Abstract
Although four of the five Salmonella pathogenicity islands (SPIs) have been characterized in detail for Salmonella enterica serovar Typhimurium, and the fifth has been characterized for Salmonella enterica serovar Dublin, there have been limited studies to examine them in detail in a range of pathogenic serovars of S. enterica. The aim of this study was to examine these regions, shown to be crucial in virulence, in pathogenic serovars to identify any major deletions or insertions that may explain variation in virulence and provide further understanding of the elements involved in the evolution of these regions. Multiple strains of each of the 13 serovars were compared by Southern blot hybridization using a series of probes that together encompassed the full length of all five SPIs. With the exception of serovar Typhimurium, all strains of the same serovar were identical in all five SPIs. Those serovars that differed from serovar Typhimurium in SPI-1 to SPI-4 and from serovar Dublin in SPI-5 were examined in more detail in the variant regions by PCR, and restriction endonuclease digestion and/or DNA sequencing. While most variation in hybridization patterns was attributable to loss or gain of single restriction endonuclease cleavage sites, three regions, in SPI-1, SPI-3, and SPI-5, had differences due to major insertions or deletions. In SPI-1 the avrA gene was replaced by a 200-base fragment in three serovars, as reported previously. In SPI-5, two serovars had acquired an insertion with similarity to the pagJ and pagK genes between pipC and pipD. In SPI-3 the genes sugR and rhuM were deleted in most serovars and in some were replaced by sequences that were very similar to either the Escherichia coli fimbrial operon, flanked by two distinct insertion sequence elements, or to the E. coli retron phage ΦR73. The distribution of these differences suggests that there have been a number of relatively recent horizontal transfers of genes into S. enterica and that in some cases the same event has occurred in multiple lineages of S. enterica. Thus, it seems that insertion sequences and retron phages are likely to be involved in continuing evolution of the pathogenicity islands of pathogenic Salmonella serovars.
Clusters of chromosomal virulence genes, termed Salmonella pathogenicity islands (SPIs), found only within the genus Salmonella and not in Escherichia coli, provide a molecular basis for understanding Salmonella pathogenicity. Most of the SPIs are adjacent to tRNA genes and have a G+C composition different from that of the rest of the chromosome, suggesting acquisition by horizontal transfer, though the origin of the SPIs and the mechanisms of their transfer are still unclear. At least five pathogenicity islands (SPI-1 to -5) have been found in a range of serovars of Salmonella enterica, with a further five islands with characteristics of SPIs identified in the complete genome of S. enterica serovar Typhi (23).
SPI-1 is a 40-kb chromosomal locus at centisome (cs) 63 in S. enterica serovar Typhimurium and is required for the invasion of host cells and induction of macrophage apoptosis (7). The genes within this island encode a type III secretion system, including the secretion apparatus components, effectors, chaperones, and regulators (11, 25). Salmonella bacteria containing mutations in these genes are attenuated in mice infected orally but fully virulent after intraperitoneal administration (12).
A second type III secretion system is encoded within SPI-2 at cs 30. SPI-2 is adjacent to the tRNAval gene and harbors genes required for systemic infection and replication within macrophages. The attenuation of SPI-2 mutants after intraperitoneal and oral inoculation indicates that the role of this system is in interactions subsequent to epithelial entry (32).
SPI-3, SPI-4, and SPI-5 are also found next to tRNA genes. SPI-3 harbors 10 genes, including the mgtCB operon, which is regulated by PhoPQ and is required for survival in macrophages and growth in low-Mg2+ environments (3). SPI-4 encodes 18 genes and is suspected to be required for intramacrophage survival (38). The fifth pathogenicity island is located next to serT and contains six genes, four of which have been shown to be involved in enteritis in calves (39).
The molecular genetic relationships between Salmonella species have been investigated using multilocus enzyme electrophoresis, by comparisons of sequences of specific genes, and by examining the distribution of some genes within SPI-1, -2, and -3 using Southern hybridization (3, 4, 22, 31). These studies have established that there are eight subspecies within the genus, with the major pathogens of mammals and birds falling into subspecies I. Southern hybridization using a probe containing spaLMNOPQRST from SPI-1 detected a positive signal in both S. enterica (subspecies I, II, IIIA and B, IV, VI, and VII) and Salmonella bongori (subspecies V), but a probe containing spiABCR of SPI-2 did not detect a signal in S. bongori. The stability of only SPI-1 genes in both Salmonella species suggests that SPI-2 was probably acquired by horizontal transfer after the ancestor of S. enterica diverged from that of S. bongori (22). However, another study of SPI-2 genes showed that a number of SPI-2 genes could be found within S. bongori (16). The gene function, base composition and codon usage and the distribution of the genes among Salmonella spp. suggest that SPI-2 has a mosaic structure which probably resulted from multiple horizontal transfer events (16).
A study of the phylogenetic distribution of SPI-3, using nine probes that encompassed part of each of the genes within SPI-3, showed that this SPI varied between serovars within S. enterica. Southern hybridization established that the probes derived from sugR and rhuM, which are at the left end of SPI-3 (Fig. 1), revealed the most variable patterns among the subspecies of S. enterica, and no hybridizing fragments were detected for S. bongori (3).
