Abstract
The gene coding for inulosucrase in Leuconostoc citreum CW28, islA, was cloned, sequenced, and expressed in Escherichia coli. The recombinant enzyme catalyzed inulin synthesis from sucrose like the wild-type enzyme. Inulosucrase presents an unusual structure: its N-terminal region is similar to the variable region of glucosyltransferases, its catalytic domain is similar to fructosyltransferases from various microorganisms, and its C-terminal domain presents similarity to the glucan binding domain from alternansucrase, a glucosyltransferase from Leuconostoc mesenteroides NRRL B-1355. From sequence comparison, it was found that this fructosyltransferase is a natural chimeric enzyme resulting from the substitution of the catalytic domain of alternansucrase by a fructosyltransferase. Two different forms of the islA gene truncated in the C-terminal glucan binding domain were successfully expressed in E. coli and retained their ability to synthesize inulin but lost thermal stability. This is the first report of an inulosucrase bearing structural features of both glucosyltransferases and fructosyltransferases.
Glycosyltransferases are enzymes that catalyze the transfer of glycosyl residues from a donor molecule to a particular acceptor (22). Lactic acid bacteria produce a wide variety of a particular group of glycosyltransferases: glucosyltransferases (GTFs) and fructosyltransferases (FTFs), which synthesize glucose and fructose polymers, respectively, from sucrose without the need of cofactors (9, 26). An interesting feature of FTFs and GTFs is their ability to synthesize oligosaccharides of different polymerization degrees when efficient acceptor molecules like maltose or lactose are added to sucrose in the reaction medium (8, 16, 24). This is known as the acceptor reaction, and the enzymes vary in their efficiency to perform this reaction.
GTFs have been the subject of intensive research, particularly those involved in industrial dextran production such as dextransucrase (DS) or alternansucrase produced by Leuconostoc spp. (28), whereas GTFs from Streptococcus spp. are important due to their role in dental plaque formation (1). Depending on its specificity, a GTF may transfer glucose, building polymers with the main chain joined by α1-6, α1-3, or α1-4 linkages (7). However, no crystal structure is available for GTFs with the exception of amylosucrase (30), a GTF producer of an amylose-type polysaccharide. GTFs present a large size, between 155 and 200 kDa, and are organized in three domains, starting in the N-terminal end by the signal peptide and a variable region with an unknown function, followed by the catalytic domain, where the residues implicated in catalysis have been located. Finally, a C-terminal domain is involved in binding to the synthesized glucan.
It has been proposed by sequence analysis and analogy with α-amylases that GTFs present a circular permutation of a (β/α)8 barrel in the catalytic domain (5). This contrasts with FTFs, which on average have one third the molecular mass of GTFs and are not organized in domains. Moreover, Pons et al. (27) predicted a β-propeller model for FTFs. Among FTFs, inulosucrase produces inulin, a fructan polymer with β2-1 linkages, whereas levansucrase catalyzes a similar reaction resulting in a β2-6-linked fructose polymer known as levan. In both cases, branching can occur in β2-6 and β2-1. In spite of the growing importance of inulin and fructooligosaccharides in the food industry, little is known about the biochemistry and molecular biology of these enzymes (4). Up to now, more than 19 bacterial FTFs have been reported in GenBank, but no crystal structure is available. FTFs are also common in fungi and plants.
The inulosucrase of Leuconostoc citreum is a cell-associated enzyme. It has been characterized both in its cell-associated insoluble form and after solubilization by urea treatment. Unexpectedly, this FTF has a molecular mass of around 165 kDa, the highest reported for FTFs. In its cell-associated form, it is highly specific for polymer synthesis, with low levels of fructose transferred to maltose and lactose when added to the reaction medium. The synthesized polymer has an inulin-like structure with β2-1 glycosidic linkages, as demonstrated by 13C nuclear magnetic resonance (25).
In the present study, we report the identification and functional characterization of islA, a gene encoding for inulosucrase from Leuconostoc citreum CW28. From the analysis of the nucleotide and the predicted amino acid sequence, it was concluded that this is a natural chimeric enzyme with three domains: the first and third with high identity with alternansucrase, a GTF from Leuconostoc mesenteroides NRRL B-1355 able to produce alternan, a dextran-type polymer with alternating α1-3 and α1-6 linkages (2). The second domain, which is the catalytic one, has similarity with levansucrase and inulosucrase of several microorganisms. No similarities have been reported between GTFs and FTFs in spite of the already mentioned analogies.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The bacterial strain L. citreum CW28 (Facultad de Química, Universidad Nacional Autonoma Mexico, México) was grown at 30°C and 200 rpm as previously reported (25), with 20 g of either sucrose, as the carbon source, or glucose per liter in order to avoid enzyme and polymer synthesis, as these interfere with DNA extraction.
