Abstract
Plasmid pAADB1 for the overexpression of the alcohol-aldehyde dehydrogenase (aad) gene and downregulation of the coenzyme A transferase (CoAT) using antisense RNA (asRNA) against ctfB (the second CoAT gene on the polycistronic aad-ctfA-ctfB message) was used in order to increase the butanol/acetone ratio of Clostridium acetobutylicum ATCC 824 fermentations. Acetone and butanol levels were drastically reduced in 824(pCTFB1AS) (expresses only an asRNA against ctfB) compared to 824(pSOS95del) (plasmid control). Compared to strain 824(pCTFB1AS), 824(pAADB1) fermentations exhibited two profound differences. First, butanol levels were ca. 2.8-fold higher in 824(pAADB1) and restored back to plasmid control levels, thus supporting the hypothesis that asRNA downregulation of ctfB leads to degradation of the whole aad-ctfA-ctfB transcript. Second, ethanol titers in 824(pAADB1) were ca. 23-fold higher and the highest (ca. 200 mM) ever reported in C. acetobutylicum. Western blot analysis confirmed that CoAT was downregulated in 824(pAADB1) at nearly the same levels as in strain 824(pCTFB1AS). Butyrate depletion in 824(pAADB1) fermentations suggested that butyryl-CoA was limiting butanol production in 824(pAADB1). This was confirmed by exogenously adding butyric acid to 824(pAADB1) fermentations to increase the butanol/ethanol ratio. DNA microarray analysis showed that aad overexpression profoundly affects the large-scale transcriptional program of the cells. Several classes of genes were differentially expressed [strain 824(pAADB1) versus strain 824(pCTFB1AS)], including genes of the stress response, sporulation, and chemotaxis. The expression patterns of the CoAT genes (ctfA and ctfB) and aad were consistent with the overexpression of aad and asRNA downregulation of ctfB.
Metabolic engineering of Clostridium acetobutylicum, a gram-positive, spore-forming, obligate anaerobe, aims to redirect carbon and energy fluxes for improved solvent production or to impart desirable cellular traits. Our laboratory has been investigating antisense RNA (asRNA) as both an investigative and metabolic engineering tool. Generally, asRNA binds target RNA and prevents RNA translation by hindering ribosome-binding site interactions with the translational machinery of the cell (i.e., ribosomes) or by altering the structure of the target RNA such that ribonucleases can then degrade the target RNA (23). Desai and Papoutsakis (6) were the first to examine the effectiveness of asRNA for the metabolic engineering of C. acetobutylicum (6). More recently, we have reported (22) the effect of asRNA structural properties on downregulation efficacy in C. acetobutylicum. Using computational algorithms, three different asRNAs directed toward acetoacetate decarboxylase, an enzyme in the acetone formation pathway, along with previously reported clostridial asRNAs, were examined for structural features (free nucleotides and components). Free nucleotides are defined as nucleotides in an asRNA molecule that are not involved in intramolecular bonding and thus are thought to provide potential sites with which the asRNA and target mRNA might interact (18). Components are structural features that contain regions of high complementarity within an asRNA molecule. When normalized metrics of these structural features were plotted against the percent downregulation, only the component/nucleotide ratio correlated well with in vivo asRNA effectiveness, which suggested that this ratio could be used as a selection parameter for the design of future asRNA molecules. Despite significant downregulation of acetoacetate decarboxylase in these strains, there were no concomitant effects on acetone formation and, therefore, using the component/nucleotide ratio as a design parameter, we next targeted the coenzyme A transferase (CoAT), the first enzyme of the acetone formation pathway. Compared to the plasmid control strain, the strains expressing asRNA against CoAT (CoAT-asRNA) produced substantially lower levels of acetone and CoAT. Interestingly, butanol levels in CoAT-asRNA-expressing strains were also significantly decreased, suggesting that expression of the gene responsible for butanol production, alcohol-aldehyde dehydrogenase (aad), might also be altered by CoAT-asRNA. This is not surprising in view of the fact that the aad resides on the same polycistronic message as ctfA and ctfB. These results suggested that the asRNA in these strains is resulting in partial degradation of the tricistronic mRNA.
To overcome this likely degradation of aad mRNA in 824(pCTFB1AS), we hypothesized that overexpressing aad, while still suppressing acetone formation with the asRNA produced by pCTFB1AS (ctfb1-asRNA), will increase butanol production substantially compared to 824(pCTFB1AS) and thereby increase the butanol/acetone ratio of C. acetobutylicum. Here we report the construction of vector pAADB1, which combines the ctfb1-asRNA cassette of pCTFB1AS with aad overexpression. To evaluate the effectiveness of this approach, we performed pH controlled bioreactor experiments with strains 824(pAADB1), 824(pSOS95del) (the plasmid control strain), and 824(pCTFB1AS). Detailed product formation, metabolic flux, Western, and microarray analyses of these strains were used to characterize the effects of ctfb1-asRNA by itself and in combination with aad overexpression on the C. acetobutylicum metabolism and transcriptome.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in the present study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Sourceb or reference |
|---|---|---|
| Strains | ||
| E. coli ER2275 | NEB | |
| E. coli OneShot chemically competent Top10 | Invitrogen | |
| C. acetobutylicum ATCC 824 | ATCC | |
| Plasmids | ||
| pAN1 | Cmr; carries the Φ3T I gene | 15 |
| pTAAD | Ampr Tetr; aad | 10 |
| pSOS95del | Ampr Mlsr; repL; ColE1; thl promoter | 22 |
| pCTFB1AS | Ampr Mlsr; repL; ColE1; thl promoter; antisense ctfb1 | 22 |
| pAADB1 | Ampr Mlsr; repL; ColE1; thl promoter; antisense ctfb1; aad | This study |
Cmr, chloramphenicol resistant, B. subtilis phage Φ3T I methyltransferase gene; Ampr, ampicillin resistant; Tetr, tetracycline resistant; aad, alcohol-aldehyde dehydrogenase gene; Mlsr, macrolide-lincosamide-streptogramin B resistant; repL, pIM13 origin of replication; ColE1, ColE1 origin of replication; th1 promoter, promoter region for the thiolase gene (thl) of C. acetobutylicum ATCC 824; ctfb1, DNA fragment containing approximately 39% of the ctfB structural gene.
