting step in protein folding (22), and multiple studies of in vitro folding reactions support a role for PPIases in folding (for reviews, see references 12 and 31). However, since mutations of PPIases generally do not affect folding in vivo (10), the function of PPIases in protein synthesis is less clear. The refolding of denatured proteins in vitro can be catalyzed by trigger factor (14, 18). However, it appears that this refolding reaction is facilitated primarily through the chaperone function of trigger factor, involving binding to hydrophobic regions exposed in the partially folded protein to prevent aggregation rather than promotion of prolyl isomerization (18). Thus, the role of the PPIase activity of trigger factor is not understood.
The PPIase domain was included in the region removed by the large central deletion in trigger factor that was analyzed for S. pyogenes (26). Since the resulting mutant secreted a defective protease, these data support a model in which the PPIase activity of trigger factor introduces essential information into the nascent polypeptide prior to its secretion. Furthermore, this model suggests that the molecular nature of this information involves prolyl isomerization. However, it is possible that, besides altering PPIase activity, the large deletion introduced into the S. pyogenes trigger factor may have altered its conformation, stability, and possible interdomain interactions. Thus, a specific requirement for PPIase activity remains to be determined. The model also predicts that there is at least one prolyl residue in the protease polypeptide whose conformation is critical for maturation. If correct, this would provide a novel model for understanding the contribution of prolyl isomerization to protein folding in vivo.
In the present study, analysis of specific RopA mutants provided evidence to support an essential role for the PPIase activity of trigger factor in protease maturation. Data to implicate the isomerization state of a single prolyl residue in directing the protease polypeptide into a productive or nonproductive maturation pathway are also presented. The location of this residue in the protease prodomain suggests a model in which prolyl isomerization influences the conformation of the prodomain, which in turn directs the protease into one of several alternative folding pathways.
MATERIALS AND METHODS
Bacterial strains.
Molecular cloning experiments utilized E. coli DH5α (Gibco-BRL), and E. coli strain BL21(DE3) (36) was used for protein expression. Strain HSC130 contains ropA from wild-type S. pyogenes strain HSC5 with an in-frame deletion of the region encoding residues 83 to 297 (ropAΔ82-297) (26). The construction of additional HSC5 mutants is described below. Routine culture of S. pyogenes strains utilized Todd-Hewitt medium (BBL) supplemented with 0.2% yeast extract (Difco). When appropriate, antibiotics were added to media at the following concentrations: kanamycin, 50 μg/ml for E. coli and 500 μg/ml for S. pyogenes; erythromycin, 750 μg/ml for E. coli and 1 μg/ml for S. pyogenes; ampicillin, 100 μg/ml for E. coli. All mutations in S. pyogenes were stably maintained to the extent that culture for all functional assays did not require the addition of antibiotics.
Plasmid construction.
Plasmids for expression of trigger factor were based on the expression vector pET24D+ (Novagen) and were constructed as follows. Oligonucleotide primers startNcoI (AATGA CTCCA TGGCT ACATC ATTTG AAAAC TTTCG ATG) and endXhoI (CCTTA ATACT CGAGC TTAAC GCTTG CTGTG CTTGT AATCA C) were used to amplify ropA by PCR from the plasmid pBL40 (see below). The resulting DNA fragment was inserted between the NcoI and XhoI sites of pET24D+ by using the NcoI and XhoI sites embedded in the primer sequences (underlined). The same strategy was used to construct expression plasmids for each of the ropA alleles engineered to encode a substitution of an alanine residue at selected positions (see below). The fidelity of the resulting plasmids, pROP4 (wild type), pROP5 (ropAD180A), pROP6 (ropAF200A), and pROP7 (ropAF235A), was confirmed by determination of their DNA sequences. Plasmids were constructed so as to place six-His affinity tags at the carboxy termini of the expressed RopA proteins.
Purification of recombinant RopA.
Plasmids were introduced into E. coli expression strain BL21(DE3), and cultures for expression were prepared by inoculation of 500 ml of Luria-Bertani medium supplemented with ampicillin with growth from an overnight cultures to an optical density at 600 nm (OD600) of 0.05. Cultures were incubated at 37°C with agitation (250 rpm) until an OD600 of 0.6 was reached, at which time isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.2 mM. Following an additional 3-h incubation, cultures were harvested by centrifugation (10 min, 6,000 × g, 4°C) and resuspended in 1 ml of a nondenaturing lysis buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 10 mM imidazole, 1 mg of lysozyme/ml). After incubation on ice for 30 min, bacteria were lysed by sonication (three pulses, 10 s each; model 185; Branson) with a microprobe at the highest-power setting. Debris was removed by centrifugation (16,000 × g, 20 min, 4°C), and the six-His-tagged proteins were purified from the supernatant fluids by chromatography over a nickel affinity resin (Ni-nitrilotriacetic acid) according the recommendations of the manufacturer (Qiagen). Purity of preparations was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and staining with Coomassie brilliant blue and was consistently greater than 95%. Protein concentrations were determined by a bicinchoninic acid assay (Sigma).
Prolyl-isomerase activity assays.
