Abstract
Transcriptional factor CarD is the only reported prokaryotic analog of eukaryotic high-mobility-group A (HMGA) proteins, in that it has contiguous acidic and AT hook DNA-binding segments and multifunctional roles in Myxococcus xanthus carotenogenesis and fruiting body formation. HMGA proteins are small, randomly structured, nonhistone, nuclear architectural factors that remodel DNA and chromatin structure. Here we report on a second AT hook protein, CarDSa, that is very similar to CarD and that occurs in the bacterium Stigmatella aurantiaca. CarDSa has a C-terminal HMGA-like domain with three AT hooks and a highly acidic adjacent region with one predicted casein kinase II (CKII) phosphorylation site, compared to the four AT hooks and five CKII sites in CarD. Both proteins have a nearly identical 180-residue N-terminal segment that is absent in HMGA proteins. In vitro, CarDSa exhibits the specific minor-groove binding to appropriately spaced AT-rich DNA that is characteristic of CarD or HMGA proteins, and it is also phosphorylated by CKII. In vivo, CarDSa or a variant without the single CKII phosphorylation site can replace CarD in M. xanthus carotenogenesis and fruiting body formation. These two cellular processes absolutely require that the highly conserved N-terminal domain be present. Thus, three AT hooks are sufficient, the N-terminal domain is essential, and phosphorylation in the acidic region by a CKII-type kinase can be dispensed with for CarD function in M. xanthus carotenogenesis and fruiting body development. Whereas a number of hypothetical proteins homologous to the N-terminal region occur in a diverse array of bacterial species, eukaryotic HMGA-type domains appear to be confined primarily to myxobacteria.
In eukaryotes, the high-mobility-group A (HMGA) subfamily of HMG proteins [previously HMGI(Y) (9)] are small, relatively abundant, nonhistone chromosomal proteins that regulate gene expression and are implicated in a variety of cellular functions (8, 10, 16). HMGA proteins serve as DNA architectural factors that remodel chromatin to aid in the assembly of specific nucleoprotein complexes that are essential for transcription, replication, recombination, or repair (48). The primary structure of HMGA proteins is characterized by multiple repeats of a conserved RGRP sequence (the AT hook motif), embedded within basic and proline residues, and a contiguous highly acidic region (3). The AT hooks have random structure when free but adopt a defined conformation on binding specifically to the narrow minor groove of AT-rich sequences 4 to 8 bp in length and present in at least two appropriately spaced tracts (20, 25, 43). Kinases such as casein kinase II (CKII) and Cdc2 phosphorylate HMGA proteins and modulate DNA binding as well as protein stability (14, 32, 36, 41, 47, 49, 50). This occurs in a cell cycle- and differentiation-dependent manner and fine-tunes HMGA activity in vivo.
The only reported prokaryotic example of an HMGA-type protein is CarD in the bacterium Myxococcus xanthus (28, 29, 31). Like HMGA proteins, CarD has multifunctional roles in vivo and has been shown to be involved in at least two processes: light-induced carotenogenesis and fruiting body formation (29). The randomly structured HMGA-like C-terminal domain of CarD is made up of a basic region of multiple AT hooks and a flanking highly acidic segment. This domain is very similar to human HMGA proteins in its physical, structural and DNA-binding properties (28, 31). CarD is, however, considerably larger and contains an N-terminal stretch of around 180 amino acids that is absent in eukaryotic HMGA proteins. This N-terminal domain, whose function remains to be elucidated, has defined secondary and tertiary structure, in contrast to the C-terminal HMGA-like region (31).
HMGA proteins are ubiquitous in higher eukaryotes (3, 8). However, as pointed out above, M. xanthus CarD is the only such protein identified so far in a prokaryote. The objective of this study was to examine whether HMGA-type proteins occur in other bacteria. As a first step we have done so for the bacterium Stigmatella aurantiaca, which belongs to the same taxonomic subgroup as M. xanthus and exhibits similar behavioral and developmental characteristics (40). We have identified a gene in S. aurantiaca that codes for a protein highly similar to M. xanthus CarD. Like CarD, its S. aurantiaca counterpart (CarDSa) contains an HMGA-like C-terminal region, as well as the N-terminal stretch of approximately 180 residues that is absent in eukaryotic HMGA proteins. The N-terminal regions of the two proteins are almost identical in sequence, while the HMGA-like C-terminal regions are less so. In CarDSa the latter region has one fewer AT hook and only one CKII phosphorylation site in the acidic part. In vitro analysis using purified proteins showed that CarDSa exhibits the same DNA-binding specificity as CarD and that CKII phosphorylation of CarDSa occurs but to a considerably lower extent than in CarD. In vivo, CarDSa can replace CarD in carotenogenesis and fruiting body formation. Our findings have been exploited to gain additional insights into the molecular bases that govern CarD activity in M. xanthus. We present evidence that the existence of HMGA domains in bacteria appears to be restricted to some other myxobacteria. The N-terminal domain in CarDSa and CarD, on the other hand, exists in various other bacteria as an independent module.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The M. xanthus wild-type strains used in this study (Table 1) are DK1050 (37) and DK1622 (23). Vegetative growth was carried out in the rich medium CTT at 33°C; TPM medium and CF agar were used to induce fruiting body formation (7, 17). S. aurantiaca DW4/3-1 (a streptomycin-resistant derivative of DW4) (33), a generous gift of H. U. Schairer, was grown at 30 to 32°C in media containing 1% Bacto Tryptone (Difco) and 0.02% MgSO4 · 7H2O per liter of medium (pH 7.2). Myxococcus (Corallococcus) coralloides, Cystobacter fuscus, Nannocystis exedens, and Polyangium cellulosum were obtained from the culture collection of Deutsche Sammlung von Mikroorganism und Zellkulturen GmbH, Braunschweig, Germany, and grown in the following media: C. fuscus and M. coralloides, 0.1% each raffinose, sucrose, and galactose, 0.25% Casitone, 0.5% starch, 0.05% MgSO4 · 7H2O, and 0.025% K2HPO4 (pH 7.4); N. exedens, 0.5% dry baker's yeast, 0.14% CaCl2 · 2H2O, and 0.5 μg of vitamin B12 per ml (pH 7.2); and P. cellulosum, 0.1% KNO3, 0.1% K2HPO4, 0.03% MgSO4 · 7H2O, 0.013% CaCl2 · 2H2O, 0.003% FeCl3 · 6H2O, and small strips of filter paper (pH 7.2). The growth temperature was 30°C in all cases except for P. cellulosum, which was grown at 26°C. Culture conditions for the Bdellovibrio sp. strain CP1, kindly provided by A. Sánchez-Amat, were as previously described (39). Escherichia coli strain DH5α was used for plasmid constructions, and strain BL21(DE3) was used for protein overexpression (44); both were grown in Luria broth medium (38). E. coli strains Q358 (2) and Y1090 (51) were used for infection by the λDASH and λgt11 phage libraries, respectively.
TABLE 1.
M. xanthus strains used in this study
| Strain | Phenotypea | Relevant genotypeb | Reference or sourcec |
|---|---|---|---|
| DK1050 | Car+ Fru+ | carD (wild type) | 35 |
| DK1622 | Car+ Fru+ | carD (wild type) | 21 |
| MR1900 | Car− Fru− | carD3 (ΔcarD) | This study (pMR2603 × DK1622) |
| MR1901 | Car+ Fru+ | carD′ | This study (pMR2696 × MR1900) |
| MR1902 | Car+ Fru+ | carDSa | This study (pMR2698 × MR1900) |
| MR1903 | Car+ Fru+ | carDSa(S198A) | This study (pMR2745 × MR1900) |
| MR1904 | Car− Fru− LacZ− Kmr | carD3 carQ::lacZ | pDAH217 × MR1900 |
| MR1905 | Car+ Fru+ LacZi Kmr | carD′ carQ::lacZ | pDAH217 × MR1901 |
| MR1906 | Car+ Fru+ LacZi Kmr | carDSacarQ::lacZ | pDAH217 × MR1902 |
| MR1907 | Car+ Fru+ LacZi Kmr | carDSa(S198A) carQ::lacZ | pDAH217 × MR1903 |
Only the phenotypes relevant to this work are indicated. LacZi, light-inducible synthesis of β-galactosidase; LacZ−, low basal levels of β-galactosidase synthesis in the dark or in the light.