Escherichia coli DH5α, the host for cloning, was grown at 37°C and 250 rpm in Luria-Bertani (LB) medium and, when appropriate, supplemented with 50 μg of kanamycin per ml in order to maintain the plasmid construction. Agar (1.5%, wt/vol) was added for growth in solid medium.
General molecular techniques.
Plasmid and genomic DNAs of L. citreum CW28 were extracted from cells according to Ausubel et al. (3). E. coli plasmid DNA was isolated with a QiaPrep spin plasmid kit (Qiagen, Inc., Chatsworth, Calif.) as recommended by the manufacturer. Restriction and DNA-modifying enzymes were from New England Biolabs (Beverly, Mass.) and Roche Biochemical (Basel, Switzerland). DNA was amplified with PCR techniques in a Robocycler Gradient 96 (Stratagene, La Jolla, Calif.). E. coli transformations were performed by electroporation in 0.2-mm cuvettes with a Bio-Rad Micropulser apparatus (Bio-Rad Laboratories, Hercules, Calif.) at 2.5 kV, 25 μF, and 200 Ω. DNA fragments were isolated from the agarose gel with a Qiagen extraction kit (Qiagen, Inc., Chatsworth, Calif.). In all cases, the biological reagents were applied following the instructions of the supplier.
For Southern hybridization, DNA was restricted with endonucleases, separated by agarose gel electrophoresis, and transferred to a Hybond-N+ membrane (Amersham Pharmacia Biotech, Buckinghamshire, England.). The probes were labeled with 32P with a Rediprime kit (Amersham Pharmacia Biotech, Buckinghamshire, England).
Isolation of IslA.
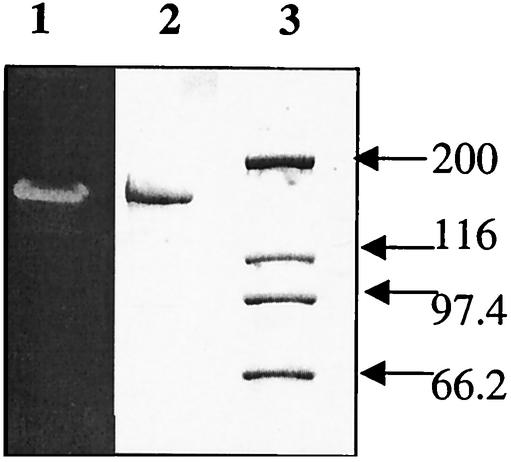
In order to isolate the islA gene, first the FTF from L. citreum CW28 was purified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (18) and located through activity assay in the gel (Fig. 1). L. citreum cells were treated with 8 M as already described, and the extracted protein was applied to the gel in the sample buffer containing SDS and 2-mercaptoethanol and incubated at 90°C for 5 min. After separation, the protein was excised from the gel and digested, and two of the resulting peptides were sequenced (Institute Pasteur, Paris, France). From the resulting inulosucrase peptide sequences (28-MDVWDSWALQDSK and 29-TAPYGEAGAQFVDYV), the degenerate primers 28IS and 29IS were designed (Table 1).
FIG. 1.
SDS-PAGE analysis of L. citreum proteins. The in situ polymer produced by inulosucrase is observed in lane 1 after incubation with raffinose, while in lane 2 the urea-extracted proteins are stained with Coomassie blue. Lane 3 contains the molecular size standards (sizes shown in kilodaltons).
TABLE 1.
Primers designed for isolation of the inulosucrase gene islA from L. citreuma
| Primer | Sequence (5′ to 3′) | Use |
|---|---|---|
| 28IS | GATGTKTGGGATAGYTGGGCKTTRCAAGAT | PCR |
| 29IS | ATCRACAAAYTGMGCMCCMGCTTCACCATATGG | PCR |
| 539IS | CCATATGGTGAAGCGGGGGCGCAGTTTGTCGAT | PCR |
| 868IS | CCCTTAAGCTTGCAAAGCACGCTTATCAATCCA | PCR |
| 128rIS | ATCTTGTAACGCCCAACTATCCCACACATC | Inverse PCR |
| 42rIS | GCAAGTGGTCTTTGCTTTAATGGGAGC | Inverse PCR |
| 11PIS | GCTATGGCGTCATGCAGGAACCACTTTATC | PCR |
| 385IS | CTAATTTAAATCGCGTGAAAAGCTAATGGC | PCR |
| 384IS | CCTAAGAGTGATCATCATCTCCCCAAACCC | PCR |
K = G or T; R = A or G; M = A or C; and Y = C or T.