The presence of PPIase activity was determined by a protease coupling assay described by Fischer et al. (11). This assay measures the conformation-specific cleavage of succinyl-Ala-Ala-Pro-Phe-4-nitroanilide (Bachem) by α-chymotrypsin (Sigma), which occurs only when the substrate is in the trans conformation. The substrate exists in an equilibrium such that at pH 7.8 approximately 20% is in the cis conformation, and the rate of conversion of this fraction of substrate to a cleavable form in the presence or absence of PPIase is determined. A stock solution of substrate (10 mg/ml) was prepared in dimethyl sulfoxide and was diluted in reaction buffer (0.035 M HEPES, pH 7.8) containing α-chymotrypsin (0.5 mg/ml) and 1.0 μM PPIase. The PPIases included RopA, mutant RopAs, and a well-characterized PPIase, recombinant human FK-binding protein (Sigma). For kinetic analysis, the change in absorbance at 390 nm over time was monitored at 10°C in a spectrophotometer (Beckman DU-80). To measure solely the cis isomerization reaction without PPIase-independent cleavage of the trans form, measurements were performed following the initial cleavage of the trans population, as determined through control reactions lacking PPIase activity. In the presence of PPIase, the kinetic equation is kobs = ku + 2(kcat/Km), where kobs is the first-order rate constant for isomerization. The variable ku represents the first-order rate constant for uncatalyzed isomerization, and kcat/Km is the catalytic activity. Thus, the enzyme-catalyzed rate of isomerization (kcat/Km) can be calculated by the equation kcat/Km = (kobs − ku)/2.
Construction of mutant streptococcal strains.
Streptococcal strains that expressed RopA containing single amino acid substitutions for alanine at selected residues were constructed by alteration of the single chromosomal copy of ropA as follows. Oligonucleotide primers RopA 5′ XbaI (GCCAT AGTCA TCCGT CTAGA AATGC) and RopA 3′ PstI (CTCAT ATCAC TGCAG CTTGA CAAAT C) were used to amplify by PCR the region containing the ropA open reading frame from the HSC5 chromosome; this region was then inserted between the XbaI and PstI sites of the E. coli/streptococcal shuttle vector pJRS233 (29) by using restriction sites embedded in the primer sequences (underlined). The resulting plasmid (pBL40) was used as the template in an “inside-out” PCR with primers RopA 180D/A up (CACCA TCAAC GGATC CAACA AAGTC AATC) and RopA 180D/A down (TTGTT GGATC CGTTG ATGGT GTTGA GTTTG CTGGC). Digestion of this amplification product with BamHI (sites underlined), followed by religation, resulted in the replacement of the aspartate codon with an alanine codon in ropA. The altered allele (ropAD180A) was used to replace the resident chromosomal allele as described elsewhere (30), and replacement was confirmed by PCR amplification and digestion of amplified products with BamHI to probe for the unique BamHI site introduced at the mutated codon. The resulting S. pyogenes mutant was designated RopA11. Additional alterations were made by the same method. Primers pairs RopA 200F/A start (ATTGT CCGGA TCCAA GTTCA AGAGA GAAG) and RopA 200F/A end (AACTT GGATC CGGAC AATTT ATCCC AGGTG CTGAA) and RopA 235F/A start (TTAGC GGCGG ATCCT GCAAG ATCTT CTGC) and RopA 235F/A end (CTTGC AGGAT CCGCC GCTAA AGCTA TGACA AC) were used for substitution of alanine for phenylalanine at positions 200 (ropA200F/A) and 235 (ropA235F/A), respectively. The resulting plasmids were designated pROP2 and pROP3, and the corresponding mutant strains were designated RopA12 and RopA13.
Streptococcal strains with the proline at residue 78 replaced by glycine were constructed by alteration of the single chromosomal copy of speB as follows: Oligonucleotide primers −1000speBBamHI (GAATG CCTAA TGGAT CCAAC GGTTT CACAA) and SpeBstopBamHI (GGATA GCTTA ACTGC TGGAT CCGCA TAGGG) were used to amplify by PCR the region containing the speB open reading frame from the HSC5 chromosome; this region was then inserted at the BamHI site of the E. coli/streptococcus shuttle vector pJRS233 (29) by using restriction sites embedded in the primer sequences (underlined). The resulting plasmid (pSpeB112) was then used as the template in an inside-out PCR with primers SpeBP78G start (CCTAG AATTT CTCCA GATCT TTTAT CTCC) and SpeBP78Gend (GGAGA TAAAA GATCT GGAGA AATTC TAGGA TAC). Digestion of this amplification product with BglII (sites underlined) followed by religation resulted in the replacement of the codon that codes for proline with a glycine codon in speB. The altered allele (speBP78G) was used to replace the resident chromosomal allele as described elsewhere (30), and replacement was confirmed by PCR amplification and digestion of amplified products with BglII to probe for the unique BglII site introduced at the mutated codon. The resulting S. pyogenes mutant was designated SpeB14.
Measurement of protease activity.