NEB, New England Biolabs (Beverly, Mass.); Invitrogen, Invitrogen Corporation (Carlsbad, Calif.); ATCC, American Type Culture Collection (Manassas, Va.).
Plasmid DNA isolation.
An alkaline lysis method was used for plasmid isolation from Escherichia coli strains (14). For plasmid isolation from recombinant C. acetobutylicum strains, another alkaline lysis method was used (10).
DNA manipulation.
All commercial enzymes utilized in the present study (restriction enzymes, T4 DNA ligase, calf intestinal alkaline phosphatase, and Vent DNA polymerase) were used under supplier-recommended conditions. DNA fragments were isolated from agarose gels by using the GFX PCR DNA and gel band purification kit (Amersham Pharmacia Biotech, Piscataway, N.J.).
Construction of pAADB1.
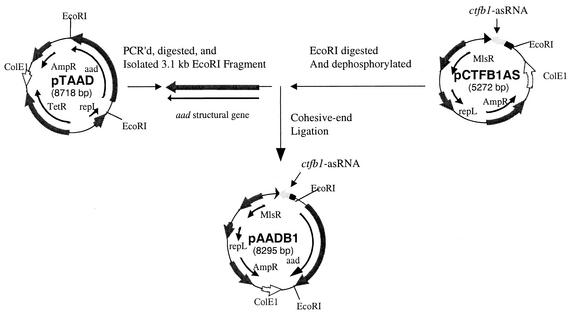
To combine asRNA and gene overexpression technologies, we constructed a vector that combined ctfb1-asRNA (22) and aad overexpression in one plasmid: pAADB1. We first PCR amplified aad with terminal EcoRI sites, along with its promoter (20), by using the primers UPECOAAD (5′-TTCTAAATATACTGAGAATTCCTAAATA-3′) and DSECOAAD (5′-CTAATTATTTTAGAATTCATTTTAATCC-3′) and pTAAD (10) as a template DNA. After EcoRI digestion, the 3.1-kb PCR fragment containing aad and its promoter was cohesive-end ligated to EcoRI-digested, dephosphorylated pCTFB1AS (22) to form pAADB1 (Fig. 1).
FIG. 1.
Construction of pAADB1. Arrows indicate the locations and directions of transcription of relevant genes and relevant restriction sites are marked. Abbreviations: MlsR, macrolide-lincosamide-streptogramin B resistance gene; repL, pIM13 origin of replication; AmpR, ampicillin resistance gene; TetR, tetracycline resistance gene; aad, alcohol-aldehyde dehydrogenase gene; ColE1, ColE1 origin of replication. Genes and plasmids are not drawn to scale.
Cell transformation.
Plasmid pAADB1 was constructed in E. coli and then transformed into C. acetobutylicum. Transformation with ligation mixtures was done by using E. coli Top10 OneShot competent cells from Invitrogen Corporation (Carlsbad, Calif.). E. coli(pAN1) and C. acetobutylicum were electrotransformed as described previously (15, 19).
Growth and maintenance of strains.
Maintenance and bioreactor experiments of all strains were carried out as described previously (22) except for each sample, RNA samples were also collected for microarray analysis. For all growth experiments, growth was monitored by measuring the absorbance at 600 nm (A600) of appropriate dilutions with a BioMate3 spectrophotometer (Thermo Spectronic, Rochester, N.Y.). For RNA sampling, 15 ml of culture suspension at an A600 of <0.5, 10 ml for samples with an A600 of 0.5 to 2.0, and 5 ml for samples with an A600 of >2.0 were centrifuged at 4,000 × g for 15 min at 4°C. After the supernatants were removed, the cell pellets were resuspended in 200 μl of SET+lysozyme (20 mg/ml) buffer (11) and incubated at 37°C for 5 min. Upon addition of 1 ml of TRIzol (Invitrogen), the lysed cells were vortexed in the TRIzol for ca. 15 s and then stored at −85°C.
For butyrate addition bioreactor experiments, when the butyrate concentrations, as determined by high-pressure liquid chromatography (HPLC), began to decrease to ca. 20 to 40 mM, filter-sterilized n-butyric acid (Sigma Chemical Co., St. Louis, Mo.) was added to increase the final concentration of butyrate in the bioreactor to the desired concentration.
Product analysis.
Culture supernatants were analyzed for product concentrations as described previously (3) by using a Waters (Milford, Mass.) HPLC system (model no. 717 plus autosampler, 1515 HPLC pump, 2410 refractive index detector, and in-line vacuum degasser) and Water's Breeze software. The standard error of the mean for product concentrations was determined from replicate analyses.
Western blot analysis.
Crude extract preparation, immunoblotting, and quantification of CoAT subunit levels were performed as described previously (22). Briefly, 5 μg of total protein for each sample, as determined by the RC DC protein assay (Bio-Rad Laboratories, Hercules, Calif.), was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis with Ready-Gels (12% Tris-HCl polyacrylamide, 4% stacking) from Bio-Rad Laboratories. The MagicMark Western protein standard (Invitrogen) was used as the protein standard for all blots. After electrophoresis, proteins were transferred to 0.45-μm-pore-size Hybond-P polyvinylidene difluoride membranes (Amersham Pharmacia Biotech). Membranes were then blocked with Tris-buffered saline plus Tween (TBST) plus 4% enhanced chemiluminescence (ECL) blocking reagent (Amersham Pharmacia Biotech) and hybridized to anti-CoAT immunoglobulin G at a 1:10,000 dilution in TBST plus 4% ECL blocking reagent (4). Donkey anti-sheep IgG (Sigma) conjugated with horseradish peroxidase (1:10,000 dilution in TBST plus 4% ECL blocking reagent) was then used as a secondary antibody. The fluorescence on each membrane was generated according to the instructions of the ECL-Plus kit (Amersham Pharmacia Biotech) and detected by using a Storm 860 Imager (Molecular Dynamics, Sunnyvale, Calif.). ImageQuant version 5.0 (Molecular Dynamics) was then used to view and quantify Western blots. The percent downregulation by asRNA was determined by calculating the percent decrease in gel band intensity of the protein of interest in the asRNA-producing strain with respect to the protein of interest's gel band intensity in the plasmid control strain for each growth phase. The standard error of the mean for the percent downregulation was determined from replicate (two to four) Western blots and represents the cumulative error for the total Western analysis assay.