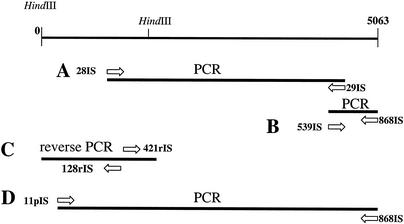
A PCR fragment of 3,000 bp was amplified with the L. citreum CW28 chromosomal DNA as the template with Vent polymerase (Fig. 2A). The analysis of its nucleotide sequence confirmed the similarities found between the two peptides with FTF and alternansucrase. Based on the 3,000-bp sequence and on the alternansucrase homology primers, 539IS and 868IS were designed in order to isolate the C terminus (Table 1). An amplification product with the expected size (about 445 bp) was obtained from L. citreum CW28 genomic DNA and sequenced (Fig. 2B). On the other hand, and based also on the 3,000-bp sequence, primers 128rIS and 421rIS were designed in order to isolate the N-terminal region by inverse PCR (Table 1). L. citreum CW28 genomic DNA was digested with HindIII and ligated, yielding circular DNA that was used as the template for a PCR with the diverging primers 128rIS and 421rIS, yielding an amplification product of about 1,600 bp (Fig. 2C). The complete sequence of the islA gene, including the promoter region, was deduced from the three fragments. Finally, a 5,063-bp fragment containing the whole gene was amplified with primers 11pIS and 868IS (Fig. 2D).
FIG. 2.
Strategy for isolation of the islA gene from L. citreum CW28 chromosomal DNA. Panels A to D are explained in the text.
Construction of plasmids for expression in E. coli of the islA gene and the truncated versions.
For the construction of an expression plasmid for the complete islA gene, primer 11pIS, located at nucleotide 56, and primer 868IS, at the end of the gene, were designed. A PCR product of 5,007 bp was amplified and cloned with the pCR4-TOPO vector in E. coli strain DH5α (Invitrogen, Carlsbad, Calif.). The correct construction of the plasmid was confirmed by sequence analysis of single DNA strands from the insert.
For the construction of a truncated version of the protein (ISE2), primer 385IS was designed and used together with primer 11pIS (Table 1). An amplification product of 3,333 bp without the region with similarity to alternansucrase was obtained. This product was cloned with the pCR4-TOPO vector in E. coli DH5α. A third version of the protein was constructed by deleting the complete C-terminal domain that is the alternansucrase-like region as well as 602 bp which are part of a transition region, a lower similarity module located between the high-homology alternansucrase region and the FTF catalytic domain. For this construction, primers 384IS and 11pIS were designed (Table 1). This product was cloned with the pCR4-TOPO vector in E. coli DH5α.
Preparation of E. coli cell extracts and enzyme activity assay.
E. coli DH5α harboring the aforementioned constructs was grown overnight and harvested by centrifugation (10 min at 4°C and 5,000 × g). The resulting pellet was washed twice with 50 mM sodium phosphate buffer (pH 6.5). Afterwards, the cells were resuspended in the same buffer and broken by ultrasonication with four 10-s pulses of 10 μE at 3-min intervals. Cell debris was removed by centrifugation for 40 min at 4°C and 10,000 × g, and the supernatant was assayed for activity. Initial rates of the inulosucrase reaction were measured at 30°C in 50 mM potassium phosphate buffer (pH 6.5) in the presence of 292 mM sucrose. The inulosucrase activity was measured by determination of glucose and fructose release from sucrose by the 3,5-dinitrosalicylic acid method (31). One unit (U) of activity is defined as the amount of enzyme that produces 1 μmol of glucose equivalent per minute. In order to determine only the transferase and hydrolytic activities, a determination of glucose and fructose by high-pressure liquid chromatography is required. All free fructose is the product of hydrolysis, so after subtracting the amount of free fructose from the total glucose, the residual glucose is equivalent to the transferase activity.
N-terminal amino acid sequencing.
In order to determine the N-terminal amino acid sequence, the protein was extracted from L. citreum cells by 8 M urea treatment and then separated in an SDS-PAGE gel as already described (18). After separation, the proteins were transferred to a polyvinylidene difluoride membrane (3), and after staining with Ponceau S solution, the inulosucrase protein band was cut from the membrane and subjected to amino acid sequence analysis (Harvard Medical School, Harvard, Mass.).
SDS-PAGE activity staining.