Only the genotypes relevant to this work are indicated. Relative to that for the wild-type carD gene in DK1622 (or DK1050), an additional 6 bp separates the initiator ATG codon and the ribosomal binding site in strains MR1901, MR1902, and MR1903 and those derived from them (MR1905, MR1906, and MR1907) (see Materials and Methods).
All of the strains except the first two were generated by electroporation of the integrative plasmid into the respective strain as indicated here and described in the text.
Immunoblot analysis.
M. xanthus strains DK1622 and MR1900 (Table 1) and S. aurantiaca strain DW4/3-1 (33) were grown to late logarithmic phase in rich media, and cells from 1 ml of culture were harvested by centrifugation. The cell pellet was resuspended in 80 μl of the following mix: 100 μl of 5× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) lysis buffer (38), 100 μl of protease inhibitor cocktail (Sigma), and 300 μl of buffer A (50 mM Tris pH 7.5, 5% glycerol, 1 mM EDTA, 7 mM β-mercaptoethanol) containing 100 mM NaCl. The cell suspension was lysed by boiling for 5 min, and 30 μl of each sample was run in an SDS-10% polyacrylamide minigel. Proteins were transferred to nitrocellulose by using a semidry electroblotting unit, and Western immunoblot analysis was carried out with anti-CarD antibodies obtained previously (31) and the ECL kit (Amersham Biosciences).
Southern hybridization analysis.
Genomic DNA was isolated by using the Promega Wizard genomic DNA purification kit. Southern analysis was done by standard procedures (38). A 1.13-kb DNA fragment containing the entire carD gene (948 bp) or a fragment spanning nucleotides 354 to 948 of this gene was used as a probe. Low-stringency hybridization was performed at 59°C in the following buffer: 6× SSC (0.9 M sodium chloride plus 0.09 mM sodium citrate), 0.1% SDS, 5× Denhardt's solution, and 5% dextran sulfate. Blots were washed twice at room temperature and once at 59°C in 2× SSC-0.1% SDS.
Cloning of the S. aurantiaca gene homologous to carD.
Two independent genomic DNA libraries constructed in phages λDASH and λgt11 (generous gifts from H. U. Schairer, University of Heidelberg) were used to screen for the gene homologous to carD in S. aurantiaca (which is referred to as carDSa). The λDASH and λgt11 libraries were constructed with genomic DNA digested with SalI and HpaII, respectively. About 16,000 plaques generated from E. coli strain Q358 (for the λDASH phage library) or Y1019 (for the λgt11 phage library) were screened under low-stringency conditions with the 1.13-kb DNA probe containing carD by standard procedures (38). The most intense positive signals were selected and subjected to a second round of hybridization. Five positive clones were chosen from the λDASH library, and three were chosen from the λgt11 library. Phage DNA from each of the selected clones was isolated, cut with SalI (for λDASH) or EcoRI (for λgt11), electrophoresed, and tested again with the carD probe. Each of the five λDASH clones gave a single positive hybridization band of ∼5 kb which was purified and cloned into the vector pUC9-2 to generate plasmid pMAR541. Single hybridization bands, but of different sizes, were obtained with the three λgt11 clones, and of these, one of ∼2 kb was cloned into pUC19 to generate plasmid pMAR542. A DNA fragment of about 1.2 kb from each clone was sequenced twice along both strands.
Construction of an M. xanthus strain with gene carD deleted.
pMAR975 is a pBJ114 derivative (22) which lacks the EcoRI site and contains a Kmr gene for positive selection and a galactose sensitivity (Gals) gene for negative selection (46). A 3.3-kb DNA fragment containing carD and about 1.2 kb of flanking DNA on each side was cloned into pMAR975 to obtain plasmid pMR2598. To generate a complete in-frame deletion of carD, pMR2598 was used as template for inverse PCR with the Expand long-template PCR system (Roche Applied Science) and two oligonucleotide primers with EcoRI overhangs (underlined): carD-DEL1 (5′-AAAGGAATTCGTCCCCCTCACGGGTGAGGT-3′), and carD-DEL2 (5′-AAAGGAATTCTGACAGCCCCATGGACCGAC-3′). The PCR-amplified fragment was cut with EcoRI and self-ligated to generate plasmid pMR2603, in which the entire carD gene is precisely deleted (from the ATG start codon to the nucleotide immediately upstream of the stop codon) and replaced by an EcoRI site. This in-frame deletion of carD is referred to as the carD3 allele. pMR2603 was electroporated into M. xanthus (strain DK1622), where it can be maintained only after integration into the chromosome by homologous recombination. Stable kanamycin-resistant transformants are thus merodiploids carrying wild-type carD as well as the carD3 allele. Cells having lost one of the two copies and the vector DNA through intramolecular recombination events were selected for on CTT plates supplemented with 10 mg of galactose per ml. Several Galr Kms colonies were picked and tested by PCR for the presence or absence of the carD deletion to isolate the carD deletion strain MR1900.
Complementation analyses.
For complementation analyses the following plasmids were constructed. (i) pMR2698 contains the carDSa gene inserted into the EcoRI site of pMR2603. The insert (initiator ATG to stop codon) was generated by PCR with S. aurantiaca genomic DNA as the template and the following primers with EcoRI overhangs (underlined): SaCD-RI-N (5′-AAAGAATTCATGCCAGAAGGACTCCAGCTC-3′) and SaCD-RI-C (5′-AAAGAATTCTCACTACTCGGTCTCACCCTC-3′). (ii) pMR2745 is derived from pMR2698 by site-directed mutagenesis of the codon for Ser198 in carDSa to that for Ala by using the PCR overlap extension method (18). Compared to the normal arrangement for carD as in pMR2598, both pMR2698 and pMR2745 have the initiator ATG codon separated from the carD ribosomal binding site by an additional 6 bp (the EcoRI site). Consequently, in the complementation analyses carried out with pMR2698 and pMR2745, the following positive control plasmid, pMR2696, was used. (iii) pMR2696 contains the carD′ allele, that is, the carD gene from the initiator ATG to the stop codon, inserted into the EcoRI site of pMR2603 (and thus separated from the ribosomal binding site by an additional 6 bp, as in constructs pMR2698 and pMR2745). The insert was generated by PCR with pMR2598 as template DNA and the following primers with EcoRI overhangs (underlined): MxCD-RI-N (5′-AAAGAATTCATGCCTGAAGGGTCCGCGTCA-3′) and MxCD-RI-C (5′-AAAGAATTCTCAGCTCTCACCCTCGGGCGG-3′). (iv) pMR2768 is a derivative of pMR2598 containing a carD allele (carDΔN) in which the DNA region coding for the N-terminal residues 2 to 178 of the protein has been deleted by following the same protocol described above for pMR2603. The PCR primers used in this case were carD-NDEL1 (5′-AAAGAATTCCATGTCCCCCTCACGGGTGAG-3′) and carD-NDEL2 (5′-ACCGCAGCCGAATTCCAGCGCG-3′, where GAA in the EcoRI site shown underlined replaces the wild-type GAG, both triplets coding for Glu179). All PCR-derived constructs described in this and other sections were verified by DNA sequencing.
Complementation analyses were performed with the carD deletion strain MR1900 (see above) as the recipient for electroporation with each of the four plasmids described above, none of which replicate in M. xanthus but which can integrate into the bacterial genome by homologous recombination. The resulting merodiploids (Kmr) bear both the carD deletion copy and the incoming allele. When complementation was observed, the merodiploids were further processed by using the same procedure described above for strain MR1900 to generate a strain with a copy of the incoming allele alone (strains MR1902 and MR1903 and the control strain MR1901 [Table 1]). These Kms strains could then be electroporated with pDAH217 (Kmr), which carries a lacZ transcriptional probe fused to the carD-dependent light-inducible carQRS promoter (19, 29), in order to quantify the ability of the complementing gene to replace carD. Promoter activity with and without illumination was assessed qualitatively by monitoring lacZ reporter gene expression through colony color formation on plates containing 40 μg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactoside) per ml and quantitatively by measurements of β-galactosidase activity as described previously (4).
Overexpression and purification of CarDSa.