Metabolic flux analysis.
Metabolic flux analysis involves the calculation of specific in vivo fluxes (measured in millimoles hour−1 g of cell−1) from substrate and product data by using a system of linear equations developed from metabolic reaction stoichiometry (17) and the CoAT constraint (5). The most relevant pathway fluxes that are considered in this model are as follows: rGLY1, pyruvate formation via the glycolytic pathway; rGLY2, conversion of pyruvate to acetyl-CoA; rBUOH, butanol formation; rETOH, ethanol formation; rPTAAK, acetate formation; rPTBBK, butyrate formation; rBYUP, butyrate uptake via the acetone formation pathway; rACUP, acetate uptake via the acetone formation pathway; and rACETONE, acetone formation that represents the sum of rACUP and rBYUP. The calculated time course profiles were examined to reveal the time-dependent patterns of metabolic activity. For these analyses, the fermentation time was rescaled to account for variable lag phases. The normalized scale, TN, is set such that TN = 0 h at A600 = 1.0. The uncertainties in the calculated fluxes are approximately as follows: rPTBBK, 10%; rPTAAK, 15%; rACUP, 8%; rBYUP, 6%; rETOH, 10%; rBUOH, 10%; rGLY1, 15%; and rGLY2, 15% (10).
Microarray analysis.
RNA isolation and purification, cDNA labeling and hybridization, microarray data analysis, and printing of cDNA microarrays were performed as described previously (21). Briefly, labeled cDNA was synthesized by random hexamer-primer reverse transcription reactions in the presence of Cy3-dUTP or Cy5-dUTP by using SuperScript II reverse transcriptase (Invitrogen) and purified RNA (12 μg for exponential phase samples and 15 μg for stationary-phase samples) from eight time points ranging from the early exponential phase to the stationary phase of both 824(pCTFB1AS) and 824(pAADB1) bioreactor experiments (Fig. 2B). For hybridizations, 5-μl portions of oppositely labeled probes were mixed with sonicated salmon sperm and a 2× hybridization volume (10× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 50% formamide, 0.2% sodium dodecyl sulfate) and then loaded onto the array. In the present study, cDNA microarrays were used that were printed by using the protocol of The Institute for Genomic Research (12), with spots representing 1,019 open reading frames, approximately one-fourth of the C. acetobutylicum genome, onto TeleChem (Sunnyvale, Calif.) ArrayIt Brand SuperAmine glass microarray slides. After hybridization for 18 h at 42°C in CMT hybridization chambers (Dow Corning, Corning, N.Y.), arrays were washed with TeleChem ArrayIt DNA microarray wash buffers A, B, and C according to the manufacturer's instructions. The hybridized arrays were dried and then analyzed with a GSI Lumonics scanner and ScanArray software (Perkin-Elmer Life Sciences, Boston, Mass.). Spot intensities were quantitated with QuantArray microarray analysis software (Perkin-Elmer Life Sciences). The data were normalized, and genes that showed significant differences in expression levels were determined by using a normalization and gene selection method described previously (24) and then cleaned by using principal component analysis (1). Gene clusters were visualized in TreeView (7).
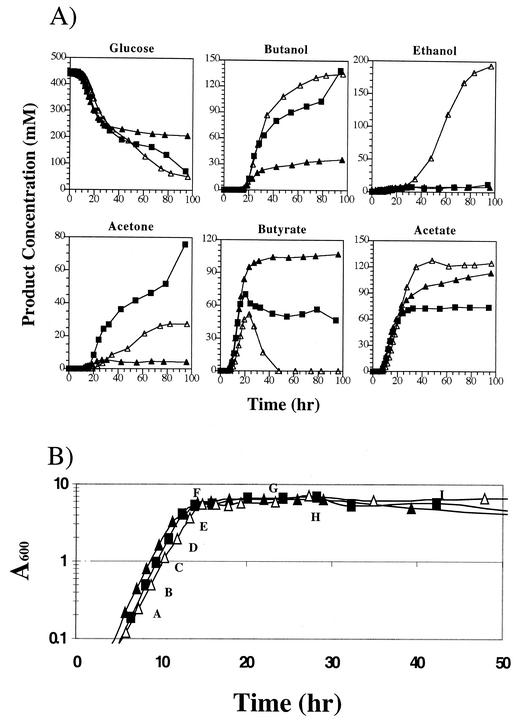
FIG. 2.
Fermentation kinetics of 824(pSOS95del), 824(pCTFB1AS), and 824(pAADB1). (A) Glucose, acid, and solvent profiles. The name of each profile in 824(pSOS95del) (▪), 824(pCTFB1AS) (▴), and 824(pAADB1) (▵) is indicated above each graph. (B) Growth curves for 824(pSOS95del) (▪), 824(pCTFB1AS) (▴), and 824(pAADB1) (▵). Sampling points for microarray analysis are labeled A through H. For Western blot analysis, samples F (transitional), H (early stationary), and I (stationary) were used.
RESULTS
Product formation in fermentations of strains 824(pAADB1), 824(pCTFB1AS), and 824(pSOS95del).