Expression of the protease was analyzed in culture supernatants as follows. Cultures in C medium were initiated using cells from overnight growth in C medium (26). The cells were washed in phosphate-buffered saline (pH 7.4) to remove any residual protease. The initial OD600s of cultures were adjusted to 0.01, samples were removed at various time points during incubation at 37°C, and cells were removed by filtration (0.45-μm-pore-size sterile Acrodisc; Gelman Sciences). The resulting supernatant fluids were diluted in fresh C medium to normalize for any differences in growth between samples based on the OD600 of the culture at time of harvest. The presence of the proprotein and processed forms of SpeB was determined in a Western blot analysis as described previously (26). The proteolytic activities of supernatants were quantitated by the method of Hauser and Schlievert (15), which measures the increase in relative fluorescence generated by the proteolytic cleavage of fluorescein isothiocyanate-casein (Sigma). The activity of uninoculated culture medium was used to derive background values, which were typically undetectable under the conditions of this assay. To ensure that all proteolytic activity was specifically the result of SpeB, the cysteine protease-specific inhibitor E-64 (final concentration, 10 mM; Sigma) was routinely added to selected samples. This treatment typically reduced activity by >95%.
RESULTS
Construction of trigger factor mutations.
The strategy for determining whether the PPIase activity of trigger factor is required for the maturation of the streptococcal protease involved the construction of streptococcal strains which expressed a mutant trigger factor that was altered at a single amino acid residue that specifically disrupted the PPIase activity of the protein. As the first step in this analysis, the S. pyogenes trigger factor (RopA) was expressed in E. coli with a six-His affinity tag and purified to homogeneity by standard methods (see Materials and Methods). Analysis of the resulting protein confirmed that it possessed an active PPIase activity for the synthetic substrate succinyl-Ala-Ala-Pro-Phe-4-nitroanilide, as determined by the protease coupling assay of Fischer et al. (11) (kcat/Km = 54.1 ± 5.3 mM−1 s−1), that was comparable to that of human FKBP (kcat/Km = 66.3 ± 4.1 mM−1 s−1), a well-studied member of the FK506 family of PPIases.
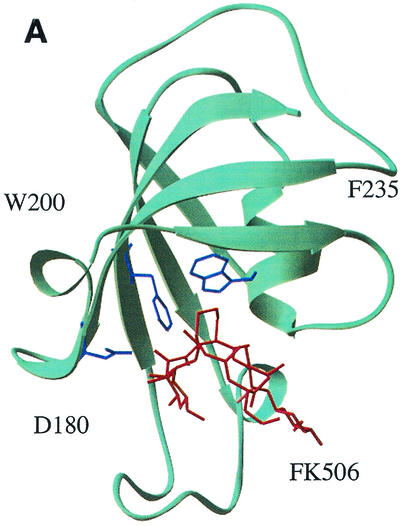
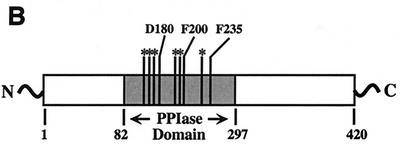
Targeting of residues in RopA for mutagenesis was assisted by the fact that the PPIase domain of trigger factor displays a high level of homology to other members of the FK506-inhibited PPIase family (16) (Fig. 1B) and that mutation of several residues in trigger factor that are conserved among FK506-binding proteins resulted in altered catalytic activity with no effect on its ability to refold certain proteins in vitro (39). In addition, the three-dimensional structures for several FK506 type PPIases in complex with FK506 have been determined. Since FK506 inhibits PPIase activity by inhibiting the binding of the substrate polypeptide, it is likely that residues important for binding FK506 will also make contact with the substrate polypeptide and that the loss of these contacts will lead to a loss of PPIase activity. An examination of the structure of FKBP12 (7, 40) identified several residues that make specific contacts with FK506 (Fig. 1A) that are similar in trigger factor (Fig. 1B). Three residues in the S. pyogenes trigger factor (D180, F200, and F235; Fig. 1B) were changed to alanine residues in order to minimize alteration of the overall conformation of the mutant protein. Purification and analysis of the proteins revealed that each mutation led to a profound decrease in PPIase activity to between 18 and 7% of the activity the wild-type protein (compare RopAD180A, RopAF200A, and RopAF235A to the wild type in Fig. 2).
FIG. 1.
Mutagenesis of RopA. (A) Structure of FKBP12 in complex with FK506. The highlighted residues D180, F200, and F235 make contact with FK506 and are conserved in the PPIase domain of trigger factor. The labels represent the residue of FKBP12 and the numbers of the conserved residues in RopA. The model shown was based on published structures (7, 40) using the molecular renderer RasMol, version 2.6 (http://www.umass.edu/microbiol/rasmol/). (B) Domain structure of trigger factor. The locations of the PPIase domain and residues (F170, G172, F179, D180, F196, I197, F200, Y223, and F235) highly conserved between trigger factor and other FK506-binding proteins are indicated. The residues named correspond to those indicated in panel A and were targeted for mutagenesis. Note that D180 and F235 are identical between trigger factor and FKBP12 and that F200 of trigger factor corresponds to W200 of FKBP12.