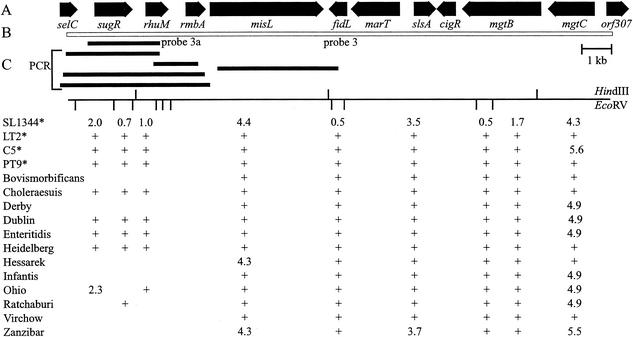
FIG. 1.
Variation between Salmonella serovars within SPI-3 as detected using Southern hybridization and chromosomal amplification analysis. (A) SPI-3 genes and their direction of transcription (modified from reference 3). (B) Extent of hybridization probe 3 and PCRs (shaded blocks) used for detailed analysis. (C) EcoRV and HindIII restriction maps for serovar Typhimurium SL1344. The numbers indicate the sizes of the HindIII and EcoRV fragments detected (in kilobases). +, presence of HindIII and EcoRV fragment of the same size as that seen in serovar Typhimurium SL1344. ∗, serovar Typhimurium strains 9806584 (PT9), C5, LT2, and SL1344.
SPI-4 and SPI-5 were found to be conserved among several pathogenic serovars within S. enterica subspecies I that were tested using Southern blot hybridization (38, 39). Two 5-kb probes, which were derived from the SPI-4 genes I, J, and K and genes O, P, Q, and R, were found to hybridize to a range of serovars, including S. enterica serovars Dublin, Enteritidis, Gallinarum, Minnesota, Paratyphi, Pullorum, Typhi, and Typhimurium (38). Three probes from SPI-5 genes (pipD, sopB, and pipAB) have been used to show that all four of these genes are present in S. enterica serovars Choleraesuis, Enteritidis, Gallinarum, Pullorum, and Typhimurium (39).
While these large-scale comparisons suggest that SPI-1 is stable, recent detailed sequence comparisons of the spaM, spaN, spaO, spaP, spaQ, invA, invE, and invH genes in SPI-1 in a range of Salmonella strains representative of the eight Salmonella subspecies have demonstrated recombinational events within SPI-1 in the inv-spa genes, principally involving S. enterica subspecies IV and VII (6). These studies suggest that more detailed studies are needed in order to fully understand the evolution of the SPIs. Furthermore many of the previous studies of variation within the SPIs have used probes covering a limited extent of the islands and thus could not be expected to have detected all the variation within the SPIs.
The aim of this study was to investigate genetic variation across the full length of the five well-characterized SPIs in a range of pathogenic serovars in S. enterica subspecies I. Southern hybridization and restriction fragment length polymorphism (RFLP) analysis were used to examine genetic variation within and between serovars, and DNA sequence analysis was used to examine the differences in more detail. Such elucidation of the distribution of SPI genes in Salmonella serovars may yield a better understanding of the evolution of pathogenic serovars and the role of SPIs in the differences in pathogenesis and epidemiology of different serovars.
MATERIALS AND METHODS
Bacterial strains.
A collection of two or three strains of each of 13 pathogenic serovars of S. enterica was used (Table 1). Most of the strains examined were obtained from the Microbiology Diagnostic Unit, The University of Melbourne (Melbourne, Australia). The exceptions were serovar Choleraesuis strains 936 (11), 930 (9), and 1198 (1) and serovar Enteritidis strain 2099, which were obtained from Faculty of Veterinary Science, Kasetsart University (Bangkok, Thailand), S. enterica serovar Ratchaburi strains 183398 (1), 183898 (5), and 184198 (6), which were obtained from the WHO National Salmonella and Shigella Centre, Bangkok, Thailand, and serovar Typhimurium strains SL1344, C5 and LT2, which were kindly donated by Richard Strugnell, Department of Microbiology and Immunology, The University of Melbourne.
TABLE 1.