Electrophoresis of the recombinant protein was performed as reported by Laemmli (18). Samples were mixed with an equal volume of 2× sample buffer (0.125 M Tris-HCl [pH 6.8], 1% SDS, 20% glycerol, 10% 2-mercaptoethanol) and incubated at 90°C for 5 min. Denatured samples were then electrophoretically separated on 6% polyacrylamide gels and stained with Coomassie brilliant blue. In order to detect the activity of the recombinant proteins in SDS-PAGE gels, the periodic acid-Schiff reagent method was performed. The proteins of the cellular extract were separated on SDS-PAGE gels, followed by incubation in 50 mM potassium phosphate buffer (pH 6.5) and 1% Tween 80. In situ polymer synthesis was then induced by overnight incubation in the same buffer including 5% raffinose, a specific substrate of FTFs. Afterwards, the gels were washed with a solution of ethanol (75%, vol/vol) and acetic acid (5%, vol/vol), for 30 min, followed by three incubations in sodium metabisulfite (0.2%, wt/vol) and acetic acid (5%, vol/vol) for 20 min. Finally, the gels were stained with Schiff reagent (Sigma-Aldrich, St. Louis, Mo.), yielding purple spots where the fructan polymer was produced. The Schiff reaction was stopped by incubation with a solution of sodium metabisulfite (0.2%, wt/vol) and acetic acid (5%, vol/vol).
In order to determine the type of polymer produced by the IS2 and IS3 forms, two SDS-PAGE activity gels were carried out in parallel. After incubation with raffinose, the buffer was changed to 50 mM acetate buffer (pH 4.5), and one gel was treated with inulinase from Aspergillus niger (Fluka) at 37°C for 48 h. After this treatment, staining by the periodic acid-Schiff method was performed.
Nucleotide sequence accession number.
The sequence of the entire islA gene has been submitted to GenBank under accession number AY191311.
RESULTS AND DISCUSSION
Purification of inulosucrase from L. citreum and isolation of islA gene.
Two peptides were sequenced from inulosucrase extracted from L. citreum cells with 8 M urea (11) and purified by SDS-PAGE (Fig. 1). With genomic DNA from L. citreum, a 3-kbp fragment was amplified by PCR with primers that were designed from the two peptides (Fig. 2A). Sequence analysis of the 3-kbp fragment indicates similarity with fructosyltransferase genes (ftf) in the amino terminus and also with the alternansucrase gene product in the C terminus. Based on the homology with alternansucrase, two additional primers were designed in order to isolate the 3′ end (Fig. 2B). The 5′ of the islA open reading frame and the promoter region of the gene were obtained with inverse PCR techniques (Fig. 2C).
Nucleotide sequence analysis.
In total, a DNA fragment of 5,063 bp was sequenced. The analysis of this sequence shows one open reading frame of 4,473 bp (ORF1), starting at position 591, and a putative ribosome-binding site with the sequence AGGGAG located 8 bp upstream from the ATG initiation codon. The same ribosome-binding site has been reported 8 bp upstream of the initiation codon for the alternansucrase in L. mesenteroides NRRL B-1355 (2). A putative promoter sequence was also identified 34 bp upstream of the ATG, with −35 (TTGTAAC) and −10 (TATAGT) sequences separated by 19 nucleotides with the program Neural Network Prediction. The gene for inulosucrase, islA, encodes a protein of 1,490 amino acids with a deduced molecular mass of 165,137 Da and a pI of 5.09. This is the largest FTF described so far.
The G+C content of the islA gene was 37.25%, which is consistent with the value reported by Kim et al. for Leuconostoc kimchii (15). The codon usage in the islA gene is also consistent with the typical codon preferences found in Leuconostoc genes.
With the complete islA gene as a probe, a Southern blot analysis was carried out in order to locate the islA gene in the L. citreum CW28 genome. The plasmid and genomic DNAs of L. citreum were run in separate lanes. After transfer and hybridization, only the lane with the genomic DNA and the control hybridized with the islA probe (data not shown), indicating that the islA gene is not located on a plasmid in L. citreum.
Amino acid sequence analysis.
The analysis of the deduced inulosucrase amino acid sequence revealed the presence of a putative gram-positive signal peptide, and a cleavage site following amino acid 39 was predicted with the program designed by Nielsen et al. (23). To confirm the cleavage site, the L. citreum CW28 enzyme was purified by SDS-PAGE and subjected to N-terminal sequence analysis. The first 10 amino acids were identified as DVNQPLLAQK; except for the 10th residue, this sequence is identical to that of the deduced amino acid sequence after the predicted signal peptide cleavage site.
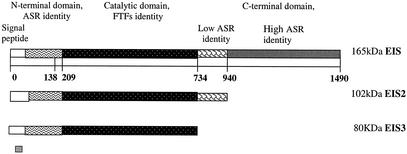
Blast searches with the deduced amino acid sequence revealed a natural chimeric enzyme with three domains, as shown in Fig. 3: the first domain in the N terminus (amino acids 1 to 138) shows similarities with alternansucrase (40% identity). This domain includes a variable region which is not conserved among GTFs and has no evident function. The second domain, which is the catalytic domain, shows similarities with Bacillus subtilis levansucrase (02730; 39% identity and 56% similarity in 433 amino acids), Clostridium acetobutylicum levansucrase (AAK79737.1; 39% identity and 55% similarities in 456 amino acids), Streptococcus salivarius FTF (Q55242; 36% identity and 52% similarities in 714 amino acids), and Streptococcus mutans FTF (P11701; 31% identity and 45% similarities in 784 amino acids). Lower similarities were found with Erwinia amylovora levansucrase (Q46656; 28% identity and 44% similarities in 438 amino acids) and Rahnella aquatilis levansucrase (O54435; 28% identity and 43% similarities in 435 amino acids).