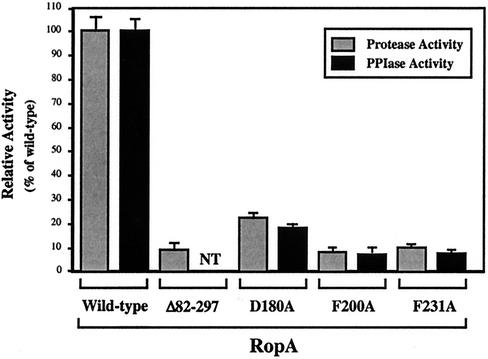
FIG. 2.
Protease maturation is defective in the absence of PPIase activity. The PPIase activity of the indicated site-specific mutant RopA proteins was compared to that of the wild type following purification of the six-His-tagged fusion proteins from E. coli. The mutant alleles lacking the regions encoding the six-His tags were used to replace the wild-type allele in single copy in the streptococcal chromosome, and the ability of the resulting strains to express proteolytic activity was compared to that of the wild type and a trigger factor mutant that contains ropA with a large in-frame deletion that includes the coding sequence for the PPIase domain (ropAΔ82-297). Data represent the means and standard errors of the means for at least two independent experiments, each of which was conducted in triplicate. NT, not tested.
Characterization of trigger factor mutants.
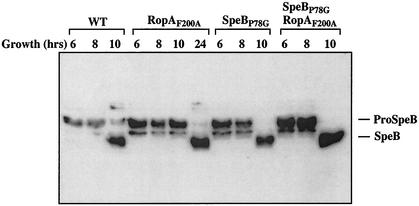
Each of the three mutations was recreated in a version of the trigger factor gene that lacked the coding sequence for the six-His affinity tag, and the mutant alleles were then used to replace the wild-type ropA allele in the chromosome of S. pyogenes HSC5. The streptococcal cysteine protease is expressed when the bacterial cells enter the stationary phase of growth, and it is secreted as an inactive 40-kDa precursor protein. In S. pyogenes HSC5, the secreted protease remains as the precursor for about 1 to 2 h after the culture reaches stationary phase and then is rapidly processed to the mature active 28-kDa protease (26). Examination of the resulting mutant strains revealed that each expressed wild-type levels of the precursor protease protein and that the protease was expressed in a temporal pattern equivalent to that for the wild type, indicating that the mutations did not disrupt transcriptional regulation of the protease gene or the trigger factor-dependent targeting activity required for secretion of the protease (data for RopAF200A are shown in Fig. 3). However, when the ability of the precursor to become processed to the 28-kDa mature form of the protease was analyzed, it was found that the protease produced by each of the PPIase-defective trigger factor mutants demonstrated the same kinetic defect in autoprocessing that was observed for the previously characterized mutant protein with the large internal ropA deletion (RopAΔ82-297) (26). A characteristic of this defect is that conversion to the 28-kDa mature form takes approximately 8 h longer than it does for the protease produced by the wild-type strain (Fig. 3, compare RopAF200A to the wild type at 10 and 24 h). A second characteristic is that the processed protease demonstrates only a fraction of the proteolytic activity of the wild-type protease (26). Comparison of proteolytic activities at a point where both wild-type and mutant proteases are fully converted to the mature form (24-h incubation; see Materials and Methods) reveals that each of the three PPIase-deficient mutants exhibited reduced proteolytic activity (Fig. 2; compare RopAD180A, RopAF200A, and RopAF235A to the wild type). The decreased level of expression closely correlated with the levels to which PPIase activity was decreased for the three mutants and approached that observed for the original deletion mutant (RopAΔ82-297; Fig. 2).
FIG. 3.
Maturation kinetics of SpeB is restored in a ropAF200A mutant through a speBP78G allelic replacement. SpeB was detected in a Western blot analysis using a SpeB-specific antiserum. The secreted SpeB zymogen (proSpeB) is 40 kDa, while the processed form is 28 kDa. Time points represent the incubation times of the culture prior to sample harvesting. Similar results were observed for the ropAD180A and ropAF235A alleles (data not shown). WT, wild type.
Identification of a target proline residue.