Salmonella isolates used in analysis of SPI variation
| Serovar | Isolate no. | Placea | Dateb | Sourcec |
|---|---|---|---|---|
| Bovismorbificans PT12 | 9805044 | VIC | 18/2/98 | Bovine |
| Bovismorbificans PT13 | 9714094 | VIC | 21/4/97 | Env |
| Bovismorbificans PT13 | 9713604 | VIC | 11/3/97 | Env |
| Choleraesuis | 936(11) | THAI | 10/2/98 | Porcine |
| Choleraesuis | 930(9) | THAI | 15/6/98 | Porcine |
| Choleraesuis | 1198(1) | THAI | 16/6/98 | Porcine |
| Derby | 9813031 | VIC | 13/7/98 | Bovine |
| Derby | 0010160 | VIC | 9/6/00 | Porcine |
| Derby | 0010158 | VIC | 9/6/00 | Porcine |
| Dublin | 9805277 | VIC | 11/3/98 | Bovine |
| Dublin | 0009131 | VIC | 15/5/00 | Bovine |
| Dublin | 0009321 | VIC | 2/6/00 | Bovine |
| Enteritidis PT4 | 2099 | THAI | 15/7/98 | Avian |
| Enteritidis PT4 | 9915159 | INDO | 5/7/99 | Avian |
| Enteritidis PT4 | 9915161 | INDO | 5/7/99 | Avian |
| Heidelberg | 547359 | VIC | 14/12/92 | Equine |
| Heidelberg | 9718249 | VIC | 4/6/97 | Env |
| Hessarek | 9820034 | VIC | 26/10/98 | Equine |
| Hessarek | 9904270 | VIC | 16/2/99 | Marsupial |
| Hessarek | 9923235 | VIC | 12/11/99 | Feline |
| Infantis | 9811189 | VIC | 15/6/98 | Avian |
| Infantis | 0007640 | VIC | 29/4/00 | Avian |
| Infantis | 0007632 | VIC | 18/4/00 | Avian |
| Ohio | 9815932 | VIC | 3/7/98 | Porcine |
| Ohio | 9714920 | VIC | 30/4/97 | Env |
| Ohio | 9714922 | VIC | 30/4/97 | Env |
| Ratchaburi | 183398(1) | THAI | 8/6/98 | Human |
| Ratchaburi | 183898(5) | THAI | 10/6/98 | Human |
| Ratchaburi | 184198(6) | THAI | 17/6/98 | Human |
| Typhimurium PT9 | 9806584 | VIC | 30/3/98 | Porcine |
| Typhimurium C5 | 18 | |||
| Typhimurium LT2 | 19 | |||
| Typhimurium SL1344 | 17 | |||
| Virchow PT34 | 0002063 | VIC | 5/2/00 | Human |
| Virchow PT8 | 0002141 | QLD | 26/1/00 | Human |
| Virchow PT34 | 0001122 | VIC | 12/1/00 | Avian |
| Zanzibar | 9800913 | VIC | 12/1/98 | Bovine |
| Zanzibar | 0009698 | VIC | 5/6/00 | Bovine |
| Zanzibar | 0009734 | VIC | 8/6/00 | Bovine |
Place of isolation. VIC, Victoria; THAI, Thailand; INDO, Indonesia; QLD, Queensland.
Date of isolation (day/month/year).
Source or reference. Env, environmental isolate.
Restriction endonuclease digestion and Southern transfer.
Genomic DNA from each of the strains was extracted using the High Pure PCR Template Preparation kit (Roche Diagnostics, Mannheim, Germany), following the method recommended by the manufacturer. Approximately 1 μg of Salmonella DNA was digested with 20 U of restriction endonucleases EcoRV, HindIII, or BglII or with 20 U each of a combination of restriction endonucleases EcoRV and HindIII or EcoRV and BglII at 37°C for 3 h. The DNA fragments were separated by electrophoresis in 0.8% agarose gels and capillary transferred to Hybond N+ membranes (27).
Preparation of PCR probes.
The DNA probes used in Southern hybridization were prepared by amplification of one or more fragments that spanned each SPI (see supplemental information at ftp://jb:jb@ftp.vet.unimelb.edu.au/). For SPI-1, the first probe, 1A, extended from fhlA to hilA and was approximately 14 kb long; the second, 1B, was 15 kb in length, overlapped probe 1A within hilA, and finished within spaP; and the third, 1C, was 10 kb in length, overlapped probe 1B, and covered spaP to invH (ftp://jb:jb@ftp.vet.unimelb.edu.au/).
The probes produced for SPI-2, 2/31 and 2/30, were approximately 26 and 16 kb in length, respectively. Probe 2/31 encompassed genes ssaU to ssrB, and probe 2/30 encompassed genes ssrB to orf48 (ftp://jb:jb@ftp.vet.unimelb.edu.au/).
For SPI-4, probes 4A and 4B were 15 and 8 kb in length, respectively. Probe 4A encompassed genes A to M, and probe 4B encompassed genes M to R (ftp://jb:jb@ftp.vet.unimelb.edu.au/). For both SPI-3 and SPI-5, single probes, 3 and 5, of 17 and 6.5 kb, respectively, were produced (ftp://jb:jb@ftp.vet.unimelb.edu.au/) (Fig. 1).
SPI primers.
PCR primers for SPI-1 were derived by examination of GenBank accession numbers U16278 and U84286, searches of the serovar Typhi sequence database at the Sanger Centre (http://www.sanger.ac.uk), and searches of the serovar Typhimurium sequence database at Washington University (http://genome.wustl.edu) (20, 23). Primers for SPI-2 were derived by examination of the sequences under GenBank accession numbers X99944, Y09357, AJ224892, U51927, Z95891, AJ224978 and X99945. The primers for SPI-3, SPI-4, and SPI-5 were derived by examination of the sequences under GenBank accession numbers AF106566, AF060869, and AF060858, respectively. The oligonucleotide sequence of each primer, the length and position of expected PCR fragments, and the amplification conditions are shown in the supplemental information on our website (ftp://jb:jb@ftp.vet.unimelb.edu.au/).