FIG. 3.
Representation of three versions of the inulosucrase proteins that were expressed in E. coli DH5α. EIS is the complete enzyme; EIS2 is the enzyme with a deletion of the high-homology C-terminal alternansucrase (ASR), and EIS3 is the enzyme with a deletion of the same region of EIS2 plus a transition region.
High similarity among FTFs and the catalytic domain of inulosucrase was found in the amino acid 209 to 734 region. In the deduced sequence, amino acid residues 293 to 734, a core of 441 amino acids, could be related to the glycosylhydrolase 68 family, which includes several bacterial levansucrases and invertase from a Zymomonas sp. (pfam02435; http//pfam.wustl.edu/). Alignments with FTFs from gram-positive bacteria showed eight blocks of conserved motifs that can be easily identified in inulosucrase (Fig. 4).
FIG. 4.
High-homology regions derived from an alignment of FTFs from S. salivarius (36% identity), S. mutans (31% identity), L. reuteri (35% identity), and L. citreum.
Surprisingly, the carboxy-terminal regions (amino acids 940 to 1490) also showed a remarkable identity (80%) with the alternansucrase C-terminal domain. This domain has been considered capable of binding polymer (glucan for alternansucrase), and it consists of a series of four related but not identical tandem repeats of 20 to 30 amino acids each, defined according to their sequences (10). In general, the glucan binding domain from GTF presents different repeat motifs (6). In inulosucrase as well as in alternansucrase, only the motif defined as A by Giffard and Jacques (10) was found (Fig. 5A). Furthermore, a seven-repeat element set only found for alternansucrase (14) but not for another glucosyltransferase was present in inulosucrase (Fig. 5B). This natural chimeric construction explains the high molecular weight of inulosucrase compared with most FTFs which are not structured by multiple domains. In this context, inulosucrase may be viewed as an alternansucrase in which the catalytic domain has been replaced with a complete FTF or as an FTF to which the alternansucrase N-terminal and C-terminal regions have been added.
FIG. 5.
(A) Alignment of the three repeat elements type A from the C terminus of L. citreum inulosucrase. The A element represents the consensus (10). (B) Seven repeat sequences from the C-terminal domain of inulosucrase of L. citreum.
The C-terminal repeat domain of streptococcal GTFs binds glucans and probably contributes to sucrose-dependent adherence. In some but not all GTFs, it is also essential for the retention of glucan synthesizing activity, as some become sucrose hydrolysing enzymes upon deletion of the repeat units (21). What are the functions of this additional domains in inulosucrase? We think that the additional C-terminal domain that is the GTFs’ glucan binding domain may be useful to inulosucrase for binding to glucans produced by other GTFs in the culture. Although a GTF in L. citreum CW28 has not been found, L. mesenteroides NRRL B-1355, producer of alternansucrase, has now been reclassified as L. citreum (personal communication from Gregory Cote, National Center for Agricultural Utilization Research, Peoria, Ill.). It is therefore not surprising that the two enzymes may have coexisted in an L. citreum ancestor, where inulosucrase of the CW28 strain resulted from recombination between an FTF and alternansucrase. The new inulosucrase could then bind to glucans supporting the growth and sucrose consumption of the strain as it is associated with the cell wall.
Expression of islA gene in E. coli and characterization of the protein.
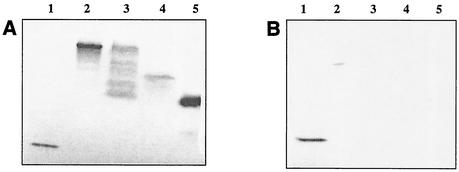
A fragment of 5,007 bp including the complete islA gene and its promoter region was cloned in vector pCR4-TOPO, resulting in the plasmid pCRIS, used to transform E. coli DH5α. The full-length construction of inulosucrase yielded many transformants. Extracts of E. coli DH5α cells containing the plasmid pCRIS showed sucrose-transforming activity, as measured by the release of reducing power from sucrose. SDS-PAGE of cell extracts showed that the inulosucrase protein was present as a band corresponding to the same molecular weight as the L. citreum enzyme. An activity gel with raffinose as the substrate was performed, showing polymer-synthesizing activity in both the L. citreum and the recombinant enzymes after staining with the Schiff reagent (Fig. 6).
FIG. 6.