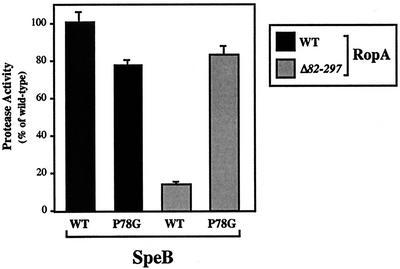
A requirement for PPIase activity in the production of an active protease implies that the protease polypeptide possesses at least one proline residue whose state of isomerization directs subsequent maturation into either a productive or a nonproductive pathway. Examination of the protease shows that it has 13 prolyl residues, including 1 in the signal sequence, 1 in the prodomain, and 11 in the mature polypeptide. Of these, the residue in the prodomain (P78, with position 1 defined as the first residue of the zymogen) was of particular interest for several reasons. First, because of its association with the translating ribosome, trigger factor may first interact with the amino terminus of the nascent polypeptide as it exits the ribosome prior to any subsequent folding (23). When analyzed in vitro, trigger factor preferentially associates with short peptides enriched in basic and aromatic residues with a positive net charge (28). Use of the algorithm of Patzelt et al. (28) indicates that a section of the proregion including P78 (S77 to F89) has a high probability of being bound by trigger factor. Thus, the prodomain proline is in a position to interact with trigger factor. Second, the prodomains of many cysteine proteases have been shown to function as intramolecular chaperones and are absolutely required for proper folding of their proteases (37). Third, the cysteine protease proregion makes intimate contacts with residues of the protease active site (6, 20). Thus, it is conceivable that the structure of the proregion itself could influence the final conformation of the active site. To test whether P78 could be a target of trigger factor, a mutant protease in which P78 was changed to a glycine residue in order to maximize the flexibility of the polypeptide at this position was first constructed. When the speBP78G allele was used to replace the wild-type allele in S. pyogenes, the resulting mutant secreted the protease polypeptide at levels equivalent to those for the wild type and produced proteolytic activity at a level about 20% reduced from that of the wild-type strain (Fig. 4; for RopAWT, compare SpeBWT to SpeBP78G). This indicates that, while P78 is required for full expression of proteolytic activity, it is not essential for maturation of the zymogen to an enzyme with activity for this substrate (casein). In the presence of a PPIase-deficient ropA allele, expression of the wild-type protease gene results in only about 10% of the proteolytic activity obtained with the wild-type ropA background (Fig. 4; for SpeBWT, compare RopAWT to RopAΔ82-297). However, when the P78G protease allele was expressed in a trigger factor PPIase-deficient background, proteolytic activity increased over that obtained with the wild-type protease allele in this background (Fig. 4; for RopAΔ82-297, compare SpeBWT to SpeBP78G) to levels equivalent to that for the P78G protease in a wild-type trigger factor background (Fig. 4; for SpeBP78G, compare RopAWT to RopAΔ82-297). Identical results were obtained when SpeBP78G was introduced into hosts expressing each of the three ropA alanine substitution alleles (data not shown). Thus, the P78G mutation makes the protease maturation step independent of the requirement for PPIase activity. The ability of the P78G mutation to suppress PPIase deficiency supports a model in which trigger factor-promoted isomerization of P78 is required for directing the maturation of the secreted protease into a productive pathway.
FIG. 4.
Mutation of P78 of the cysteine protease suppresses the requirement for PPIase activity. A site-specific mutation in the single chromosomally encoded copy of the protease gene was constructed to change the prodomain P78 to G in S. pyogenes hosts containing either wild-type trigger factor (ropA) or trigger factor defective in PPIase activity (ropAΔ82-297). Protease activity of the resulting strains was compared to that of the wild type (WT). Data represent the means and standard errors of the means for at least two independent experiments, each of which was conducted in triplicate.
DISCUSSION
Trigger factor is found in virtually all species of bacteria and is the only PPIase of Mycoplasma genitalium, the bacterium that possess the smallest known genome of any freely self-replicating organism (2). Evidence suggests that trigger factor in M. genitalium is essential for viability (19). It is present in pathogenic bacterial species such as the streptococci, and in E. coli it provides an essential activity that overlaps with that of DnaK (8, 38). These data imply that trigger factor is essential for the ability of bacteria to survive in their natural environments. However, since trigger factor mutants generally have no obvious defect for growth or for expression of any protein under laboratory conditions, the function of trigger factor is not understood.
In the present study, we have shown that the PPIase activity of trigger factor is required for the maturation of the protease of S. pyogenes following its secretion from the bacterium. Furthermore, the observation that the mutation of a specific proline residue results in a bypass of the requirement for trigger factor in protease maturation implies that this residue is the target of trigger factor's PPIase activity. When combined with the known properties of trigger factor, the trigger factor cycle (23), and our previous observation that trigger factor is also required for targeting the protease to the streptococcal secretion pathway, these data support the following model. During translation of the protease message, trigger factor binds to the N-terminal proregion of the nascent protease polypeptide as it emerges from the translating ribosome. This interaction is essential for stabilizing the preproprotease in a secretion-competent conformation, and the complex is targeted to the Sec pathway translocon for secretion across the cellular membrane. During this targeting phase, the PPIase domain of trigger factor introduces information into the nascent protease polypeptide through isomerization of the proregion proline residue at position 78. Trigger factor dissociates from the protease polypeptide as it transits through the Sec translocon and is recycled into a cytoplasmic pool for reassociation with a ribosome. The nascent protease polypeptide is now in an extracellular space, and it folds into its zymogen form via a folding pathway that is influenced by the isomerization state of P78.
A key feature of the model is that it predicts that the conformation of the proregion is influenced by the isomerization state of P78, which in turn provides steric information that influences the subsequent folding pathway. This idea is consistent with what is known concerning the roles of some proregions in the biogenesis of their cognate proteases. For example, the proregions of many proteases, including some cysteine proteases (37), are essential for the proper folding and activation of the proteases. When the serine protease subtilisin is folded in the absence of its prodomain, it can obtain a stable but inactive molten globule-like intermediate state that can be converted to an active enzyme upon addition of the prodomain (32-34). Mutation of single amino acid residues in the subtilisin proregion can produce an altered enzymatic conformation of the activated protease compared to the wild type, even though the mutant and wild-type enzymes retain identical amino acid sequences. These data have been interpreted to suggest that the proregion provides a folding template for configuration of the active-site residues. Thus, the final state of the protein is not necessarily that dictated by a global free-energy minimum directed by the primary amino acid sequence but rather is one of an ensemble of low-energy minima whose accessibility is influenced by the conformation of the proregion. Since the subtilisin proregion does not become part of the final structure, it has been referred to as an intramolecular chaperone.