The carDSa gene was PCR amplified with S. aurantiaca genomic DNA as the template to generate a fragment with an NdeI site containing the initiator Met and a BamHI site immediately downstream of the stop codon. The PCR-amplified fragment was purified and then cloned into the NdeI-BamHI sites of the overexpression vector pET11b or into pET15b to produce His6-tagged protein (44). Overexpression of CarDSa and its purification by ion-exchange chromatography, first from phosphocellulose and then by MonoS high-performance liquid chromatography (HPLC) (AKTA-Amersham Biosciences), were as described for CarD (31). His6-tagged proteins were purified by using TALON metal affinity resin and the accompanying purification protocol for native conditions (Clontech, Palo Alto, Calif.) and by MonoS HPLC. For concentrations determined from the absorbance at 280 nm, the values used for ɛ280 (in molar−1 centimeter−1) are 8,480 and 9,970 for CarD and CarDSa, respectively (30).
Analytical size exclusion chromatography.
The apparent molecular mass for CarDSa was estimated by analytical size exclusion chromatography at room temperature with a Superdex-200 HPLC column (Amersham Biosciences). One hundred microliters of protein (∼10 μM) was injected into the column, which was previously equilibrated with buffer A containing 200 mM NaCl, and its elution was tracked by the absorbance at 280, 235, and 220 nm at a flow rate of 0.3 ml/min. The molecular mass was estimated from the elution volume Ve and the calibration curve log molecular mass = 7.91 − 0.23Ve, generated as described elsewhere (31). The identity of eluted CarDSa was confirmed by Coomassie blue staining after SDS-PAGE and Western blotting with anti-CarD antibodies.
In vitro DNA-binding and CKII phosphorylation assays.
DNA binding was examined by electrophoretic mobility shift assay with a radiolabeled 169-bp DNA probe that spans positions −117 to +52 relative to the transcription start site and contains the CarD-binding site in the carQRS promoter region (28, 31). The fragment was generated by PCR with pDAH231 as the template (26) and the synthetic oligonucleotide primers carQRS5 (5′-GGGCAGGACGGGATGCTGCTG-3′) and carQRS6 (5′-CCGTCGCGAAACCGTTCCATGA-3′). The primer carQRS5 was labeled with [γ-32P]ATP and T4 polynucleotide kinase prior to its addition to the PCR mix. The electrophoretic mobility shift assay was carried out in 20-μl-total reaction volumes containing 1 to 3 pM end-labeled probe (∼13,000 cpm), 500 nM protein, and 1 μg of double-stranded poly(dA-dT), poly (dG-dC), or poly(dI-dC) as nonspecific competitor DNA in binding buffer (50 mM NaCl, 15 mM HEPES, 4 mM Tris [pH 7.9], 1 mM dithiothreitol, 10% glycerol, 1 mg of bovine serum albumin per ml, and 0.1% Nonidet P-40). After a 30-min equilibration period at 4°C, DNA binding was analyzed by electrophoresis for 1 to 1.5 h in nondenaturing 4% polyacrylamide gels (acrylamide-bisacrylamide, 37.5:1) that were prerun at 200 V and 4°C for 30 min in 0.5× TBE buffer (45 mM Tris base, 45 mM boric acid, 1 mM EDTA). Gels were dried and analyzed by autoradiography.
CKII phosphorylation of purified CarD and CarDSa was examined by using recombinant human CKII (New England BioLabs). The protein (0.3 to 1 μM) was treated with 0.5 U of CKII and 1μCi of [γ32-P]ATP in DNA-binding buffer for 1 h at 30°C. After removal of unincorporated [γ32-P]ATP through Sephadex G-50, the samples were subjected to SDS-PAGE and analyzed by autoradiography. To compare relative amounts of protein used in the assay, mock CKII phosphorylation reactions were carried out under identical conditions but without labeled ATP and examined by Coomassie blue staining after SDS-PAGE.
Sequence comparisons and analysis.
Sequence database searches were performed with the BLAST suite of programs provided at the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/BLAST), Baylor College of Medicine search launcher (http://searchlauncher.bcm.tmc.edu) and/or European Bioinformatics Institute (http://www.ebi.ac.uk). Similarity searches of the nonredundant protein sequence database were done with the gapped BLASTP program (1). Bacterial proteins with the AT hook motif were also retrieved from the SMART database (http://smart.embl-heidelberg.de). Protein sequences were aligned by using the CLUSTAL software at BCM and analyzed for phosphorylation sites by using PROSITE (http://us.expasy.org/prosite/). Open reading frames (ORFs) in the cloned DNA sequence were identified by using the ORF Finder at NCBI (http://www.ncbi.nlm.nih.gov/gorf/gorf.html).
Nucleotide sequence accession number.
The nucleotide sequence obtained from the pMAR541 clone, which contains the whole carDSa gene, has been deposited at the DDBJ/EMBL/GenBank databases (accession number AJ536154).
RESULTS
A protein very similar to M. xanthus CarD exists in S. aurantiaca.
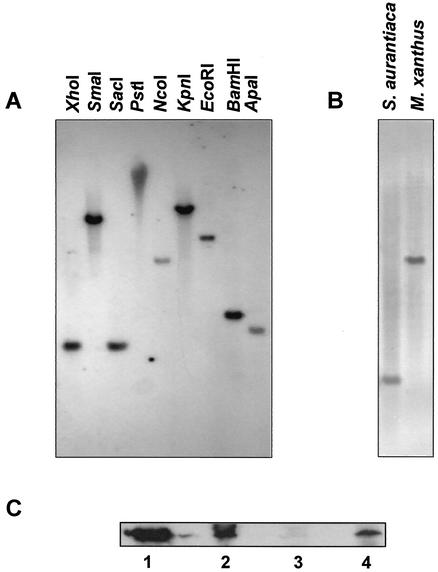
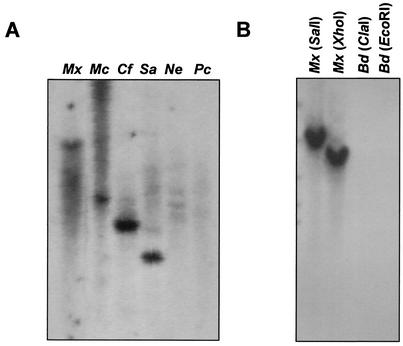
The presence of a gene homologous to carD in S. aurantiaca was examined by Southern hybridization with a 1.13-kb DNA fragment which contains the 948-bp M. xanthus carD gene as a probe. Hybridization was carried out under low-stringency conditions with S. aurantiaca genomic DNA digested with different restriction enzymes (Fig. 1A). The hybridization pattern of XhoI-digested genomic DNA from S. aurantiaca is compared with that from M. xanthus in Fig. 1B. The single hybridization band observed in every case is consistent with the existence of a carD-like gene in S. aurantiaca.
FIG. 1.
Evidence for the existence of a carD homolog in S. aurantiaca. (A) Southern analysis under low-stringency conditions of S. aurantiaca genomic DNA digested with the specific restriction enzyme indicated and with a probe corresponding to the M. xanthus carD gene. (B) Comparison of the results obtained for Southern analysis, as described for panel A, with XhoI-digested genomic DNAs from S. aurantiaca and M. xanthus. (C) Immunoblots with anti-CarD polyclonal antibodies and cell lysate samples prepared as indicated in Materials and Methods. Lane 1, purified CarD; lane 2, M. xanthus strain DK1622 (wild type); lane 3, M. xanthus strain MR1900 (ΔcarD); lane 4, S. aurantiaca strain DW4/3-1 (wild type).
We had previously obtained polyclonal anti-CarD antibodies that detected segments in both the N- and C-terminal regions of the protein (31). To examine whether these anti-CarD antibodies were also capable of detecting any protein components in S. aurantiaca, whole-cell extracts were analyzed in Western blots. Figure 1C shows that whole-cell lysates of wild-type S. aurantiaca (lane 4) yielded a band that comigrated with the one observed for the M. xanthus wild-type strain DK1622 (lane 2) and for pure CarD (lane 1) but which was not observed for MR1900, the carD deletion strain (lane 3). Thus, there exists a protein in S. aurantiaca sufficiently similar to CarD for it to cross-react with anti-CarD antibodies and whose size, moreover, closely matches that of M. xanthus CarD.