The characteristics of the C. acetobutylicum ATCC 824(pAADB1) fermentations along with those of the two control strains, 824(pSOS95del) (plasmid control) and 824(pCTFB1AS) (ctfb1-asRNA expression only) are summarized in Fig. 2. Glucose consumption for all three strains looks very similar until ca. 30 h at which point the glucose levels of 824(pSOS95del) and 824(pAADB1) continue to decrease to ca. 60 mM, while the glucose levels of 824(pCTFB1AS) decrease only to ca. 200 mM. As expected (22), acetone and butanol levels were drastically reduced and acetate and butyrate levels were dramatically increased in 824(pCTFB1AS) compared to 824(pSOS95del). When compared to 824(pCTFB1AS), 824(pAADB1) fermentations exhibited two profound differences. First, butanol levels are ca. 2.8-fold higher in 824(pAADB1) and restored back to plasmid control levels. This finding validates the hypothesis that aad overexpression combined with ctfb1-asRNA will result in dramatically higher butanol formation than with ctfb1-asRNA expression alone and further supports the suggestion (22) that partial degradation of aad mRNA by ctfb1-asRNA results in the low butanol production of 824(pCTFB1AS). Second, final ethanol titers were ∼23-fold higher in 824(pAADB1) and are the highest levels of ethanol ever reported in C. acetobutylicum, exceeding previously reported levels (10) by ca. 100%. Acetone levels increased in 824(pAADB1) by ∼4.6-fold compared to 824(pCTFB1AS) but were still ca. 63% lower than those seen in 824(pSOS95del), which resulted in the butanol/acetone ratios of 824(pSOS95del) (1.83 ± 0.05) and 824(pAADB1) (4.89 ± 0.29); these were 71 and 25%, respectively, lower than the butanol/acetone ratio of 824(pCTFB1AS) (6.48 ± 0.87). The peak butyrate concentrations in 824(pAADB1) were ca. 95 and 21% lower than those in 824(pCTFB1AS) and 824(pSOS95del), respectively. A more drastic difference can be seen in final butyrate concentration, for which no detectable levels of butyrate can be seen in 824(pAADB1), whereas both 824(pCTFB1AS) and 824(pSOS95del) have final butyrate concentrations of at least 47 mM. Peak and final acetate levels in 824(pAADB1) were also higher (ca. 11 and 68%) compared to 824(pCTFB1AS) and 824(pSOS95del), respectively. Thus, it appears that aad overexpression, in conjunction with ctfb1-asRNA expression, greatly modifies the metabolism of C. acetobutylicum compared to ctfb1-asRNA expression alone. The significant change in metabolism is best exemplified by the dramatically higher butanol and ethanol formation in 824(pAADB1) compared to 824(pCTFB1AS).
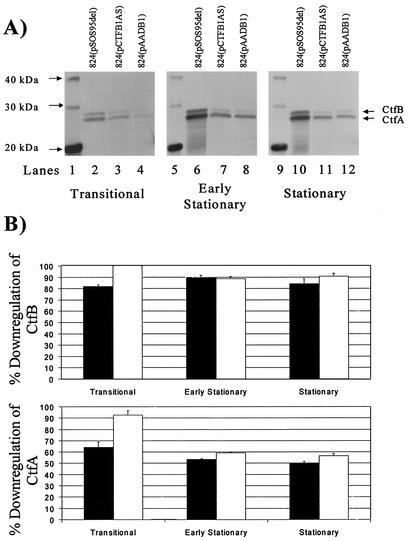
Western blot analysis of 824(pAADB1), 824(pCTFB1AS), and 824(pSOS95del).
To verify that the ctfb1-asRNA downregulated CoAT in 824(pAADB1) relative to the plasmid control strain, we carried out Western blot analyses of samples from the transitional point between the exponential and stationary phases (referred to here as the transitional phase), from the early stationary phase, and from the stationary phase of fermentations of each strain (Fig. 2B), and typical Western blots are shown in Fig. 3A. A summary of the quantitation results from these Western blots is shown in Fig. 3B. In all three phases, 824(pCTFB1AS) and 824(pAADB1) exhibited significant downregulation of both the CtfA and CtfB subunits compared to the plasmid control strain. In addition, 824(pAADB1) exhibited lower CtfA and CtfB levels compared to 824(pCTFB1AS) in the transitional phase. For CtfA, the trend of increased downregulation in 824(pAADB1) compared to 824(pCTFB1AS) appears to continue into both the early stationary and the stationary phases. Since the promoter of aad is used to overexpress aad in pAADB1, as well as to naturally express the sol operon transcript (which contains ctfA and ctfB), the increased downregulation of CoAT subunits in 824(pAADB1) is probably caused by a decrease in sol operon transcription due to dilution of transcriptional regulators by the promoter of aad on pAADB1.
FIG. 3.
Western blot analysis of CoAT. (A) Examples of CoAT Western blots from the transitional, early stationary, and stationary phases of fermentations of 824(pSOS95del), 824(pCTFB1AS), and 824(pAADB1). The CtfA and CtfB subunits of CoAT, as well as the closest marker bands, are indicated for all blots. The culture phases of the samples used on each blot are indicated below the blots. Lanes 1, 4, and 9, protein standard; lanes 2, 6, and 10, 824(pSOS95del); lanes 3, 7, and 11, 824(pCTFB1AS); lanes 4, 8, and 12, 824(pAADB1). (B) Percent downregulation of CtfA and CtfB subunits of CoAT by strains expressing CoAT-asRNA. The percent downregulation values of CtfB and CtfA in 824(pCTFB1AS) (▪) and 824(pAADB1) (□) were calculated as the percent decrease in each subunit's gel band intensity in the asRNA expressing strain compared to the plasmid control strain [824(pSOS95del)] at the same culture phase. The standard error of each measurement was calculated from two to four different blots.
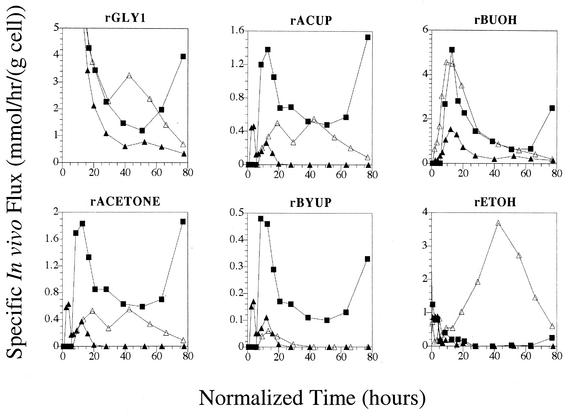
Metabolic flux analysis in strains 824(pAADB1), 824(pCTFB1AS), and 824(pSOS95del).