(A) SDS-PAGE activity analysis of L. citreum proteins cloned in E. coli after staining with Schiff. We used levansucrase of B. subtilis as a control (lane 1), wild inulosucrase from L. citreum (lane 2), recombinant EIS from E. coli (lane 3), the truncated EIS2 from E. coli (lane 4), and the truncated EIS3 from E. coli (lane 5). (B) Activity analysis of the polymers after staining with Schiff reagent after inulinase treatment. We used levansucrase from B. subtilis as a control (lane 1), inulosucrase from L. citreum (lane 2), recombinant EIS from E. coli (lane 3), the truncated EIS2 from E. coli (lane 4), and the truncated EIS3 from E. coli (lane 5).
Expression of two C-terminally (glucan binding domain) truncated versions of the islA gene in E. coli.
In order to investigate the function of the C-terminal region in the enzyme, two deletions of the carboxy-terminal region were designed (Fig. 3). In the first deletion (bp 56 to 3333), the alternansucrase identity region was eliminated. A fragment of 3,277 bp was cloned in the pCR4-TOPO vector, resulting in plasmid pCRIS2, which was used to transform E. coli DH5α. In the second deletion, a fragment of 2,731 bp was constructed, deleting the same region as before plus the transition region. The fragment was cloned in the same vector, resulting in plasmid pCRIS3, used to transform E. coli DH5α. Transformants were obtained with both plasmids.
Extracts of E. coli DH5α cells showed polymer-synthesizing activities. According to the SDS-PAGE activity gel, molecular weights of 105,000 and 83,000 were estimated for the two deletion-truncated proteins (Fig. 5A). The three enzyme forms showed activity at 30°C and pH 6.5. The effect of the additional C-terminal domain on inulosucrase activity and stability was studied in three recombinant forms, and it was concluded that while the complete enzyme expressed in E. coli (EIS) had a specific activity of 0.015 U/mg of protein, 0.016 U/mg and 0.031 U/mg were measured for EIS2 and EIS3, respectively (Table 2). It may be concluded that deletion of 750 amino acids had an impact on the enzyme activity, increasing the reaction rate, probably due to less steric hindrance in reactant transport and usually controlling the reaction rate. Interestingly, the half-life of EIS2 and EIS3 at 40°C was reduced to 20 and 12 min, respectively, while EIS1 was relatively more stable, with a half-life of 70 min (Table 2). It is therefore possible to conclude that the carboxy terminus contributes to the stability of the enzyme. Moreover, when the C terminus was deleted, the enzyme EIS3 became more hydrolytic than the complete enzyme (Table 2).
TABLE 2.
Physicochemical characterization of inulosucrase and two C-terminally truncated recombinant forms
| Enzymatic form | Molecular mass (kDa) | Total activity (U/mg) | Half-life at 40°C (min) | Hydrolysis/transfer (%) |
|---|---|---|---|---|
| EIS | 165,137 | 0.015 | 70 | 47 |
| EIS2 | 102,630 | 0.016 | 20 | NDa |
| EIS3 | 80,556 | 0.031 | 12 | 70 |
ND, not determined.
In order to determine the type of polymer produced by the EIS2 and EIS3 forms, two SDS-PAGE activity gels were carried out in parallel as already described. It was found that the three recombinant proteins synthesized a polymer after incubation with raffinose (Fig. 6A) and that this product was digested by inulinase (Fig. 6B), while the control (levan) remained in the gel. Therefore, EIS1, EIS2, and EIS3 are all capable of inulin synthesis.
The L. citreum inulosucrase is the first glycosyltransferase that has been found as a natural chimeric enzyme bearing structural and modular features of GTFs but still bearing fructosyltransferase activity. Whereas several microorganisms have been reported as FTF producers (12), only a few genera produce GTF: Streptomyces (19), which has not been studied in detail; Leuconostoc (20); Streptococcus (13); and Lactobacillus (17, 32). It is interesting that GTFs are produced by microorganisms that have also been reported to be FTF producers, while the reverse is not true. Although both groups of enzymes, GTFs and FTFs, build a polysaccharide from sucrose by transferring the glucose or fructose residue, they have been found in the same microorganism, but their reaction mechanisms, their sequences, and their structural organization are quite different. This type of recombination could be specific for microorganisms in which both GTFs and FTFs are present, such as the genera Leuconostoc and Streptococcus.
We have also identified two hypothetical proteins of Leuconostoc mesenteroides ATCC 8293 with homology to another FTF, also bearing C-terminal regions with identity with GTFs in the recently available genome of Leuconostoc (NCBI Microbial Genome Annotation Project). Actually, the FTFs reported in Leuconostoc spp. have molecular masses of more than 100 kDa, the average bacterial FTFs molecular size. In 1983, Russell et al. reported a glucan binding protein with FTF activity in S. mutans, but no explanation could be given for this property (29), so it is possible that the same recombination had taken place in S. mutans.