Amplification of SPI probes.
Target DNA for production of probes for SPI-1, -2, -3, and -4 was prepared by phenol-chloroform extraction of the chromosomal DNA of serovar Typhimurium type strain SL1344, and that for probes for SPI-5 was prepared by phenol-chloroform extraction of the chromosomal DNA of serovar Dublin. Approximately 100 to 200 ng of DNA template was used for amplification of products up to 18 kb, and for longer amplification products the amount of DNA was increased to 250 to 500 ng.
The long template PCR was prepared as two separate mixtures of 25 μl each in thin-walled PCR tubes. The first mixture contained 10 μl of 5× Expand plus PCR buffer, 2.6 U of Expand 20 kb plus enzyme mixture (Roche Diagnostics), and distilled water to a final volume of 25 μl. The second contained a 500 μM concentration (each) of deoxynucleoside triphosphates, a 0.4 μM concentration (each) of primer, the required amount of template, and distilled water to a final volume of 25 μl. The concentration of each deoxynucleoside triphosphate was decreased to 350 μM when the expected PCR product was smaller than 12 kb. The PCR mixture was overlaid with 40 μl of mineral oil when the extension time was longer than 13 min/cycle. All the reactions were performed using a Gene Amp Model 2400 thermocycler (Perkin-Elmer).
PCR cycle conditions were individually adjusted for each probe depending on the expected size of the PCR product. The two mixtures were combined and immediately incubated at 92°C for 3 min followed by 10 cycles of 92°C for 10 s, the appropriate annealing temperature for 30 s and 68°C for the appropriate elongation time, followed by another 20 cycles of 92°C for 10 s, the appropriate annealing temperature for 30 s and 68°C for the appropriate elongation time with 10 s/cycle added to the time, and a final incubation at 68°C for 7 min. The annealing temperatures and the extension times for each pair of primers are shown on our website (ftp://jb:jb@ftp.vet.unimelb.edu.au.).
The fragments amplified for probe preparation were partially sequenced using the forward and reverse PCR primers and the ABI Prism BigDye Terminator Sequencing Ready Reaction kit (Perkin-Elmer Applied Biosystems) following the manufacturer's instructions.
Southern blot hybridization.
The Southern blots were hybridized with prepared probes at 55°C in hybridization buffer (0.5 M Na2HPO4 [pH 7.2], 7% [wt/vol] sodium dodecyl sulfate [SDS], 0.01% [wt/vol] bovine serum albumin, and 0.1 mM EDTA [pH 8]) for 4 h. The membranes were washed once for 20 min in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.5% SDS, followed by three washes for 20 min each in 2× SSC-0.5% SDS at 55°C, and then subjected to autoradiography (27).
Investigation of serovars that had variant hybridization patterns.
Several serovars had bands of different sizes or no hybridization signal in specific regions. The SPI regions found to differ in these serovars were subjected to further analysis by PCR and restriction endonuclease cleavage and/or DNA sequence analysis. Differences in hybridization patterns at the left-hand side of SPI-1 were investigated using the primer pairs 1A/F and orgC/R, orgC/F and prgK/R, sitD/F and orgC/R, sitD/F and sprB/R, and sprB/F and orgC/R to amplify the DNA sequences within this region (ftp://jb:jb@ftp.vet.unimelb.edu.au/). PCR products that were derived by amplification between fhlA and orgC (using primers 1A/F and orgC/R) were digested with EcoRV, and their restriction endonuclease cleavage patterns were verified using Southern blot hybridization with probe 1A. The products derived by amplification between sitD and sprB were investigated further by nucleotide sequencing. The regions between spaS and spaO of SPI-1 (ftp://jb:jb@ftp.vet.unimelb.edu.au/) of serovar Ratchaburi and serovar Typhimurium SL1344 were amplified using primers spaS/F and spaO/R (ftp://jb:jb@ftp.vet.unimelb.edu.au/). These PCR products were digested with EcoRV, and the sizes of the fragments were compared.
To investigate differences observed in the hybridization pattern of SPI-2 in S. enterica serovar Derby, the region between ssrA and ssrB was amplified using the primer pair ssrA/F and ssrB/R (ftp://jb:jb@ftp.vet.unimelb.edu.au/). The regions between ssaV and sscA, and between ssaE and sseF, were amplified using the primer pairs ssaV/F and sscA/R and ssaE/R and sseF/F. The resultant PCR products were examined by digestion with EcoRV or HindIII.