For a more detailed analysis of the alteration of metabolism by overexpressing aad and ctfb1-asRNA in 824(pAADB1), we performed a metabolic flux analysis of the fermentation data from each strain (Fig. 4). A major difference in all three strains is seen in their uptake of glucose. The flux that corresponds to the formation of pyruvate from glucose, rGLY1, is relatively the same for both 824(pSOS95del) and 824(pAADB1) until ca. TN = 28 h (TN = 0 is between 9 and 12 h of real fermentation time) at which point the flux for 824(pAADB1) increases until ca. TN = 42 h and then begins a gradual decrease throughout the rest of the fermentation. In contrast, rGLY1 for 824(pSOS95del) decreases until ca. TN = 51 h and then increases throughout the rest of the fermentation process. This suggests that 824(pAADB1) is better able to uptake glucose in the middle of fermentations, whereas 824(pSOS95del) is better suited for utilizing glucose in the later stages of the fermentation. In 824(pCTFB1AS), the rGLY1 flux is significantly lower than the fluxes of both 824(pAADB1) and 824(pSOS95del) from ca. TN = 15 h throughout the rest of the fermentation, which suggests that this strain is not able to utilize glucose as well as the two other strains. These same trends are also seen (data not shown) in rGLY2, the flux from pyruvate to acetyl-CoA.
FIG. 4.
Metabolic flux analysis of 824(pSOS95del), 824(pCTFB1AS), and 824(pAADB1). The flux for each profile of 824(pSOS95del) (▪), 824(pCTFB1AS) (▴), and 824(pAADB1) (▵) is indicated above each graph.
The acetone formation flux, rACETONE, for 824(pAADB1) is considerably higher than that for 824(pCTFB1AS) but still far lower than the flux for 824(pSOS95del). In addition, the shape of the rACETONE profile for 824(pCTFB1AS) is strikingly different compared to 824(pSOS95del) and 824(pAADB1). 824(pCTFB1AS)'s rACETONE flux has a peak at ca. TN = 5 h and a second peak at ca. TN = 12 h and then decreases and becomes zero at TN = 20 h. On the other hand, both 824(pAADB1) and 824(pSOS95del) have rACETONE values above zero throughout the fermentation. This shows that acid uptake via the acetone formation pathway is discontinued at TN = 20 h for 824(pCTFB1AS), whereas it continues throughout the fermentation for both 824(pAADB1) and 824(pSOS95del). The rACETONE profiles of 824(pSOS95del) and 824(pAADB1) are also quite different, suggesting that acid uptake via the acetone formation pathway is also significantly diminished in 824(pAADB1) compared to 824(pSOS95del). For all three strains, the flux representing acetate uptake via the acetone formation pathway, rACUP, has levels and profile patterns very similar to those of the corresponding rACETONE flux profiles. In both 824(pCTFB1AS) and 824(pAADB1), rBYUP is small and, therefore, acetone formation in these strains due to butyrate uptake appears to be minimal. In contrast, butyrate uptake (rBYUP) via the acetone formation pathway is significant in strain 824(pSOS95del).
For the butanol formation flux, rBUOH, both 824(pSOS95del)and 824(pAADB1) generally show significantly higher levels of rBUOH compared to 824(pCTFB1AS). Although final titers of butanol are nearly the same for 824(pAADB1) and 824(pSOS95del), the rBUOH fluxes for both 824(pSOS95del) and 824(pAADB1) exhibit some differences. It appears that butanol formation is initiated earlier in 824(pAADB1) than in 824(pSOS95del), as shown by the higher rBUOH seen in 824(pAADB1) for TN values less than ca. 10 h. In addition, from TN = 14 to 28 h, rBUOH is higher in 824(pAADB1) than in 824(pSOS95del), which corresponds to the region where the butanol concentration profiles of fermentations (Fig. 2A) of these two strains begins to differ. Also, after ca. TN = 60 h, the rBUOH for 824(pSOS95del) increases, whereas rBUOH for 824(pAADB1) decreases, which also correlates well with a difference seen in butanol concentration profiles. These results further suggest that aad overexpression in 824(pAADB1) results in restoration of butanol formation to plasmid control levels. The rETOH profiles of 824(pSOS95del) and 824(pCTFB1AS) are very similar in both their levels and their shapes. The rETOH in 824(pAADB1) is much higher than in the other two strains and has a peak well into the stationary phase, indicating that aad overexpression in combination with ctfb1-asRNA greatly enhances the ethanol formation pathway.
These data show that, as previously suggested (10, 16), AAD can catalyze the formation of both butanol and ethanol. We also note that rBUOH becomes virtually zero when butyrate is depleted around TN = 48 h of the 824(pAADB1) fermentation and that, in contrast, rETOH is sustained much longer. These data suggest that the intracellular concentration of the acyl-CoA species (butyryl-CoA versus acetyl-CoA) is determining the ratio of butanol versus ethanol formation. Thus, butanol formation appears to be severely limited by the availability of butyryl-CoA. To test this hypothesis, namely, that an increased availability of butyryl-CoA would alter the butanol/ethanol ratio, we added butyrate to the fermentation broth as described below.
Butyrate addition to 824(pAADB1) fermentations decreases ethanol formation and the ethanol/butanol ratio.
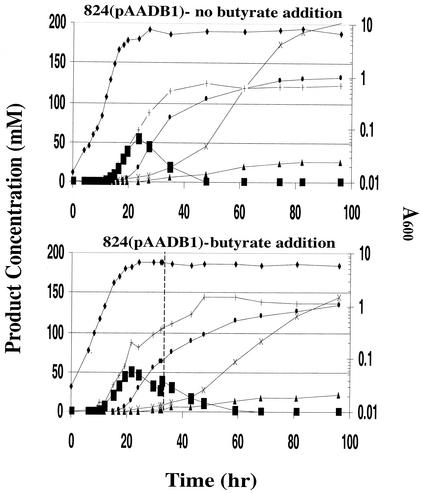
Exogenously added butyric acid to 824(pAADB1) fermentations is meant to sustain a higher intracellular butyryl-CoA concentration and thus increase the butanol/ethanol ratio. This is based on the key assumption that the rate of formation of each alcohol via AAD is determined by the relative concentration of the corresponding acyl-CoA species. At first, we increased the butyrate concentration of 824(pAADB1) fermentations by ca. 30 mM when the butyrate concentration had decreased to ca. 40 mM. However, this resulted in premature termination of the fermentation, presumably due to fast and excessive intracellular acidification (13) caused by uptake of butyric acid from the medium. Thus, a new feed strategy was employed, in which butyrate was added to increase the concentration of the fermentation by ca. 10 mM. Product profiles for these butyrate addition bioreactor experiments, as well as the profile of 824(pAADB1) fermentations without butyrate addition, are shown in Fig. 5.