The unique aspect of the present study is that, unlike previous mutational studies that have altered the proregion sequence (34), different protease conformations were derived from interactions with proregions of identical sequences. The only difference was the presence or absence of PPIase activity in trigger factor, an accessory protein required for targeting the protease to the secretory pathway. The streptococcal cysteine protease is a member of the papain-like cysteine protease family, and, while the proregions of family members are diverse in regards to sequence and length, they can be divided into two major subfamilies based on structural similarities to cathepsin L or to cathepsin B. It is not uncommon for members of both subfamilies to have a proline residue approximately 15 to 20 residues from the carboxy terminus of the proregion, as for cathepsin B and cathepsin L (P49 and P83, respectively (6, 27). The three-dimensional structures of the two subfamily prototypes, procathepsin B and procathepsin L, revealed that their proregions have several common structural features, including proregions that loop through the entire substrate binding groove in the opposite direction to that of the substrate and proregion prolines that are positioned in the immediate vicinity of the cysteine and histidine residues that comprise the catalytic machinery (6). The location of the proline residue suggests that its configuration could influence active-site conformation.
The recently determined three-dimensional structure of the streptococcal protease zymogen revealed that, while the protease has the canonical papain family structure, the proregion adopts a unique structure which itself surrounds a substantial hydrophobic core (20). The latter feature implies that the proregion can fold independently into a stable structure. Furthermore, instead of the extended loop structure used by other cysteine protease zymogens to inhibit proteolytic activity, the proregion of the streptococcal zymogen inserts an alpha-helical region into the active-site cleft so that it displaces the catalytic His (H195) from the active site to prevent any interaction of this residue with the catalytic Cys (C47). Activation of the streptococcal protease apparently involves removal of the proregion loop and a substantial reorganization of the active site, including a rotation of H195 into the active site. The fact that the proregion actively interacts with residues in the active site, combined with its apparent stability, suggests that the structure of the proregion could directly influence the final conformation of the active site. Exactly how P78 could participate in this process is less clear. Unlike what is found in many other cysteine proteases, this Pro residue does not enter the active-site cleft. In addition, in the recently determined structure P78 is in the trans conformation. However, it remains to be determined whether this is the native state of isomerization for this residue for several reasons, among which are that the structure was derived from a mutant protease with the catalytic Cys residue replaced and that the protease was expressed and purified from E. coli, a process that is notorious for producing a streptococcal protease with low specific activity.
Problems associated with production of the streptococcal protease in the gram-negative bacterium E. coli illustrate some interesting contrasts with the problem of protein secretion in gram-positive bacteria. While the processes look similar at first glance and involve many of the same pathways and accessory proteins such as trigger factor, there are some fundamental differences. For example, proteins that are secreted by the general secretory pathway in gram-negative bacteria translocate across the cellular membrane in an unfolded conformation and are released into the periplasmic space. They then fold into their final conformations in a controlled environment rich in folding catalysts including specific chaperones, thiol-disulfide oxioreductases, and multiple PPIases. In contrast, gram-positive bacteria lack a clearly defined periplasmic space, few extracellular folding catalysts have been identified, and it is unknown whether the organization of their outer surfaces provides any type of protected environment to facilitate folding. Thus, the mechanisms by which gram-positive bacteria fold secreted proteins are unclear.
Also unclear is why the PPIase activity of trigger factor is required for maturation of the secreted streptococcal protease. Replacement of P78, the apparent target of the PPIase activity, has an effect on the ability of the protease precursor to form an active protease; however, the mutant protease retains substantial activity. The fact that a proline residue has been conserved at this position implies that its role in protease biogenesis is more substantial than was revealed by these studies. It is possible that the in vitro conditions used for folding and analyses of proteolytic activity do not reflect the conditions that S. pyogenes encounters during infection. Also, it is possible that the casein substrate used to assess proteolytic activity was more permissive than the native substrate(s), which has not been identified. Further studies on the folding of this protease and the role of accessory factors such as trigger factor will be important for understanding the function of this protease in infection and the process of protein folding in gram-positive bacteria.
Acknowledgments
This work was supported by Public Health Service grant AI46433 from the National Institutes of Health.