The regions at the left-hand end of SPI-3 were examined in further detail by Southern blot hybridization with probe 3A, a 2.7-kb PCR product encompassing sugR to rhuM in serovar Typhimurium SL1344. The probe was radiolabeled with [α32-P]dATP and hybridized at 60°C to EcoRV- and HindIII-digested chromosomes. The membrane was washed three times with 0.1× SSC-0.5% SDS at 60°C for 20 min each. DNA products were amplified from strains using five different primer pairs, sugR/F and rhuM/R, 3/F and rhuM/R, rhuM/F and rmbA/R, 3/F and misL/R, and selC/F and misL/R (ftp://jb:jb@ftp.vet.unimelb.edu.au/). PCR products encompassing selC to misL were digested with EcoRV, Southern blotted, and hybridized to radiolabeled probe 3 or 3A. These PCR products were also used for DNA sequence analysis.
The primer pair (4B/F and 4B/R) (ftp://jb:jb@ftp.vet.unimelb.edu.au/) used to produce probe 4B was used to amplify the corresponding region from genomic DNA of S. enterica serovars Derby, Dublin, Heidelberg, Hessarek, Ohio, and Ratchaburi. The resultant PCR products were digested with EcoRV. The primer pair 4K/F and 4A/R were used for amplification of the region between genes K and L in S. enterica serovars Derby, Dublin, Enteritidis, Heidelberg, Infantis, Ohio, Ratchaburi, Virchow, and Zanzibar.
The SPI-5 regions from a representative strain of each of the 13 serovars were amplified using primers 5/F and 5/R (ftp://jb:jb@ftp.vet.unimelb.edu.au/). The PCR was prepared as described for SPI-5 probe preparation but using the enzyme Expand High Fidelity PCR (Roche Diagnostics) and 5 μl of 10× Expand High Fidelity PCR buffer with 15 mM MgCl2. Immediately after mixing, the PCR was incubated at 93°C for 3 min, followed by 10 cycles of 93°C for 30 s, 60°C for 1 min, and 68°C for 5 min and another 20 cycles of 93°C for 30 s, 60°C for 1 min, and 68°C for 5 min with 10 s added each cycle using a Hybaid OmniGene thermocycler (Hybaid). The PCR fragments were separated in a 0.8% agarose gel and purified using a QiaexII gel extraction kit (Qiagen). Approximately 300 ng of purified PCR products were digested with 10 U of either EcoRV or DraI or with 10 U of both enzymes, and the sizes of the fragments were compared.
Two primer pairs, sopB/F and sopB/R and pipC/F and pipB/R, were used to amplify the sopB gene and the region between pipC and pipB, respectively, from SPI-5 in selected serovars. The resultant PCR products were digested with 10 U of EcoRV and/or DraI, and fragments were separated in a 2% agarose gel and further analyzed by DNA sequencing to identify the sequences that differed from serovar Dublin.
Nucleotide sequencing and phylogenetic analysis.
The purified PCR products of each variant region were sequenced by primer walking using an ABI BigDye terminator sequencing kit, except that the long insertions in serovar Derby and serovar Hessarek within SPI-3 were subcloned into pUC19, and purified plasmids were used as a template for sequencing.
DNA sequences were assembled and edited using the software package GeneWorks (Oxford Molecular Group, Inc.), the programs EclustalW (34) and DNA Strider 1.2 (by Christian Marck, Service de Biochimie, Départment di Biologie, Institut di Recherche Fondamentale, CEA, Coif-sur-Yvette, France), and the GCG package run on the Australian National Genomic Information Service. Database searching was done using programs available through the Australian National Genomic Information Service, including BlastX (36), BlastN (1), and FastA (24).
The phylogenetic analysis of the 13 Salmonella serovars was performed using restriction endonuclease cleavage data obtained for the five pathogenicity islands. The EcoRV restriction sites in all five islands and the HindIII sites in SPI-1 and -2 were used. HindIII sites in SPI-3 and -4 were not used because not all sites could be accurately mapped. The analysis did not include regions of the SPIs that were not present in all serovars.
The phylogenetic inference was performed using the Restml program in PHYLIP (Phylogeny Inference Package) version 3.6 for the PowerMac, distributed by J. Felsenstein, Department of Genetics, University of Washington, Seattle (9). Global rearrangement was used to search for the best tree, but the input order was not randomized. Statistical analysis of the tree topology was performed using bootstrap resampling of the data using the Seqboot program (in PHYLIP 3.6). One hundred resamplings were then analyzed using Restml as described above. Consense (in PHYLIP 3.6) was then used to derive a consensus tree.
Nucleotide sequence accession numbers.
The sequences of the insertions in SPI-3 in serovars Ratchaburi and Derby were assigned the GenBank accession numbers AY144489 and AY144490, respectively. The sequences of the insertions in SPI-5 in serovar Ohio and serovar Derby were assigned the GenBank accession numbers AY 144491 and AY144492, respectively.
RESULTS
Variation in SPI-1 to SPI-5 within each serovar.
With the exception of serovar Typhimurium, all strains within the same serovar had the same Southern hybridization pattern with each of the SPI probes (data not shown). This implies that classification using serotyping, which differentiates salmonellae on the basis of their lipopolysaccharide and flagellum antigens, was generally representative of the genotype, even within the more variable SPI regions.
Variation between Salmonella serovars within SPI-1.