When the C-terminal domain was deleted (EIS3), the resulting enzyme was more active and more hydrolytic than the wild-type enzyme. The presence of the additional domains increased its stability and reduced the hydrolytic activity of sucrose in favor of the inulin-synthesizing activity, probably by limiting water access to the catalytic domain. Nevertheless, FTFs could also recombine in these genera to acquire useful GTF properties such as binding to glucans. A phylogenetic study of various strains and glycosyltransferases is in progress.
Acknowledgments
This project was supported by Dirección General de Estudios de Posgrado (DGEP) UNAM and Consejo Nacional de Ciencia y Tecnología (CONACyT) no. 118116 and 25281-B.
We thank Brenda Valderrama, Miguel Lara, and Guadalupe Espin for important comments on the project and on the manuscript; we also thank T. L. Fernando Gonzalez for technical support. Finally, we thank Eugenio López and Paul Gaytan for primer synthesis.
REFERENCES
- 1.Abo, H., T. Matsumura, T. Kodama, H. Ohta, K. Fukui, K. Kato, and H. Kagawa. 1991. Peptide sequence for sucrose splitting and glucan binding within Streptococcus sobrinus glucosyltransferase (water-insoluble glucan synthetase). J. Bacteriol. 173:989-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Argüello-Morales, M., M. Remaud-Simeon, S. Pizzut, P. Sarçabal, R. Willemot, and P. Monsan. 2000. Sequence analysis of the gene encoding alternansucrase, a sucrose glucosyltransferase from Leuconostoc mesenteroides NRRL B-1355. FEMS Microbiol. Lett. 182:81-85. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2001. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 4.Batista, F. R., L. Hernandez, J. Fernández, J. Arrieta, C. Méndez, R. Gómez, Y. Tambara, and T. Pons. 1999. Substitution of ASP-309 by ASN in the Arg-Asp-Pro (RDP) motif of Acetobacter diazotrophicus levansucrase affect sucrose hydrolysis, but not enzyme specificity. Biochem. J. 337:503-506. [PMC free article] [PubMed] [Google Scholar]
- 5.Devulapalle, K., D. Goodman, Q. Gao, A. Hemsley, and G. Mooser. 1997. Knowledge-based model of a glucosyltransferase from the oral bacterial group of mutans streptococci. Protein Sci. 6:2489-2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferretti, J. J., M. L. Gilpin, and R. Russell. 1987. Nucleotide sequence of a glucosyltransferase gene from Streptococcus sobrinus MFe28. J. Bacteriol. 169:4271-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Funane, K., T. Ishii, M. Matsushita, K. Hori, K. Mizuno, H. Takahara, Y. Kitamura, and M. Kobayashi. 2001. Water-soluble and water-insoluble glucans produced by Escherichia coli recombinant dextransucases from Leuconostoc mesenteroides NRRL B-512F. Carbohydr. Res. 334:19-25. [DOI] [PubMed] [Google Scholar]
- 8.Funane, K., T. Ookuta, and M. Kobayashi. 1998. Glucan binding regions of dextransucrase from Leuconostoc mesenteroides NRRL B-512F. Biosci. Biotechnol. Biochem. 62:123-127. [DOI] [PubMed] [Google Scholar]
- 9.Funane, K., M. Shiraiwa, K. Hashimoto, E. Ichishima, and M. Kobayashi. 1993. An active-site peptide containing the second essential carboxyl group of dextransucrase from Leuconostoc mesenteroides by chemical modifications. Biochemistry 32:13696-13702. [DOI] [PubMed] [Google Scholar]
- 10.Giffard, P., and N. Jacques. 1994. Definition of a fundamental repeating unit in streptococcal glucosyltransferase glucan-binding regions and related sequences. J. Dent. Res. 73:1133-1141. [DOI] [PubMed] [Google Scholar]
- 11.Hamada, S., T. Horikoshi, T. Minami, N. Okahashi, and T. Koga. 1989. Purification and characterization of cell-associated glucosyltransferase synthesizing water-insoluble glucan from serotype c Streptococcus mutans. J. Gen. Microbiol. 135:335-344. [DOI] [PubMed] [Google Scholar]
- 12.Hettwer, U., M. Gross, and K. Rudolph. 1995. Purification and characterization of an extracellular levansucrase from Pseudomonas syringae pv. phaseolicola. J. Bacteriol. 177:2834-2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacques, N. A. 1995. Extracellular sucrose metabolism by Streptococcus salivarius. Dev. Biol. Stand. 85:315-322. [PubMed] [Google Scholar]