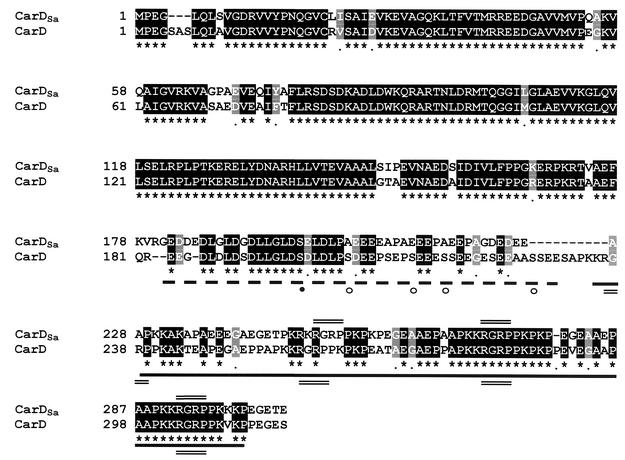
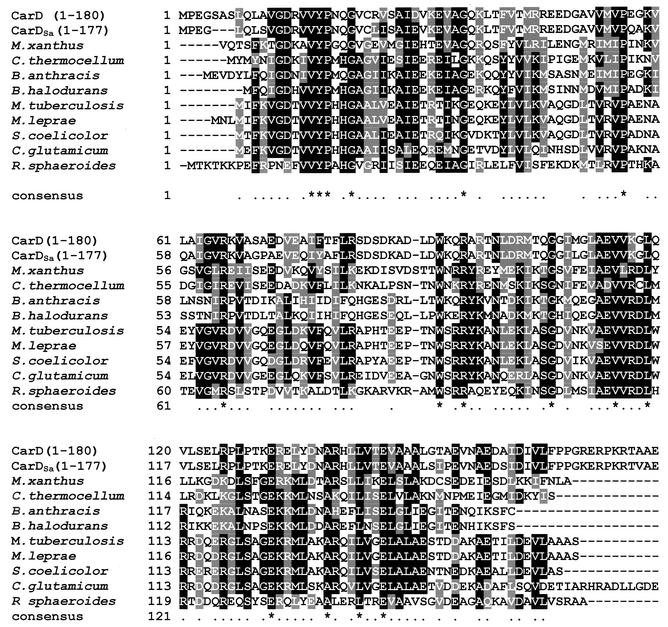
The gene homologous to carD in S. aurantiaca, designated carDSa, was cloned by using phage genomic DNA libraries (see Materials and Methods). Both strands of a 1.2-kb stretch were sequenced, and potential ORFs were identified in computer-aided searches, taking into account the bias for G or C at the third codon position (owing to the high GC content of myxobacterial DNA [6, 12]). One ORF of 915 bp that would yield a gene product of 305 amino acids with high sequence similarity to the 316-residue-long CarD was identified. This would be in accord with the results from the immunoblot and Southern hybridization analyses described above (Fig. 1). The sequence alignment between CarD and its S. aurantiaca counterpart (Fig. 2) indicates that the characteristic HMGA-type domain is also present in CarDSa. It is, however, precisely in this domain where most of the differences in the primary structures of CarDSa and CarD appear. One significant difference is the presence of only three AT hooks (the RGRP DNA-binding motif) in CarDSa as opposed to the four in CarD; another is that only one consensus CKII phosphorylation site, S198, can be identified in the acidic region of CarDSa by using PROSITE, in contrast to the five predicted in CarD. In fact, other than Ser198, no other Ser or Thr appears in the entire acidic region of CarDSa. By contrast, the approximately 180-residue N-terminal segment that precedes the HMGA-like domain in CarDSa shows high sequence identity (86%) with that in CarD. This segment includes a region that may be involved in CarD dimerization (31).
FIG. 2.
Sequence alignment of proteins CarDSa and CarD. Identical residues in CarDSa (top line) and CarD (second line) are shaded in black and indicated by an asterisk in the bottom line, while similar residues are shaded gray. The thick dashed line indicates the highly acidic region, and the thick solid line indicates the flanking basic AT hook region. The RGRP AT hooks (three in CarDSa and four in CarD) are marked by double lines. CKII target sites predicted in the acidic region are indicated by circles, with the filled one being the only such site in CarDSa that is also present in CarD.
Comparison of purified CarDSa and CarD.
CarDSa was overexpressed and purified by using procedures essentially identical to those used for CarD. Expression levels of CarDSa were typically lower than those of CarD under identical conditions and may reflect a lower intracellular stability for CarDSa. The identity of the purified protein was confirmed in Western blots with polyclonal anti-CarD antibodies. CarDSa exhibited the anomalous mobility in SDS-PAGE that was observed for CarD (31). Both have an apparent molecular mass in SDS-PAGE of around 40 to 41 kDa, compared to values calculated from their sequences of 33.1 kDa for CarDSa and 33.9 kDa for CarD. This anomalous mobility in SDS-PAGE has also been reported for the mammalian and insect HMGA proteins and appears to be related to the presence of the AT hooks (11, 50). CarDSa and CarD also exhibit similar hydrodynamic behavior, with both proteins eluting as a single symmetrical peak from a Superdex-200 analytical gel filtration HPLC column. The apparent molecular mass of 117 kDa calculated for CarDSa from these data suggests a tetramer or an extended molecule, as for CarD (apparent molecular mass, 129 kDa). A detailed analysis of the domain organization in CarD had indicated that whereas the C-terminal portion was extended and monomeric like in HMGA1a, the N-terminal domain was compact and dimeric (31). It is reasonable to expect that this will also apply to CarDSa given that its N-terminal region is nearly identical to that in CarD and that the two proteins exhibit similar physical properties.
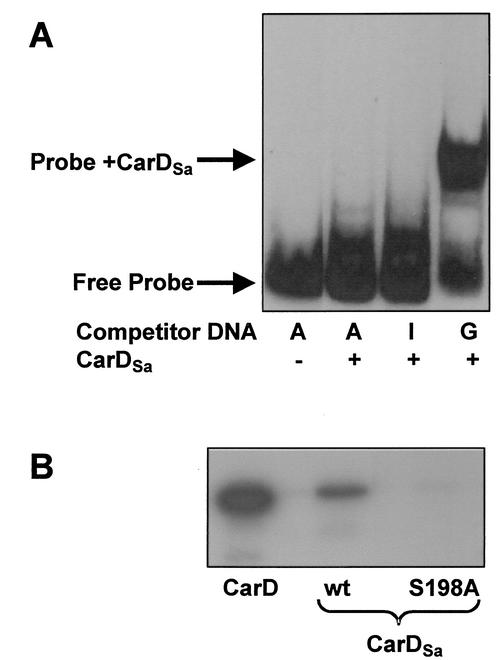
CarD is essential for the expression of the light-inducible carQRS operon, a key regulatory gene cluster in M. xanthus carotenogenesis (29). A site containing two appropriately spaced AT-rich tracts upstream of the promoter of carQRS is required in vivo for its expression, and in vitro, CarD binds to this site with the characteristic HMGA minor-groove binding specificity (28, 31). Figure 3A shows that CarDSa binds to a 169-bp double-stranded DNA probe that includes the M. xanthus carQRS promoter region and the CarD-binding site. Moreover, the retarded band is observed in the presence of poly(dG-dC) as a nonspecific competitor (lane 4) but not with poly(dA-dT) or poly(dI-dC) (lanes 2 and 3). This behavior has been used to infer that HMGA1a binds to the minor groove of AT-rich tracts (43), and this is also observed with CarD (31). Therefore, our results show that CarDSa has the same AT hook DNA-binding specificity as CarD (or HMGA1a).
FIG. 3.
DNA binding and CKII phosphorylation of CarDSa. (A) Binding of CarDSa (0.5 μM) to a 169-bp 32P-labeled DNA probe containing the PQRS promoter and the CarD-binding region (∼2 pM; 13,000 cpm) in the presence of 1 μg of poly(dA-dT) (lanes A), 1 μg of poly(dI-dC) (lane I), or 1 μg of poly(dG-dC) (lane G) under the solution conditions indicated in the text. (B) In vitro CKII phosphorylation of CarD, wild-type CarDSa (lane wt) and mutant CarDSa (lane S198A) carried out in the presence of [γ-32P]ATP under the reaction conditions described in Materials and Methods.