FIG. 5.
Product profiles of 824(pAADB1) fermentations with or without butyrate addition. The type of fermentation is indicated above each graph. The different products are represented by the following symbols: A600 (♦), acetone (▴), butanol (•), ethanol (×), butyrate (▪), and acetate (+). The vertical dashed line represents the time at which butyrate was added.
Butyrate addition not only increased the butyrate concentration of the fermentation but also extended the presence of butyrate in the fermentation by ca. 10 h. Butyrate addition decreased ethanol formation by ca. 28% (144 mM) without reducing butanol formation (134 mM), thus decreasing the ethanol/butanol ratio from ca. 1.5 to ca. 1.0. Without butyrate addition, butanol and ethanol product concentrations cross at ca. 61 h at a concentration of 119 mM. With butyrate addition, this happens much later (ca. 85 to 90 h) and at a higher concentration (ca. 130 mM). These results are also consistent with fermentations by using another butyrate feed strategy, whereby three additions of butyrate were implemented to increase the butyrate concentration in the fermentation medium by ca. 10 mM per addition. Although this strategy resulted in premature termination of the fermentation (again, probably due to sudden intracellular acidification due to butyric acid uptake), butanol and ethanol concentration profiles did not cross, and ethanol formation was severely reduced by 79%, whereas butanol formation was reduced less (by 29%) compared to fermentations without butyrate addition. These results show that butyrate addition enhances the butanol/ethanol ratio, most likely by increasing the ratio of butyryl-CoA versus acetyl-CoA and thus limiting the ability of AAD to convert acetyl-CoA to ethanol.
Microarray analysis of 824(pAADB1) and 824(pCTFB1AS).
To assess the global impact of overexpressing aad in combination with ctfb1-asRNA expression on the transcriptome, we used DNA microarrays for a comparative analysis of strains 824(pCTFB1AS) and 824(pAADB1). Samples from eight time points varying from the early exponential phase to the early stationary phase of fermentations (Fig. 2B) were analyzed on 18 arrays. All duplicate arrays (n = 4 for the fourth time point) were hybridized with reverse-labeled samples [e.g., 824(pAADB1)-Cy3/824(pCTFB1AS)-Cy5 and 824(pCTFB1AS)-Cy3/824(pAADB1)-Cy5]. Array data were normalized (24), and genes, identified as differentially expressed for at least one array at the 95% confidence level, were cleaned by principal component analysis (1) and further analyzed by average-linkage hierarchical clustering (7). A total of 273 differentially expressed genes were identified (data not shown). Contrary to our expectation, overexpression of aad changed the transcriptional profile of 824(pCTFB1AS) drastically. More specifically, several classes of genes (data not shown) involved in the stress (heat shock) response, sporulation, and chemotaxis were dramatically affected by aad overexpression. Most stress genes especially groES and groEL were upregulated during the exponential phase in 824(pAADB1). However, a large number of stress genes including dnaK, dnaJ, and clpC were strongly downregulated in 824(pAADB1) in the stationary phase. These results suggest that aad overexpression results in an earlier activation of the stress response. Sporulation also appears to be affected by aad overexpression, as shown by the downregulation of early sporulation genes such as sigF and sigE in 824(pCTFB1AS). Chemotaxis also appears to be significantly altered by aad overexpression: nearly all chemotaxis genes were upregulated in 824(pCTFB1AS), thus suggesting that 824(pCTFB1AS) is more motile than 824(pAADB1).
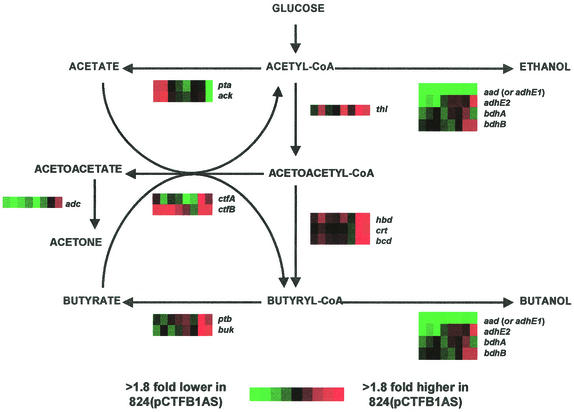
Differences in transcriptional levels may not necessarily result in concomitant differences in protein level (the translation efficiency and protein stability varies greatly among genes or proteins), let alone in concomitant flux differences (some fluxes are determined by protein levels, but many are not). Nevertheless, we thought that it might be interesting to examine whether differences in transcriptional profiles of genes directly involved in the solvent formation pathways might correlate with differences in product formation trends (Fig. 6). First, we looked at the impact of ctfb1-asRNA on ctfB gene expression levels. Because arrays were spotted with double-stranded DNA, the ctfB gene expression profile represents the relative abundance of both ctfB sense mRNA and ctfb1-asRNA expression. However, since ctfA and ctfB are both part of the sol operon and have been shown to be expressed at very similar levels (21), we can assume the portion of the ctfB gene expression profile that is due to just ctfB mRNA expression is very similar to the ctfA mRNA expression profile. Thus, the difference in the ctfA and ctfB expression profiles can be assumed to represent ctfb1-asRNA expression. Using these assumptions, ctfA and ctfB gene expression in 824(pCTFB1AS) is slightly higher throughout the exponential phase and then lower as the fermentation enters the stationary phase relative to 824(pAADB1). This is consistent with Western blot data that shows lower CtfA and CtfB levels in 824(pAADB1) compared to 824(pCTFB1AS) at the transitional and early stationary phases. On the other hand, ctfb1-asRNA expression appears to be substantially lower in 824(pAADB1). This is most likely due to a decrease in plasmid copy number in 824(pAADB1) because of the difference in the sizes of pAADB1 (8.3 kb) and pCTFB1AS (5.3 kb). As expected, aad shows much higher expression in 824(pAADB1). The second alcohol-aldehyde dehydrogenase gene (8), adhE2, shows higher expression in 824(pAADB1) during the exponential phase, but the levels are lower in 824(pAADB1) when ethanol formation takes place; this suggests adhE2 is not as crucial to ethanol formation in strain 824(pAADB1) under our fermentation conditions. The butanol dehydrogenase genes, bdhA and bdhB, show higher expression in 824(pAADB1) during the exponential phase, but as the fermentation enters stationary phase, the trend is reversed, and they are higher in 824(pCTFB1AS). Similar to ctfA expression, adc gene expression was mostly higher in 824(pAADB1). This is somewhat surprising in view of the fact that we are only overexpressing aad in pAADB1. All of the genes involved in the conversion of acetyl-CoA to butyryl-CoA showed nearly the same gene expression pattern, namely, overexpression in the stationary phase in 824(pCTFB1AS) compared to 824(pAADB1) which is similar to the pattern observed in their respective pathway fluxes (data not shown). Both genes in each of the acid formation pathways show a similar gene expression pattern, which is what was expected in view of the fact that the genes for each pathway are part of the same operon. Consistent with acetate profiles (Fig. 2A), expression of the genes responsible for acetate formation (phosphotransacetylase [pta] and acetate kinase [ack]) was upregulated in exponential phase and then downregulated in the stationary phase for 824(pCTFB1AS). For butyrate formation, the phosphotransbutyrylase (ptb) and butyrate kinase (buk) genes were upregulated in the stationary phase in 824(pCTFB1AS), a finding which correlates well with the much higher levels of butyrate in 824(pCTFB1AS) than in 824(pAADB1) (Fig. 2A).