- 14.Janecek, S., B. Svensson, and R. Russell. 2000. Location of repeat elements glucansucrases of Leuconostoc and Streptococcus species. FEMS Microbiol. Lett. 192:53-57. [DOI] [PubMed] [Google Scholar]
- 15.Kim, J., J. Chun, and H. U. Han. 2000. Leuconostoc kimchii spp. nov., a new species from kimchi. Int. J. Syst. E vol. Microbiol. 50:1915-1919. [DOI] [PubMed] [Google Scholar]
- 16.Koepsell, H., H. Tsuchiya, N. Hellman, A. Kazenko, C. Hoffman, E. Sharpe, and R. Jackson. 1953. Enzymatic synthesis of dextran. J. Biol. Chem. 200:793-801. [PubMed] [Google Scholar]
- 17.Kralj, S., G. Van Geel-Shutten, H. Rahaoui, R. Leer, E. Faber, Van der M. Maarel, and L. Dijkhuizen. 2002. Molecular characterization of a novel glucosyltransferase from Lactobacillus reuteri strain 121 synthesizing a unique, highly branched glucan with α-(1→4) and α-(1→6) glucosidic bonds. Appl. Environ. Microbiol. 68:4283-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 19.Mateju, J., J. Cudlin, N. Steinerova, M. Blumauerova, and Z. Vanek. 1978. Isolation of glucosyltransferase from Streptomyces aureofaciens. Folia Microbiol. (Prague) 23:337-340. [DOI] [PubMed] [Google Scholar]
- 20.Miller, A., S. H. Eklund, and J. Robyt. 1986. Milligram to gram scale purification and characterization of dextransucrase from Leuconostoc mesenteroides NRRL B-512F. Carbohydr. Res. 147:119-133. [DOI] [PubMed] [Google Scholar]
- 21.Monchois, V., A. Reverte, R.-M. Simeon, P. Monsan, and R. Willemot. 1998. Effect of Leuconostoc mesenteroides NRRL B-512F dextransucrase carboxy-terminal deletions on dextran and oligosaccharide synthesis. Appl. Environ. Microbiol. 64:1644-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monsan, P., and Paul, F. 1995. Enzymatic synthesis of oligosaccharides. FEMS Microbial Rev. 16:187-192. [Google Scholar]
- 23.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage site. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 24.Ohtsuka, K., S. Hino, T. Fukushima, O. Osawa, T. Kanematsu, and T. Uchida. 1992. Characterization of levansucrase from Rahnella aquatilis JCM-1683. Biosci. Biotechnol. Biochem. 56:1373-1377. [Google Scholar]
- 25.Olivares-Illana, V., W.-C. Rodarte, S. LeBorgne, and L.-A. Munguía. 2002. Characterization of a cell-associated inulosucrase from a novel source: A Leuconostoc citreum strain isolated from Pozol, a fermented corn beverage of Mayan origin. J. Ind. Microbiol. Biotechnol. 28:112-117. [DOI] [PubMed] [Google Scholar]
- 26.Pérez, M. A., L. Güereca, and L.-A. Munguía. 1996. Properties of levansucrase from Bacillus circulans. Appl. Microbiol. Biotechnol. 45:465-471. [Google Scholar]
- 27.Pons, T., L. Hernández, F. Batista, and G. Chinea. 2000. Prediction of a common β-propeller catalytic domain for fructosyltransferases of different origin and substrate specificity. Protein Sci. 9:2289-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Remaud-Simeon, M., R. Willemot, P. Sarçabal, G. Potoki, and P. Monsan. 2000. Glucansucrases: molecular engineering and oligosaccharide synthesis. J. Mol. Catal. B 10:117-128. [Google Scholar]
- 29.Russell, R. R., A. C. Donald, and C. Douglas. 1983. Fructosyltransferase activity of a glucan binding protein from Streptococcus mutans. J. Gen. Microbiol. 129:3243-3250. [DOI] [PubMed] [Google Scholar]
- 30.Skov, L., O. Mirza, A. Henriksen, G. Potocki de Montalk, M. Remaud-Simeon, P. Sarcabal, R. Willemot, P. Monsan, and M. Gajhede. 2000. Crystallization and preliminary X-ray studies of recombinant amylosucrase from Neisseria polysaccharea. Acta Crystallogr. D 56:203-205. [DOI] [PubMed] [Google Scholar]
- 31.Summer, J. B., and S. F. Howell. 1935. A method for determination of invertase activity. J. Biol. Chem. 108:51-54. [Google Scholar]
- 32.van Hijum, S., G.-H. van Schutten, H. Rahaoui, M. van der Maarel, and L. Dijkhuizen. 2002. Characterization of a novel fructosyltransferase from Lactobacillus reuteri that synthesized high-molecular-weight inulin and inulin oligosaccharides. Appl. Environ. Microbiol. 68:4390-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]