The acidic regions of CarD and HMGA1a can be phosphorylated in vitro by CKII. Sequence analysis had predicted a total of 10 CKII phosphorylation sites in CarD. Five of these were in the N-terminal region, and the other five were in the acidic portion of the HMGA-type segment. However, in vitro, phosphorylation by CKII mapped only to those in the HMGA part (31). Four of the five CKII phosphorylation sites predicted in the N-terminal region of CarD are also present in CarDSa. Of the five predicted CKII phosphorylation sites in the acidic region of CarD, however, only one (S198) is present in CarDSa (Fig. 2). Consistent with this, CarDSa was phosphorylated by CKII in vitro to a considerably lower degree than CarD for equivalent protein concentrations (Fig. 3B). Furthermore, when S198 in CarDSa was replaced by Ala by using site-directed mutagenesis, CKII phosphorylation in vitro was effectively eliminated (Fig. 3B). Thus, phosphorylation of CarDSa by CKII in vitro also maps to the HMGA-type domain, and more specifically to S198 in the acidic region.
Complementation of a carD deletion by carDSa.
The considerable similarity between CarD and CarDSa led us to examine if the latter would substitute for CarD function in M. xanthus. For this, we first constructed an M. xanthus strain (MR1900) with a precise in-frame deletion of carD (see Materials and Methods). As expected, this mutant strain was defective both in light-induced carotenogenesis and in fruiting body formation. To check for complementation by the carDSa gene, plasmid pMR2698 (Kmr) was introduced into strain MR1900 by electroporation. Electroporants should arise by integration of the plasmid into the M. xanthus chromosome by homologous recombination. The resulting Kmr colonies would therefore be merodiploids bearing both the carD deletion allele (carD3) and the carDSa gene. On plates, all of the Kmr electroporants showed light-induced carotenogenesis, as judged by the intense red color developed on illumination with blue light. Several of these electroporants were also picked on TPM and CF agar plates to monitor multicellular development, and the fruiting bodies were examined for the presence of spores as previously described (29). In all cases, development proceeded at the same rate as for the wild-type M. xanthus strain and yielded the same number of mature fruiting bodies. The behavior was equivalent to that observed in control experiments where MR1900 was electroporated with plasmid pMR2696 (Kmr), which has the carD′ gene instead of carDSa. All of these results are summarized in Table 2. Restoration of the wild-type phenotype for both carotenogenesis and fruiting body development on introduction of the carDSa gene into the M. xanthus strain MR1900 clearly indicates that carD can be replaced by carDSa in vivo.
TABLE 2.
Summary of Results of complementation of a carD deletion by carDSa or carDSa (S198A)
| Genotype | Carotenogenesisa | Fruiting body formationa |
|---|---|---|
| Wild type | + | + |
| carD3 | − | − |
| carD′/carD3 | + | + |
| carDSa/carD3 | + | + |
| carDSa(S198A)/carD3 | + | + |
| carDΔN/carD3 | − | − |
+, complementation (restoration of wild-type phenotype); −, lack of complementation.
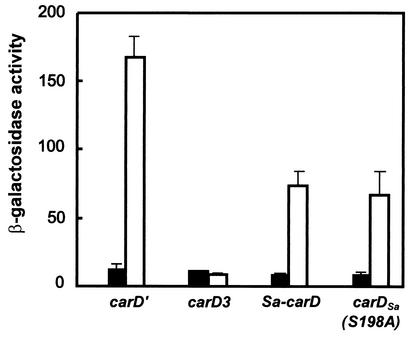
To quantitate the ability of CarDSa to replace CarD, we electroporated strain MR1902 as well as the control strains MR1900 and MR1901 (Table 1) with plasmid pDAH217 (Kmr). This plasmid cannot replicate in M. xanthus, but on integration into the bacterial chromosome by homologous recombination, it provides a transcriptional lacZ probe for the light-inducible carD-dependent carQRS promoter (PQRS). As expected, MR1901- and MR1902-derived Kmr colonies, but not MR1900-derived ones, exhibited blue colony color formation in light due to induction of the lacZ reporter gene in pDAH217. Although the intensity of the blue color that developed on plates was apparently indistinguishable for electroporants from strains MR1901 and MR1902, measurements of β-galactosidase activity indicated that CarDSa was about half as effective as CarD in activating PQRS in M. xanthus (Fig. 4). This result may reflect differences between CarD and CarDSa in their intracellular stabilities in M. xanthus and/or in their interactions with other proteins or DNA.
FIG. 4.
Quantitative measure of complementation from β-galactosidase assays with strains MR1904 (carD3), MR1905 (carD′), MR1906 (carDSa), and MR1907 [carDSa(S198A)] bearing the reporter lacZ gene fused to the CarD-dependent promoter PQRS. Cell cultures were grown in the dark to exponential phase, divided in two, and grown for a further 6 h with one half in the dark and the other in light. Samples were then collected, and β-galactosidase activities (in nanomoles of o-nitrophenol produced per minute per milligram of protein) were measured. Values for cells grown in the dark (filled bars) and those exposed to light (empty bars) and the corresponding standard deviations are shown.
Effects of the absence of the conserved N terminus or CKII phosphorylation sites in vivo.
CarDSa, as we have shown in the preceding section, can substitute for CarD function in vivo in both carotenogenesis and fruiting body formation. The near identity between CarD and CarDSa in their N-terminal domains led us to examine the importance of this region for protein function in vivo. For this, we constructed plasmid pMR2768, which bears a truncated version of carD (carDΔN) coding for the C-terminal segment spanning residues 179 to 316. We have previously shown that this CarD fragment is stably overexpressed in E. coli and that it retains the in vitro DNA-binding and CKII phosphorylation properties of CarD, but it appears to be largely monomeric, unlike the whole protein (31). Plasmid pMR2768 was electroporated into strain MR1900 to obtain merodiploids bearing the carD3 allele as well as the carDΔN truncated form. These Kmr electroporants were deficient in both light-induced carotenogenesis and fruiting body formation (Table 2), even though the corresponding CarD C-terminal fragment was stably expressed, as verified by Western blots of whole cell extracts (data not shown). This demonstrates that the 180-residue N-terminal region that is so highly conserved in CarD and CarDSa (and absent in HMGA proteins) is essential for the protein to carry out its in vivo roles in carotenogenesis and fruiting body formation; the presence of the HMGA-like domain alone is not sufficient.
Of the five predicted CKII phosphorylation sites in the acidic region of CarD, only S198 is present in CarDSa (Fig. 2), and the mutation S198A eliminates in vitro phosphorylation of CarDSa by CKII, as described earlier. We exploited this and the ability of CarDSa to substitute for CarD function to examine the role, if any, of phosphorylation by CKII-type kinases in vivo. For this, plasmid pMR2745, containing the carDSa(S198A) mutant gene, was introduced into strain MR1900. This restored the wild-type phenotype for carotenogenesis and multicellular development, just as do the plasmids pMR2698 (containing carDSa) and pMR2696 (containing carD′) (Table 2). Moreover, light induction of PQRS by the CarDSa(S198A) mutant was essentially identical to that brought about by CarDSa (Fig. 4). These results suggest that phosphorylation of the HMGA domain by a CKII-type kinase is not a necessary regulatory event for normal carotenogenesis and fruiting body formation in M. xanthus.
carD-like genes in other bacteria.
M. xanthus CarD had been the only protein with an HMGA-type domain identified in a prokaryote until the description in this study of the homologous CarDSa protein in S. aurantiaca. Based on 16S RNA analysis, the myxobacteria form a monophyletic grouping consisting of three distinct subgroups (Myxococcus, Chondromyces, and Nannocystis), and both M. xanthus and S. aurantiaca fall into the Myxococcus subgroup (42). To examine if the occurrence of HMGA-type proteins in myxobacteria is a general phenomenon, a DNA fragment corresponding to nucleotides 354 to 948 of carD (coding for its entire HMGA part) was used to probe under low-stringency conditions the genomic DNAs from M. coralloides and C. fuscus (Myxococcus subgroup), P. cellulosum (Chondromyces subgroup), and N. exedens (Nannocystis subgroup). As with M. xanthus and S. aurantiaca, a strong hybridization band was observed for M. coralloides and C. fuscus but not for P. cellulosum and N. exedens (Fig. 5A). Given that the hybridization was done under low-stringency conditions, the faint and diffuse signals observed for the last two strains could stem from the high GC content of the genomic DNA of myxobacteria. Consequently, these results suggest that the existence of HMGA domain-containing proteins in myxobacteria may be largely confined to the members of the Myxococcus subgroup.