FIG. 6.
Differential expression profiles [strain 824(pCTFB1AS) versus strain 824(pAADB1)] of genes involved in acid and solvent formation pathways. Red and green indicate upregulation and downregulation in 824(pCTFB1AS), respectively. The name of the gene for each profile is indicated to the right of the profile. Abbreviations: aad (adhE1), alcohol-aldehyde dehydrogenase; adc, acetoacetate decarboxylase; ack, acetate kinase; bdhA, bdhB, butanol dehydrogenase A and B; bcd, butyryl-CoA dehydrogenase; hbd, β-hydroxybutyryl dehydrogenase; buk, butyrate kinase; crt, crotonase; ctfA, CoAT subunit A; ctfB, CoAT subunit B; pta, phosphotransacetylase; ptb, phosphotransbutyrylase; thl, thiolase.
DISCUSSION
The present study suggests that combination of asRNA and gene overexpression technologies can be effectively used for beneficial alterations of cell metabolism. asRNA downregulation at levels >80% offers ease of implementation and flexibility not available by gene knockout technologies, especially for organisms such as C. acetobutylicum, for which genomic integration remains a challenge (11). This might prove especially beneficial for functional genomic studies.
We successfully overexpressed aad while downregulating CoAT by using asRNA, and this led to a significantly altered transcriptome and phenotype compared to ctfB1-asRNA expression alone. Overexpression of aad restored butanol production in strain 824(pCTFB1AS), thus supporting the hypothesis that ctfb1-asRNA expression leads to degradation of the tricistronic transcript aad-ctfA-ctfB. The fact that ctfb1-asRNA in 824(pCTFB1AS) does not suppress butanol formation nearly as severely as acetone formation suggests that there may exist a larger number of transcripts with the aad portion intact than transcripts with an intact ctfA-ctfB portion. The data of Fig. 3B might also suggest that there is probably less degradation of ctfA than of ctfB. Taken together, these data suggest that nascent portions of the tricistronic aad-ctfA-ctfB transcript remain unaffected by the ctfb1-asRNA molecule until the ctfB portion of the aad-ctfA-ctfB mRNA has been synthesized, thus accounting for the likely differential expression of AAD, CtfA, and CtfB.
It was surprising to us that a simple genetic change such as aad overexpression on a plasmid that expresses CoAT-asRNA can create such major changes in the transcriptional program of the cells. We did not expect that this specific simple change in a pathway with no known feedback mechanisms or interactions with other cellular programs could have such a profound effect. In perspective, there are several possible reasons for that. Overexpression of aad from its native promoter on a multicopy plasmid might have led to titration of Spo0A (11) and other transcription factors. Also, by increasing the levels of AAD, we have altered product concentrations (butanol, ethanol, butyrate, and acetate) significantly, and such changes may have a large impact on the transcriptional program of the cell. Possible mechanisms might include an increased stress response due to elevated alcohol accumulation, which might lead to accelerated sporulation (2).
Butyrate depletion in the 824(pAADB1) fermentation and the subsequent cessation of butanol formation to the benefit of ethanol formation suggested that butyryl-CoA is limiting butanol formation. Upon butyrate depletion, acetyl-CoA appears to be generated (Fig. 4) at much higher rates than butyryl-CoA due to both a sustained acetate uptake via the CoAT and direct formation from pyruvate. This apparently leads to much higher rates of ethanol formation compared to butanol formation. Whereas butyrate addition appears to confirm this hypothesis, butyrate addition apparently inhibits product formation and is far from practical for enhancing butanol formation. A mechanism to enhance endogenous butyryl-CoA formation and at the same time reduce the levels of acetyl-CoA is needed. The idea is to direct the flux toward butyryl-CoA while at the same time suppress acetate accumulation, which might become a source of acetyl-CoA for ethanol formation. Two metabolic engineering strategies could achieve that. First, this can be possibly accomplished by introducing an asRNA molecule against one of the genes involved in acetate formation on pAADB1. Also, one could introduce a modified version of pAADB1 (i.e., a version that contains a selective marker other than erythromycin) into the strain PJC4PTA (a strain that has the phosphotransacetylase gene [pta] knocked out) (9). At the same time, one will need to enhance the biosynthetic capability to generate butyryl-CoA from acetyl-CoA by overexpressing the protein or proteins that limit carbon flux through this pathway.