Three probes (1A, 1B, and 1C) were used to examine variation between different Salmonella serovars in SPI-1. Hybridization of probe 1A to EcoRV-digested chromosomes resulted in a similar pattern in most of the strains tested (ftp://jb:jb@ftp.vet.unimelb.edu.au/). The 5.9-kb fragment seen in serovar Typhimurium, encompassing genes sitD, avrA, sprB, sprA, orgC, orgD, and orgA of SPI-1, was absent in S. enterica serovar Bovismorbificans, serovar Virchow, serovar Choleraesuis, serovar Ohio, and serovar Ratchaburi, but fragments of 8, 8, 5.1, 7.2, and 7.2 kb, respectively, were seen. These five serovars were subjected to further analysis of the sequences within this region.
To verify the different hybridization patterns at the left-hand end of SPI-1 for serovars Bovismorbificans and Virchow, the primers 1A/F and orgC/R were used to amplify chromosomal DNA from both serovars and from serovar Typhimurium SL1344. The PCR products obtained from all three were 7.9 kb in size but had differing EcoRV cleavage patterns.
Four pairs of primers, sitD/F and orgC/R, orgC/F and prgK/R, sitD/F and sprB/R, and sprB/F and orgC/R, were used to amplify fragments of SPI-1 from the five serovars mentioned above. In all five serovars the products encompassing sprB to orgC and orgC to prgK were 2.1 and 3.1 kb in size, respectively, and the 3.1-kb products had identical EcoRV cleavage patterns. The PCR products encompassing sitD to sprB from serovar Bovismorbificans, serovar Virchow, and strain SL1344 were 1.5 kb in size, whereas those from serovars Choleraesuis, Ohio, and Ratchaburi were 600 bp in size. Sequence analysis of the PCR products showed that these last three serovars lacked approximately 900 bp, corresponding to avrA. The nucleotide sequences at each end of the 600-bp PCR product were identical to those of serovar Typhimurium, but the sequence that would encode AvrA was replaced by a 200-bp fragment flanked by sitD and sprB. The nucleotide sequences were similar for all three serovars (ftp://jb:jb@ftp.vet.unimelb.edu.au/) and contained an open reading frame (ORF) that would encode a protein, the sequence of which was not significantly similar to any protein sequence in the GenBank database.
The hybridization patterns obtained using probes 1B and 1C on Southern blots of chromosomes digested with HindIII or EcoRV were nearly identical for all isolates. The one obvious difference was detected at the border of SPI-1, next to invH, with either a 2.8- or 2.9-kb HindIII fragment seen. All other differences were attributable to the loss or gain of cleavage sites.
Variation between Salmonella serovars within SPI-2.
Two probes, 2/31 and 2/30 (ftp://jb:jb@ftp.vet.unimelb.edu.au/), derived from SPI-2 at the cs 31 and cs 30 regions, respectively, were used to probe Salmonella chromosomal DNA digested with either HindIII or EcoRV. The only differences detected were attributable to the loss or gain of restriction endonuclease cleavage sites, not insertions or deletions.
Variation within SPI-3 between Salmonella serovars.
The 17-kb probe derived from SPI-3 detected much more variable hybridization patterns in the serovars examined than did the other SPI probes (Fig. 1). Digestion of the chromosomes with HindIII revealed the most-variable hybridization pattern, with many fragments seen in S. Typhimurium SL1344 absent in other serovars. The absence of many HindIII fragments resulted from the absence of HindIII cleavage sites within SPI-3 in different serovars. Hybridization with either EcoRV- or BglII-digested chromosomes revealed less-variable patterns. SPI-3 hybridization patterns of chromosomes digested with EcoRV or with EcoRV and HindIII showed that most serovars tested had fragments that corresponded with genes rmbA, misL, fidL, marT, slsA, cigR, mgtB, and mgtC in the middle and on the right-hand side of SPI-3 (Fig. 1). The left-hand end of SPI-3, containing sugR and rhuM, was found to be the most variable region. Of the 13 serovars tested, serovars Choleraesuis, Dublin, Enteritidis, Heidelberg, and Typhimurium possessed the fragments of 2041, 738, and 1064 bp that encompassed the genes sugR and rhuM, but the other 8 serovars, including serovars Bovismorbificans, Derby, Hessarek, Infantis, Ohio, Ratchaburi, Virchow, and Zanzibar, had fragments of different sizes or appeared to lack fragments within this region (Fig. 1). Probe 3A (2.7 kb), encompassing sugR and rhuM of serovar Typhimurium SL1344, revealed that the hybridization pattern in this region was identical to that obtained using probe 3 (17 kb) and that many serovars did not have positive signals for the sugR and rhuM genes.
The genomic DNAs from the eight variant serovars and strain SL1344 were further amplified using primer pairs sugR/F and rhuM/R, 3/F and rhuM/R, rhuM/F and rmbA/R, 3/F and misL/R, and selC/F and misL/R. Only serovar Typhimurium and serovar Ohio yielded specific PCR products using every primer pair, and most of the serovars examined yielded no specific PCR products for most of the primer pairs (Table 2). However, primer pair selC/F and misL/R did generate specific PCR products for all serovars examined, and primer pair sugR/F and rhuM/R produced a specific product from serovar Ohio (Table 2). The results indicated that the DNA sequence following the selC tRNA gene was variable, and this region was subjected to further analysis by DNA sequencing.