FIG. 5.
Southern hybridization analysis for the presence of carD-like genes in other myxobacteria and in bdellovibrios. (A) XhoI-digested genomic DNAs from M. xanthus (Mx), M. coralloides (Mc), C. fuscus (Cf), S. aurantiaca (Sa), N. exedens (Ne), and P. cellulosum (Pc) probed with a DNA fragment corresponding to nucleotides 354 to 948 of the carD gene. (B) Genomic DNAs from M. xanthus (Mx) and Bdellovibrio sp. strain CP41 (Bd) digested with the indicated restriction enzyme and probed with the same DNA fragment as for panel A.
The bdellovibrios, like myxobacteria, lie within the δ subdivision of the proteobacteria (42). Therefore, we also performed a Southern hybridization analysis with the same probe used above and ClaI- or EcoRI-digested genomic DNA from Bdellovibrio sp. strain CP1. As shown in Fig. 5B, no hybridization signal was detected in this member of the same taxonomic subdivision as the myxobacteria.
Whether bacteria other than myxobacteria contain proteins with HMGA-type domains was further addressed by similarity searches of the nonredundant protein sequence database and of the microbial genome protein database at NCBI (77 complete and 33 partial bacterial genome sequences at the time of this analysis, covering a wide range of taxonomic groups but no myxobacteria), using the gapped BLASTP program. Use of the AT hook segment of CarD or CarDSa alone as the query in a search for short, nearly exact matches in the nonredundant sequence database yielded as best hits eukaryotic HMGA or HMGA-like proteins. We also encountered one hit to a hypothetical protein in the bacterium Ralstonia metallidurans (accession no. ZP_00021411) which has one PGRP sequence, a less frequent core AT hook motif (3). That AT hooks are very rare among bacteria was further confirmed by searching the microbial genome database. Use of the AT hook region of CarD or CarDSa as the search query provided just one hypothetical protein in the bacterium Rhodopseudomonas palustris (accession no. ZP_00008414), which has two closely spaced RGRP sequences and one PGRP motif. However, neither the R. metallidurans hypothetical protein nor that in R. palustris contains a highly acidic region, which invariably lies adjacent to the AT hook segment in HMGA proteins. The SMART database with the AT hook (SMART accession number SM0384) as the query in a domain search of bacterial proteins lists 17 additional proteins. Interestingly, the majority of these are putative DNA-binding proteins, some of which could be implicated in transcriptional control, for example, transcriptional regulators of the TetR type in Caulobacter crescentus (accession no. AAK22321) and Streptomyces coelicolor (accession no. T36295) and of the WhiB type in Mycobacterium tuberculosis (accession no. NP_337818). Among the 17 proteins, a single RGRP AT hook is found in 13, one PGRP motif is found in another, and 2 have the very uncommon AGRP and LGRP motifs (3). The remaining protein has an RGRP AT hook and the very infrequent VGRP motif (Saccharopolyspora erythraea, accession no. AAL78056). Again, in none of these cases is the AT hook portion associated with an adjacent highly acidic region. Thus, based on presently available data, it appears that prokaryotic HMGA-type proteins are largely restricted to myxobacteria.
By contrast, 25 proteins found exclusively in bacteria showed significant similarity to the N-terminal domain of CarD or CarDSa in a search of the NCBI microbial genome protein database. All of these are hypothetical proteins grouped in the conserved-domain protein family pfam02559 (5), whose defining element is the CarD segment between residues 9 and 158. Figure 6 displays the multiple sequence alignment of the N-terminal portions of CarD and CarDSa and the eight proteins showing the highest similarity (sequence identities of 26 to 33%; similarities of 50 to 59%). It should be noted that the similarity to the 180-residue N-terminal segment of CarD and CarDSa extends over the entire length of the primary sequence of the homologs, whose sizes range from 153 to 165 residues, except for the 198-residue Corynebacterium glutamicum protein. Interestingly, a search of M. xanthus sequences in the Cereon microbial genome database allowed us to identify a 164-residue hypothetical protein with 35% identity and 58% similarity to the CarD/CarDSa N terminus, which is thus a new member of the pfam02559 protein family (Fig. 6). These results therefore uncover a distinct prokaryotic domain that exists as an independent module in various bacteria but which is linked to an HMGA-type DNA-binding domain in CarD and CarDSa.
FIG. 6.
Sequence alignment of the 180-residue N-terminal regions of CarDSa, CarD, and the nine bacterial proteins with the highest similarity drawn from the microbial genome sequence database. Accession numbers and total polypeptide lengths are, respectively, JC6146 and 316 residues for M. xanthus CarD; no accession number presently available and 164 residues for M. xanthus; ZP_00061994 and 160 residues for Clostridium thermocellum; NP_657551 and 158 residues for Bacillus anthracis; NP_244803 and 153 residues for Bacillus halodurans; NP_218100 and 162 residues for M. tuberculosis; NP_301347 and 165 residues for Mycobacterium leprae; NP_628406 and 160 residues for Streptomyces coelicolor; NP_601860 and 198 residues for C. glutamicum; and ZP_00005573 and 169 residues for Rhodobacter sphaeroides. The sequence for the 305-residue CarDSa is from this study. Residues are shaded only when they are identical or similar in the majority (≥6) of the 11 aligned sequences described above. Of these, identical residues are shaded black only if they appear in at least six of the sequences. An asterisk in the line corresponding to the consensus indicates identical residues when conserved in all of the aligned sequences.
DISCUSSION
The S. aurantiaca CarD-like protein.
We have identified and characterized CarDSa, a protein in the bacterium S. aurantiaca that is highly similar to the M. xanthus HMGA-type protein CarD. CarDSa would therefore be the second such protein identified in a prokaryote. Like in its M. xanthus counterpart, the HMGA-like domain in CarDSa (composed of a highly acidic region situated toward the N terminus of the adjacent basic AT hooks) spans the final C-terminal segment between residues 180 and 305. In human HMGA1a, by contrast, the acidic region is toward the C terminus of the adjacent AT hook portion. In addition, CarD and CarDSa also share a nearly identical N-terminal region of 180 residues that is not present in HMGA proteins. This segment, which is compact, dimeric, and with a defined structure in CarD, would very likely be so in CarDSa (31). On the other hand, the HMGA-type domain in CarDSa would be expected to lack a defined structure, given its constituent amino acids and their distribution along the sequence, as has been shown for CarD and HMGA1a (20, 31). Our in vitro assays demonstrate that CarDSa binds DNA with a specificity similar to that of CarD, suggesting that the one fewer AT hook in CarDSa and other sequence variations between the two proteins in their HMGA domains may not affect DNA-binding specificity. This is in line with the behavior reported for human HMGA1a, where both the native protein with three AT hooks and a truncated form with only two AT hooks exhibit the same specific DNA binding with nanomolar affinities (11, 20). CarDSa is also phosphorylated in vitro by CKII in its C-terminal region but to a lesser degree than CarD. This is consistent with only one predicted CKII phosphorylation site, S198, being present in its acidic region compared to the five in CarD. Moreover, mutation of this serine to alanine eliminates CKII phosphorylation of CarDSa in vitro. We have found that an M. xanthus strain that stably expresses only the HMGA domain of CarD (residues 179 to 316) showed the same Car− and Fru− phenotype as the strain in which carD is entirely deleted. Thus, even though the CarD HMGA domain exhibits the DNA-binding and CKII phosphorylation properties of the whole protein, the N-terminal domain is essential for CarD function in vivo. Nevertheless, the exact functional role of the highly conserved N-terminal segment that is present in CarDSa and CarD remains to be elucidated. Our sequence comparisons (Fig. 6) highlight several invariant residues that would be obvious targets for future site-directed mutational analysis. One possible role for this domain would be in interacting with other factors to assemble specific transcriptionally competent complexes. This possibility is strengthened by preliminary data from our ongoing examination using the yeast two-hybrid system.