Acknowledgments
We acknowledge the use of the Keck Biophysics and Center for Genetic Medicine facilities at Northwestern University. We thank G. N. Bennett for the donation of CoAT primary antibodies, Payam Roshandel for technical assistance, and Abbott Laboratories for the donation of clarithromycin.
This work was supported by National Science Foundation grant BES-9905669.
REFERENCES
- 1.Alter, O., P. O. Brown, and D. Botstein. 2000. Singular value decomposition for genome-wide expression data processing and modeling. Proc. Natl. Acad. Sci. USA 97:10101-10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bahl, H., H. Muller, S. Behrens, H. Joseph, and F. Narberhaus. 1995. Expression of heat shock genes in Clostridium acetobutylicum. FEMS Microbiol. Rev. 17:341-348. [DOI] [PubMed] [Google Scholar]
- 3.Buday, Z., J. C. Linden, and M. N. Karim. 1990. Improved acetone-butanol fermentation analysis using subambient HPLC column temperature. Enzymol. Microb. Technol. 12:24-27. [Google Scholar]
- 4.Cary, J. W., D. J. Petersen, E. T. Papoutsakis, and G. N. Bennett. 1990. Cloning and expression of Clostridium acetobutylicum ATCC 824 acetoacetyl-coenzyme A:acetate/butyrate:coenzyme A-transferase in Escherichia coli. Appl. Environ. Microbiol. 56:1576-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desai, R. P., L. K. Nielsen, and E. T. Papoutsakis. 1999. Stoichiometric modeling of Clostridium acetobutylicum fermentations with non-linear constraints. J. Biotechnol. 71:191-205. [DOI] [PubMed] [Google Scholar]
- 6.Desai, R. P., and E. T. Papoutsakis. 1999. Antisense RNA strategies for the metabolic engineering of Clostridium acetobutylicum. Appl. Environ. Microbiol. 65:936-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fontaine, L., I. Meynial-Salles, L. Girbal, X. H. Yang, C. Croux, and P. Soucaille. 2002. Molecular characterization and transcriptional analysis of adhE2, the gene encoding the NADH-dependent aldehyde/alcohol dehydrogenase responsible for butanol production in alcohologenic cultures of Clostridium acetobutylicum ATCC 824. J. Bacteriol. 184:821-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green, E. M., Z. L. Boynton, L. M. Harris, F. B. Rudolph, E. T. Papoutsakis, and G. N. Bennett. 1996. Genetic manipulation of acid formation pathways by gene inactivation in Clostridium acetobutylicum ATCC 824. Microbiology 142:2079-2086. [DOI] [PubMed] [Google Scholar]
- 10.Harris, L. M., R. P. Desai, N. E. Welker, and E. T. Papoutsakis. 2000. Characterization of recombinant strains of the Clostridium acetobutylicum butyrate kinase inactivation mutant: need for new phenomenological models for solventogenesis and butanol inhibition? Biotechnol. Bioeng. 67:1-11. [PubMed] [Google Scholar]
- 11.Harris, L. M., N. E. Welker, and E. T. Papoutsakis. 2002. Northern, morphological, and fermentation analysis of spo0A inactivation and overexpression in Clostridium acetobutylicum ATCC 824. J. Bacteriol. 184:3586-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hegde, P., R. Qi, K. Abernathy, C. Gay, S. Dharap, R. Gaspard, J. E. Hughes, E. Snesrud, N. Lee, and J. Quackenbush. 2000. A concise guide to cDNA microarray analysis. BioTechniques 29:548-562. [DOI] [PubMed] [Google Scholar]
- 13.Husemann, M. H. W., and E. T. Papoutsakis. 1988. Solventogenesis in Clostridium acetobutylicum fermentations related to carboxylic acid and proton concentrations. Biotechnol. Bioeng. 32:843-852. [DOI] [PubMed] [Google Scholar]
- 14.Lee, S. Y., and S. Rasheed. 1990. A simple procedure for maximum yield of high-quality plasmid DNA. BioTechniques 9:676-679. [PubMed] [Google Scholar]
- 15.Mermelstein, L. D., and E. T. Papoutsakis. 1993. In vivo methylation in Escherichia coli by the Bacillus subtilis phage Φ3T I methyltransferase to protect plasmids from restriction upon transformation of Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 59:1077-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nair, R. V., G. N. Bennett, and E. T. Papoutsakis. 1994. Molecular Characterization of an aldehyde/alcohol dehydrogenase gene from Clostridium acetobutylicum ATCC 824. J. Bacteriol. 176:871-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papoutsakis, E. T. 1984. Equations and calculations for fermentations of butyric acid bacteria. Biotechnol. Bioeng. 26:174-187. [DOI] [PubMed] [Google Scholar]
- 18.Patzel, V., and G. Sczakiel. 1998. Theoretical design of antisense RNA structures substantially improves annealing kinetics and efficacy in human cells. Nat. Biotechnol. 16:64-68. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Thormann, K., L. Feustel, K. Lorenz, S. Nakotte, and P. Durre. 2002. Control of butanol formation in Clostridium acetobutylicum by transcriptional activation. J. Bacteriol. 184:1966-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomas, C. A., K. V. Alsaker, H. P. J. Bonarius, W. T. Hendriksen, H. Yang, J. A. Beamish, C. J. Paredes, and E. T. Papoutsakis. DNA-array based transcriptional analysis of asporogenous, non-solventogenic Clostridium acetobutylicum strains SKO1 and M5. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 22.Tummala, S. B., N. E. Welker, and E. T. Papoutsakis. 2003. Design of antisense RNA constructs for downregulation of the acetone formation pathway of Clostridium acetobutylicum. J. Bacteriol. 185:1923-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagner, E. G., and R. W. Simons. 1994. Antisense RNA control in bacteria, phages, and plasmids. Annu. Rev. Microbiol. 48:713-742. [DOI] [PubMed] [Google Scholar]
- 24.Yang, H., H. Haddad, C. A. Tomas, K. V. Alsaker, and E. T. Papoutsakis. 2003. A segmental nearest neighbor normalization and gene identification method gives superior results for DNA-array analysis. Proc. Natl. Acad. Sci. USA 100:1122-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]