TABLE 2.
PCR products derived from different SPI-3 primer pairs
| Serovar | PCR product (kb) from:
|
|||
|---|---|---|---|---|
| selC/F and misL/R | sugR/F and rhuM/R | 3/F and rhuM/R | 3/F and misL/R | |
| Bovismorbificans | 2.4 | —a | — | — |
| Derby | 13 | — | — | — |
| Hessarek | 13 | — | — | — |
| Infantis | 2.4 | — | — | — |
| Ohio | 5.8 | 2.3 | 3.7 | 5.8 |
| Ratchaburi | 5.5 | — | — | — |
| Virchow | 2.4 | — | — | — |
| Zanzibar | 2.3 | — | — | — |
| Typhimurium | 6.2 | 2.7 | 4.1 | 6.2 |
—, no specific product amplified.
The serovar Ohio PCR product of 2.3 kb, amplified using primers sugR/F and rhuM/R, showed high nucleotide similarity to sugR of serovar Typhimurium, but there was a 516-bp deletion within the gene, and the sequences at the junctions of the deletion at both ends were directly repeated (ftp://jb:jb@ftp.vet.unimelb.edu.au/). However the nucleotide sequences of rhuM of serovars Ohio and Typhimurium were 99% identical, and the gene arrangement in serovar Ohio was identical to that shown in Fig. 1.
Amplification of Salmonella genomic DNA with primer pair selC/F and misL/R revealed that there was variation in the nucleotide sequence between selC and rhuM in many serovars. Serovars Bovismorbificans, Infantis, Virchow, and Zanzibar yielded PCR products of approximately 2.4 kb. Serovar Ohio and serovar Ratchaburi yielded PCR products of 5.8 and 5.5 kb, respectively, and serovar Derby and serovar Hessarek yielded 13-kb PCR products. Strain SL1344 yielded the expected 6.2-kb product (Table 2).
The PCR products obtained using primer pair selC/F and misL/R were digested with EcoRV, and the RFLP patterns were examined. A fragment of 1.6 kb was seen in RFLPs of all serovars except serovar Zanzibar. Southern hybridization using probe 3 (17 kb of SPI-3) but not probe 3A (sugR and rhuM) detected the 1.6 kb-EcoRV fragments and a 2.3-kb fragment of serovar Zanzibar, indicating that the DNA region following rhuM was present in all strains examined.
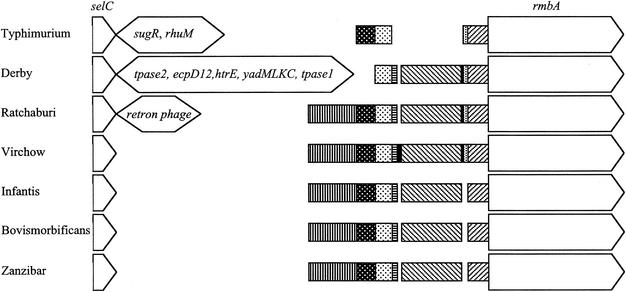
Comparison of the selC region of serovars Bovismorbificans, Infantis, Virchow, and Zanzibar with that of serovar Typhimurium showed that there was a deletion immediately after selC in these four serovars. A 4.6-kb DNA sequence of SPI-3 of serovar Typhimurium was reduced to around 0.7 kb in each of the four serovars. Southern hybridization using probe 3A showed that the genes sugR and rhuM were not present in the 0.7-kb region. Sequence analysis at the left end of SPI-3 revealed that the four serovars had nucleotide sequences similar to each other's but that serovar Zanzibar lacked an EcoRV restriction site at base 831 relative to the end of selC and that serovar Virchow had a 10-bp tandem duplication of the sequence from 454 to 463 and a 30-bp insertion between base 702 and base 732 (Fig. 2). DNA database searches showed that the sequence from base 219 to 438 of the region after selC in serovars Bovismorbificans, Infantis, Virchow, and Zanzibar was similar to the sequence between bases 4251 and 4470 of SPI-3 of serovar Typhimurium (GenBank accession no. AF106566). The nucleotide sequence after the first 0.7 kb of SPI-3 of the four serovars (from base 712 in serovar Virchow and base 693 in serovars Bovismorbificans, Infantis, and Zanzibar) was similar to SPI-3 of serovar Typhimurium (Fig. 2). The sequence from base 54 to base 190 had significant similarity to part of a putative transposase B gene of IS3 of Erwinia herbicola (GenBank accession no. AF327445). The sequence from base 439 to 691 (711 in serovar Virchow) in the four serovars had no significant similarity to any sequence in the GenBank database, but from base 466 the serovar Virchow sequence, including the 10-bp tandem repeat, had significant similarity to the serovar Typhi genome sequence at http://www.sanger.ac.uk/Projects/S_typhi/blast_server.shtml (23).