The studies reported here demonstrate that the S. aurantiaca protein CarDSa shares several of the molecular properties of CarD in vitro and can replace the latter in vivo in its dual role in M. xanthus carotenogenesis and fruiting-body formation. This strongly suggests that carDSa is the S. aurantiaca ortholog of carD. The ability of CarDSa to mimic CarD in M. xanthus has also provided two additional insights into the molecular basis for CarD activity. The first is that CarD function is not impeded when there is one fewer AT hook, as in CarDSa. The second is on the significance of in vivo phosphorylation of CarD by a CKII-type kinase. Phosphorylation of eukaryotic HMGA proteins has been linked to their multifunctional roles in vivo, especially in the cell cycle, differentiation, and development (8, 27, 36, 41, 49). For instance, CKII phosphorylation of the acidic region appears to fine-tune the inherent structural plasticity of the randomly structured HMGA proteins, and this, as a consequence, modulates their intracellular stabilities as well as DNA-binding affinities. A similar scenario could have been envisaged for CarD given its role in M. xanthus cellular differentiation and development. However, as shown in this study, phosphorylation of CKII sites in the acidic region of the HMGA domain of CarD does not appear to play such an essential regulatory role, as can be inferred from the observed functional equivalence of CarDSa and its mutant form lacking the only target for CKII phosphorylation. Although CKII-type kinases have not yet been found in M. xanthus, a number of eukaryotic-type serine/threonine protein kinases have been reported, whose substrates remain to be identified (21). Whereas our results do not exclude the possibility that a CKII-type kinase activity exists in M. xanthus, they do suggest that CarD is not an obligatory substrate.
The ability of CarDSa to function in carotenogenesis and fruiting-body formation in M. xanthus suggests that it may be involved in similar roles in its natural context. S. aurantiaca is also capable of developing fruiting bodies, but these are considerably more elaborate than those in M. xanthus (12). Interestingly, efficient fruiting-body formation in S. aurantiaca is driven by light (34). Useful leads in defining the roles that CarDSa plays in these and other processes in S. aurantiaca may come from our comparison with the properties exhibited by M. xanthus CarD.
CarD-like proteins in other bacteria.
The results obtained in this study indicate that proteins containing HMGA-like domains in prokaryotes are found primarily in myxobacteria (order Myxococcales). In particular, such proteins appear to be confined to the suborder Cystobacterineae, which includes M. xanthus, S. aurantiaca, M. coralloides, and C. fuscus. We also have no evidence for the presence of HMGA proteins in the bdellovibrios, which belong to the taxonomic group closest to myxobacteria in the δ subdivision of the proteobacteria. Our analysis of the many partial or complete available genome sequences for bacteria from other taxonomic groups yielded 19 proteins which have AT hook sequences. One of these has three AT hooks, one has two AT hooks, and the rest have no more than a single AT hook. However, a flanking highly acidic region, which is a hallmark of HMGA domains, is not apparent in any of these proteins. That proteins containing HMGA domains appear to be exclusive for one specific suborder of myxobacteria suggests two possible alternatives for the evolutionary origins of carD in myxobacteria: a de novo occurrence in the specific phylogenetic subgroup of myxobacteria or a consequence of horizontal acquisition of a eukaryotic HMGA-type gene. Nevertheless, when a protein is widely prevalent in eukaryotes but occurs in only one or very few bacterial species, horizontal transfer is the most parsimonious explanation (24). Horizontal gene transfer into the myxobacteria may be linked to their particular lifestyles, i.e., choice of habitat, feeding, and/or social behavior. In fact, the hypothesis that feeding habits may account for lateral transfer (13) has been invoked to account for the particular organization of the gene encoding the β-1-4-endoglucanase in M. xanthus and S. aurantiaca, both of which are scavengers that prey on other soil microorganisms (35). Moreover, myxobacteria have generally been considered to be phylogenetically distinct and an evolutionarily advanced group of bacteria (42). The formation of fruiting bodies and spores in myxobacteria represent rather complex developmental and cellular differentiation cycles that involve elaborate signal transduction and gene regulatory cascades. These processes, which are generally not observed with most other bacteria, are more akin to those occurring in eukaryotes. It is therefore not surprising that a number of protein factors typically found in eukaryotes have analogs in myxobacteria. In M. xanthus these include, besides CarD, the serine/threonine protein kinases referred to above and at least one associated phosphatase (45) and CarF, a recently identified regulatory protein in carotenogenesis (15).
Our comparative sequence analysis of CarD and CarDSa revealed homologs to the N-terminal non-HMGA domain, but these were all entirely prokaryotic in origin. CarD and CarDSa could therefore exemplify interkingdom domain fusion between a preexisting bacterial domain and an acquired eukaryotic domain, a phenomenon that is particularly common in Actinomycetes (24). Furthermore, the additional presence of a distinct stand-alone version of the prokaryotic domain in M. xanthus is consistent with a probable two-step evolutionary process for the origin of CarD and CarDSa, that is, capture of the eukaryotic HMGA portion with subsequent fusion to the bacterial N-terminal part by recombination (24). Consistent with this two-domain makeup is our analysis of CarD, which revealed that the N-terminal and HMGA-type C-terminal parts are structurally distinct modules (31). Any selective advantage conferred by this particular domain organization would necessarily be speculative. The GC-rich nature of most myxobacteria would clearly narrow down the search for the specific AT-rich DNA-binding sites by the HMGA domain. Any speculation with regard to the N-terminal domain, on the other hand, must await future work aimed at identifying the exact nature of the part it plays in CarD/CarDSa functions. At least for CarD, as we have shown above, the association of the N-terminal region with the HMGA domain is an essential functional requirement in carotenogenesis and multicellular development.
Acknowledgments
We thank H. U. Schairer (University of Heidelberg) for the generous gift of the phage λDASH and λgt11 S. aurantiaca DNA library, Victoriano Garre for technical advice with the library screening experiments, and J. A. Madrid for technical assistance. Christophe Hoor contributed to the construction of the carD deletion strain. We are grateful to F. Torrella and A. Sánchez-Amat (University of Murcia) for some of the bacterial species used in this work and to David Hodgson (University of Warwick, Warwick, United Kingdom) for plasmids pDAH217 and pDAH231. We are also grateful to the Monsanto Company for access to the M. xanthus sequence database (now available at the TIGR microbial database). We thank the anonymous referees for useful comments.