REFERENCES
- 1.Agashe, V. R., and F. U. Hartl. 2000. Roles of molecular chaperones in cytoplasmic protein folding. Semin. Cell Dev. Biol. 11:15-25. [DOI] [PubMed] [Google Scholar]
- 2.Bang, H., A. Pecht, G. Raddatz, T. Scior, W. Solbach, K. Brune, and A. Pahl. 2000. Prolyl isomerases in a minimal cell. Catalysis of protein folding by trigger factor from Mycoplasma genitalium. Eur. J. Biochem. 267:3270-3280. [DOI] [PubMed] [Google Scholar]
- 3.Burns, E. H., S. Lukomski, J. Rurangirwa, A. Podbielski, and J. M. Musser. 1998. Genetic inactivation of the extracellular cysteine protease enhances in vivo internalization of group A streptococci by human epithelial and endothelial cells. Microb. Pathog. 24:333-339. [DOI] [PubMed] [Google Scholar]
- 4.Crooke, E., L. Brundage, M. Rice, and W. Wickner. 1988. ProOmpA spontaneously folds in a membrane assembly competent state which trigger factor stabilizes. EMBO J. 7:1831-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crooke, E., and W. Wickner. 1987. Trigger factor: a soluble protein that folds Pro-OmpA into a membrane-assembly-competent form. Proc. Natl. Acad. Sci. USA 84:5216-5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cygler, M., and J. S. Mort. 1997. Proregion structure of members of the papain superfamily. Mode of inhibition of enzymatic activity. Biochimie 79:645-652. [DOI] [PubMed] [Google Scholar]
- 7.DeCenzo, M. T., S. T. Park, B. P. Jarrett, R. A. Aldape, O. Futer, M. A. Murcko, and D. J. Livingston. 1996. FK506-binding protein mutational analysis: defining the active-site residue contributions to catalysis and the stability of ligand complexes. Protein Eng. 9:173-180. [DOI] [PubMed] [Google Scholar]
- 8.Deuerling, E., A. Schulze-Specking, T. Tomoyasu, A. Mogk, and B. Bukau. 1999. Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature 400:693-696. [DOI] [PubMed] [Google Scholar]
- 9.Diatchenko, L., Y. F. C. Lau, A. P. Campbell, A. Chenckik, F. Moqadam, B. Huang, S. A. Lukyanov, K. A. Lukyanov, N. Gurskaya, E. D. Sverdlov, and P. D. Siebert. 1996. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. USA 93:6025-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolinski, K., S. Muir, M. Cardenas, and J. Heitman. 1997. All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable for viability in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:13093-13098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer, G., B. Wittmann-Liebold, K. Lang, T. Kiefhaber, and F. X. Schmid. 1989. Cyclophilin and peptidyl-prolyl cis-trans isomerase are probably identical proteins. Nature 337:476-478. [DOI] [PubMed] [Google Scholar]
- 12.Göthel, S. F., and M. A. Marahiel. 1999. Peptidyl-prolyl cis-trans isomerases, a superfamily of ubiquitous folding catalysts. Cell. Mol. Life Sci. 55:423-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Göthel, S. F., R. Schmid, A. Wipat, N. M. Carter, P. T. Emmerson, C. R. Harwood, and M. A. Marahiel. 1997. An internal FK506-binding domain is the catalytic core of the prolyl isomerase activity associated with the Bacillus subtilis trigger factor. Eur. J. Biochem. 244:59-65. [DOI] [PubMed] [Google Scholar]
- 14.Göthel, S. F., C. Scholz, F. X. Schmid, and M. A. Marahiel. 1998. Cyclophilin and trigger factor from Bacillus subtilis catalyze in vitro protein folding and are necessary for viability under starvation conditions. Biochemistry 37:13392-13399. [DOI] [PubMed] [Google Scholar]
- 15.Hauser, A. R., and P. M. Schlievert. 1990. Nucleotide sequence of the streptococcal pyrogenic exotoxin type B gene and relationship between the toxin and the streptococcal proteinase precursor. J. Bacteriol. 172:4536-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hesterkamp, T., and B. Bukau. 1996. Identification of the prolyl isomerase domain of Escherichia coli trigger factor. FEBS Lett. 385:67-71. [DOI] [PubMed] [Google Scholar]
- 17.Hesterkamp, T., E. Deuerling, and B. Bukau. 1997. The amino-terminal 118 amino acids of Escherichia coli trigger factor constitute a domain that is necessary and sufficient for binding to ribosomes. J. Biol. Chem. 272:21865-21871. [DOI] [PubMed] [Google Scholar]
- 18.Huang, G. C., Z. Y. Li, J. M. Zhou, and G. Fischer. 2000. Assisted folding of D-glyceraldehyde-3-phosphate dehydrogenase by trigger factor. Protein Sci. 9:1254-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutchison, C. A., S. N. Peterson, S. R. Gill, R. T. Cline, O. White, C. M. Fraser, H. O. Smith, and J. C. Venter. 1999. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286:2087-2090. [DOI] [PubMed] [Google Scholar]