This work was supported by the Spanish Ministerio de Ciencia y Tecnología (grant BMC2000-1006 to F.J.M. and Programa Ramón y Cajal to S.P.), Ministerio de Educación y Cultura (fellowship to M.L.C.), and Fundación Séneca (fellowship to M.P.-M.).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appleyard, R. K. 1954. Segregation of new lysogenic types during growth of a doubly lysogenic strain derived from Escherichia coli K12. Genetics 39:440-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aravind, L., and D. Landsman. 1998. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 26:4413-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balsalobre, J. M., R. M. Ruiz-Vázquez, and F. J. Murillo. 1987. Light induction of gene expression in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 84:2359-2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bateman, A., E. Birney, E., L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bibb, M. J., P. R. Findlay, and M. W. Johnson. 1984. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene 30:157-166. [DOI] [PubMed] [Google Scholar]
- 7.Bretscher, A. P., and D. Kaiser. 1978. Nutrition of Myxococcus xanthus, a fruiting myxobacterium. J. Bacteriol. 133:763-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bustin, M. 1999. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol. 19:5237-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bustin, M. 2001. Revised nomenclature for high mobility group (HMG) chromosomal proteins. Trends Biochem. Sci. 26:152-153. [DOI] [PubMed] [Google Scholar]
- 10.Bustin, M., and R. Reeves. 1996. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog. Nucleic Acid Res. Mol. Biol. 54:35-100. [DOI] [PubMed] [Google Scholar]
- 11.Claus, P., E. Schulze, and J. R. Wisniewski. 1994. Insect proteins homologous to mammalian high mobility group proteins I/Y (HMG I/Y). J. Biol. Chem. 269:33042-33048. [PubMed] [Google Scholar]
- 12.Dawid, W. 2000. Biology and global distribution of myxobacteria in soils. FEMS Microbiol. Rev. 24:403-427. [DOI] [PubMed] [Google Scholar]
- 13.Doolittle, W. F. 1998. You are what you eat: a gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet. 14:307-311. [DOI] [PubMed] [Google Scholar]
- 14.Ferranti, P., A. Malorni, G. Marino, P. Pucci, G. H. Goodwin, G. Manfioletti, and V. Giancotti. 1992. Mass spectrometric analysis of the HMGY protein from Lewis lung carcinoma. Identification of phosphorylation sites. J. Biol. Chem. 267:22486-22489. [PubMed] [Google Scholar]
- 15.Fontes, M., L. Galbis-Martínez, and F. J. Murillo. 2003. A novel regulatory gene for light-induced carotenoid synthesis in the bacterium Myxococcus xanthus. Mol. Microbiol. 47:561-571. [DOI] [PubMed] [Google Scholar]
- 16.Grosschedl, R. K., K. Giese, and J. Pagel. 1994. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 10:94-100. [DOI] [PubMed] [Google Scholar]
- 17.Hagen, D. C., A. P. Bretscher, and D. Kaiser. 1978. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev. Biol. 64:284-296. [DOI] [PubMed] [Google Scholar]
- 18.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 19.Hodgson, D. A. 1993. Light-induced carotenogenesis in Myxococcus xanthus: genetic analysis of the carR region. Mol. Microbiol. 7:471-488. [DOI] [PubMed] [Google Scholar]
- 20.Huth, J. R., C. A. Bewley, M. S. Nissen, J. N. S. Evans, R. Reeves, A. M. Gronenborn, and G. M. Clore. 1997. The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif. Nat. Struct. Biol. 4:657-665. [DOI] [PubMed] [Google Scholar]
- 21.Inouye, S., R. Jain, T. Ueki, H. Nariya, C. Y. Xu, M. Y. Hsu, B. A. Fernández-Luque, J. Muñoz-Dorado, E. Farez-Vidal, and M. Inouye. 2000. A large family of eukaryotic-like protein Ser/Thr kinases of Myxococcus xanthus, a developmental bacterium. Microb. Comp. Genomics 5:103-120. [DOI] [PubMed] [Google Scholar]
- 22.Julien, B., D. Kaiser, and A. Garza. 2000. Spatial control of cell differentiation in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 97:9098-9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaiser, D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 76:5952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koonin, E., K. S. Makarova, and L. Aravind. 2001. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 55:709-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maher, J. F., and D. Nathans. 1996. Multivalent DNA-binding properties of the HMG-I proteins. Proc. Natl. Acad. Sci. USA 93:6716-6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGowan, S. J., H. C. Gorham, and D. A. Hodgson. 1993. Light-induced carotenogenesis in Myxococcus xanthus: DNA sequence analysis of the carR region. Mol. Microbiol. 10:713-735. [DOI] [PubMed] [Google Scholar]
- 27.Merika, M., and D. Thanos. 2001. Enhanceosomes. Curr. Opin. Gen. Dev. 11:205-208. [DOI] [PubMed] [Google Scholar]
- 28.Nicolás, F. J., M. L. Cayuela, I. M. Martínez-Argudo, R. M. Ruiz-Vázquez, and F. J. Murillo. 1996. High mobility group I(Y)-like DNA-binding domains on a bacterial transcription factor. Proc. Natl. Acad. Sci. USA 93:6881-6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicolás, F. J., R. M. Ruiz-Vázquez, and F. J. Murillo. 1994. A genetic link between light response and multicellular development in the bacterium Myxococcus xanthus. Genes Dev. 8:2375-2387. [DOI] [PubMed] [Google Scholar]
- 30.Pace, C. N., F. Vajdos, L. Fee, G. Grimsley, and T. Gray. 1995. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4:2411-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Padmanabhan, S., M. Elías-Arnanz, E. Carpio, P. Aparicio, and F. J. Murillo. 2001. Domain architecture of a high mobility group A-type bacterial transcriptional factor. J. Biol. Chem. 276:41566-41575. [DOI] [PubMed] [Google Scholar]
- 32.Palvimo, J., and A. Linnala-Kankkunen. 1989. Identification of sites on chromosomal protein HMG-I phosphorylated by casein kinase II. FEBS Lett. 257:101-104. [DOI] [PubMed] [Google Scholar]
- 33.Qualls, G. T., K. Stephens, and D. White. 1978. Morphogenetic movements and multicellular development in the fruiting myxobacterium, Stigmatella aurantiaca. Dev. Biol. 66:270-274. [DOI] [PubMed] [Google Scholar]
- 34.Qualls, G. T., K. Stephens, and D. White. 1978. Light-stimulated morphogenesis in the fruiting myxobacterium Stigmatella aurantiaca. Science 201:444-445. [DOI] [PubMed] [Google Scholar]
- 35.Quillet, L., S. Barray, B. Labedan, F. Petit, and J. Guespin-Michel. 1995. The gene encoding the β-1-4 endoglucanase (CelA) from Myxococcus xanthus: evidence for independent acquisition by horizontal transfer of binding and catalytic domains from actinomycetes. Gene 158:23-29. [DOI] [PubMed] [Google Scholar]
- 36.Reeves, R., T. A. Langan, and M. Nissen. 1991. Phosphorylation by cdc2 kinase modulates DNA binding activity of high mobility group I nonhistone chromatin protein. Proc. Natl. Acad. Sci. USA 88:1671-1675.2000376 [Google Scholar]
- 37.Ruiz-Vázquez, R. M., and F. J. Murillo. 1984. Abnormal motility and fruiting behavior of Myxococcus xanthus bacteriophage-resistant strains induced by a clear-plaque mutant of bacteriophage Mx8. J. Bacteriol. 160:818-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 39.Sánchez-Amat, A., and F. Torrella. 1990. Formation of stable bdelloplasts as a starvation survival strategy of marine bdellovibrios. Appl. Environ. Microbiol. 56:2717-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schairer, H. U. 1993. Stigmatella aurantiaca, an organism for studying the genetic determination of morphogenesis, p. 333-346. In M. Dworkin and D. Kaiser (ed.), Myxobacteria II. American Society for Microbiology, Washington, D.C.
- 41.Schwanbeck, R., M. Gymnopoulos, I. Petry, A. Piekielko, Z. Szewczuk, T. Heyduk, K. Zechel, and J. R. Wisniewski. 2001. Consecutive steps of phosphorylation affect conformation and DNA binding of the chironomus high mobility group A protein. J. Biol. Chem. 276:26012-26021. [DOI] [PubMed] [Google Scholar]
- 42.Shimkets, L., and C. R. Woese. 1992. A phylogenetic analysis of the myxobacteria; basis for their classification. Proc. Natl. Acad. Sci. USA 89:9459-9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Solomon, M. J., F. Strauss, and A. Varshavsky. 1986. A mammalian high mobility group protein recognizes any stretch of six AT base pairs in duplex DNA. Proc. Natl. Acad. Sci. USA 83:1276-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 45.Treuner-Lange, A., M. J. Ward, and D. R. Zusman. 2001. Pph1 from Myxococcus xanthus is a protein phosphatase involved in vegetative growth and development. Mol. Microbiol. 40:126-140. [DOI] [PubMed] [Google Scholar]
- 46.Ueki, T., S. Inouye, and M. Inouye. 1996. Positive-negative KG cassettes for construction of multi-gene deletions using a single drug marker. Gene 183:153-157. [DOI] [PubMed] [Google Scholar]
- 47.Wang, D.-Z., P. Ray, and M. Boothby. 1995. Interleukin 4-inducible phosphorylation of HMG-I(Y) is inhibited by rapamycin. J. Biol. Chem. 270:22924-22932. [DOI] [PubMed] [Google Scholar]
- 48.Werner, M. H., and S. K. Burley. 1997. Architectural transcription factors: proteins that remodel DNA. Cell 88:733-736. [DOI] [PubMed] [Google Scholar]
- 49.Wisniewski, J. R., Z. Szewczuk, I. Petry, R. Schwanbeck, and U. Renner. 1999. Constitutive phosphorylation of the acidic tails of the high mobility group I proteins by casein kinase II alters their conformation, stability, and DNA binding specificity. J. Biol. Chem. 274:20116-20122. [PubMed] [Google Scholar]
- 50.Yie, J., S. Liang, M. Merika, and D. Thanos. 1997. Intra- and intermolecular cooperative binding of high-mobility-group protein I(Y) to the beta-interferon promoter. Mol. Cell. Biol. 17:3649-3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Young, R. A., and R. W. Davis. 1983. Yeast RNA polymerase II genes: isolation with antibody probes. Science 222:778-782. [DOI] [PubMed] [Google Scholar]