- 20.Kagawa, T. F., J. C. Cooney, H. M. Baker, S. McSweeney, M. Liu, S. Gubba, J. M. Musser, and E. N. Baker. 2000. Crystal structure of the zymogen form of the group A Streptococcus virulence factor SpeB: an integrin-binding cysteine protease. Proc. Natl. Acad. Sci. USA 97:2235-2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kandror, O., M. Sherman, M. Rhode, and A. L. Goldberg. 1995. Trigger factor is involved in GroEL-dependent protein degradation in E. coli and promotes binding of GroEL to unfolded proteins. EMBO J. 14:6021-6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiefhaber, T., R. Quaas, U. Hahn, and F. X. Schmid. 1990. Folding of ribonuclease T1. 2. Kinetic models for the folding and unfolding reactions. Biochemistry 29:3061-3070. [DOI] [PubMed] [Google Scholar]
- 23.Lill, R., E. Crooke, B. Guthrie, and W. Wickner. 1988. The “trigger factor cycle” includes ribosomes, presecretory proteins, and the plasma membrane. Cell 54:1013-1018. [DOI] [PubMed] [Google Scholar]
- 24.Lukomski, S., C. A. Montgomery, J. Rurangirwa, R. S. Geske, J. P. Barrish, G. J. Adama, and J. M. Musser. 1999. Extracellular cysteine protease produced by Streptococcus pyogenes participates in the pathogenesis of invasive skin infection and dissemination in mice. Infect. Immun. 67:1779-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lukomski, S., W. Sreevatsan, C. Amberg, W. Reichardt, M. Woischnik, A. Podbielski, and J. M. Musser. 1997. Inactivation of Streptococcus pyogenes extracellular cysteine protease significantly decreases mouse lethality of serotype M3 and M49 strains. J. Clin. Investig. 99:2574-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyon, W. R., C. M. Gibson, and M. G. Caparon. 1998. A role for trigger factor and an Rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 17:6263-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Padilla-Zuniga, A. J., and A. Rojo-Dominguez. 1998. Non-homology knowledge-based prediction of the papain prosegment folding pattern: a description of plausible folding and activation mechanisms. Fold. Des. 3:271-284. [DOI] [PubMed] [Google Scholar]
- 28.Patzelt, H., S. Rüdiger, D. Brehmer, G. Kramer, S. Vorderwülbecke, E. Schaffitzel, A. Waitz, T. Hesterkamp, L. Dong, J. S. Schneider-Mergener, B. Bukau, and E. Deuerling. 2001. Binding specificity of Escherichia coli trigger factor. Proc. Natl. Acad. Sci. USA 98:14244-14249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perez-Casal, J., N. Okada, M. G. Caparon, and J. R. Scott. 1995. Role of the conserved C-repeat region of the M protein of Streptococcus pyogenes. Mol. Microbiol. 15:907-916. [DOI] [PubMed] [Google Scholar]
- 30.Ruiz, N., B. Wang, A. Pentland, and M. G. Caparon. 1998. Streptolysin O and adherence synergistically modulate proinflammatory responses of keratinocytes to group A streptococci. Mol. Microbiol. 27:337-346. [DOI] [PubMed] [Google Scholar]
- 31.Schiene, C., and G. Fischer. 2000. Enzymes that catalyze restructuring of proteins. Curr. Opin. Struct. Biol. 10:40-45. [DOI] [PubMed] [Google Scholar]
- 32.Shinde, U., X. Fu, and M. Inouye. 1999. A pathway for conformational diversity in proteins mediated by intramolecular chaperones. J. Biol. Chem. 274:15615-15621. [DOI] [PubMed] [Google Scholar]
- 33.Shinde, U., and M. Inouye. 1996. Propeptide-mediated folding in subtilisin: the intramolecular chaperone concept. Adv. Exp. Med. Biol. 379:147-154. [DOI] [PubMed] [Google Scholar]
- 34.Shinde, U. P., J. J. Liu, and M. Inouye. 1997. Protein memory through altered folding mediated by intramolecular chaperones. Nature 389:520-522. [DOI] [PubMed] [Google Scholar]
- 35.Stoller, G., T. Tradler, K. P. Rucknagel, J. Rahfeld, and G. Fischer. 1996. An 11.8kDa proteolytic fragment of the E. coli trigger factor represents the domain carrying the peptidyl-prolyl cis/trans isomerase activity. FEBS Lett. 384:117-122. [DOI] [PubMed] [Google Scholar]
- 36.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct the expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 37.Tao, K., N. A. Stearns, J. Dong, Q. L. Wu, and G. G. Sahagian. 1994. The proregion of cathepsin L is required for proper folding, stability, and ER exit. Arch. Biochem. Biophys. 311:19-27. [DOI] [PubMed] [Google Scholar]
- 38.Teter, S. A., W. A. Houry, D. Ang, T. Tradler, D. Rockabrand, G. Fischer, P. Blum, C. Georgopoulos, and F. U. Hartl. 1999. Polypeptide flux through bacterial Hsp70: DnaK cooperates with trigger factor in chaperoning nascent chains. Cell 97:755-765. [DOI] [PubMed] [Google Scholar]
- 39.Tradler, T., G. Stoller, K. P. Rücknagel, A. Schierhorn, J. Rahfeld, and G. Fischer. 1997. Comparative mutational analysis of peptidyl prolyl cis/trans isomerases: active sites of Escherichia coli trigger factor and human FKBP12. FEBS Lett. 407:184-190. [DOI] [PubMed] [Google Scholar]
- 40.Van Duyne, G. D., R. F. Standaert, P. A. Karplus, S. L. Schreiber, and J. Clardy. 1991. Atomic structure of FKBP-FK506, an immunophilin-immunosuppressant complex. Science 252:839-842. [DOI] [PubMed] [Google Scholar]
- 41.Zarnt, T., T. Tradler, G. Stoller, C. Scholz, F. X. Schmid, and G. Fischer. 1997. Modular structure of the trigger factor required for high activity in protein folding. J. Mol. Biol. 271:827-837. [DOI] [PubMed] [Google Scholar]