FIG. 2.
Variant SPI-3 regions in serovars Derby, Ratchaburi, Virchow, Infantis, Bovismorbificans, and Zanzibar compared to serovar Typhimurium. The regions encoding selC and rmbA are shown as arrows indicating the direction of transcription. Narrower boxes indicate the sequences shared by some of the serovars, with shading used to distinguish the different regions. The large insertions in serovars Typhimurium, Derby, and Ratchaburi and the genes they encode are shown as double-headed arrows. The insertions are shown at 1/10 the scale of the other regions.
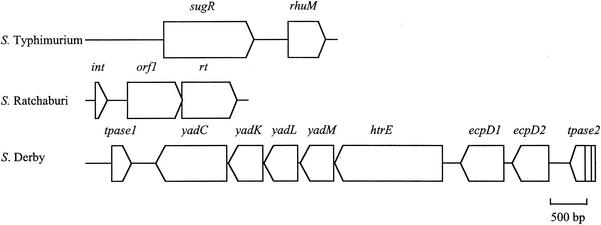
Even though the size of the PCR product from selC to misL of serovar Ratchaburi (5.5 kb) and that predicted for serovar Typhimurium (6.2 kb) were similar, the RFLP and Southern hybridization patterns of these two serovars were quite different. Only the 1.6- and 0.7-kb fragments of the serovar Ratchaburi RFLP pattern (3, 1.6, and 0.7 kb) reacted with probe 3. The hybridization pattern obtained with serovar Ratchaburi was similar to those of serovars Bovismorbificans, Infantis, Virchow, and Zanzibar, suggesting that the 3-kb fragment next to selC in serovar Ratchaburi had a nucleotide sequence different from that within SPI-3 of serovar Typhimurium. Characterization of this sequence determined that much of it was very similar to the nucleotide sequence of E. coli retron phage ΦR73 (33). The initial 334 bases were very similar to the first 334 bases of the ΦR73 phage genome inserted next to selC in E. coli and included the 5′ end of a truncated integrase gene, which extended to the end of the 334-base region (ftp://jb:jb@ftp.vet.unimelb.edu.au/). The region from base 338 to base 449 had some similarity to the region from 10061 to 10172 of the ΦR73 phage genome (GenBank accession no. ECOLAA). The following serovar Ratchaburi sequence (base 470 to base 2586) had high similarity to a region extending from 10375 to 12487 of the ΦR73 sequence and encoded ORFs homologous to the full length of an ORF of unknown function and a reverse-transcriptase (RT) gene in ΦR73 (Table 3; Fig. 3). The following region of serovar Ratchaburi (base 2792 to base 3510) was very similar to the region extending from base 1 to base 732 of SPI-3 in serovar Virchow, with the exception of the 10-bp tandem repeat that was also absent from this region in serovars Infantis, Bovismorbificans, and Zanzibar. The sequence from base 3511, as with all the other serovars, corresponded to SPI-3 of serovar Typhimurium.
TABLE 3.
ORFs predicted within the insertion in serovar Ratchaburi SPI3
| ORF | Locationa | Size (bp) | G+C (%) | E valueb | Identity (%) | Description of homologous gene (accession no.) |
|---|---|---|---|---|---|---|
| int | 162-371 | >210 | 46.7 | 3.70 × 10−36 | 95 | Integrase of φ-R73 (A42465) |
| orf | 711-1664 | 954 | 30.7 | 3.60 × 10−128 | 82.3 | ORF of φ-R73 (ECOLAA)c |
| rt | 1651-2602 | 951 | 31.9 | 2.30 × 10−173 | 76 | Reverse transcriptase of φ-R73 (E41830) |
Nucleotide position in the sequenced DNA.
Expected value obtained using BlastP and FastA (ORF only) to search the GenBank nonredundant bacterial database.
Greatest identity obtained with ORF (10,611 to 11,554 bp) of accession ECOLAA.
FIG. 3.
Insertion at left-hand end of SPI-3 in serovars Ratchaburi and Derby compared to that in serovar Typhimurium, with selC at the left side of each schematic. Open reading frames are shown as arrows indicating the direction of transcription. Lines in tpase2 of serovar Derby indicate interruptions to the reading frame by a nonsense mutation and a deletion creating a frame shift.
EcoRV digestion of the PCR products obtained using primers selC/F and misL/R showed that the regions from selC to misL in serovar Derby and serovar Hessarek were similar to each other but different from those of all other serovars examined. An insertion of approximately 10 kb replaced a 4.6-kb region in serovar Typhimurium. Southern hybridization showed that no genes within SPI-3 of serovar Typhimurium were located in this 10-kb fragment. Therefore, the genes sugR and rhuM were absent in these two serovars. The complete sequence of the insertion was determined for serovar Derby. This revealed that the insertion contained seven fimbrial genes, including structural, assembly, and chaperone genes, all with the same transcriptional orientation (ftp://jb:jb@ftp.vet.unimelb.edu.au/ [Table 4]). The order of six of these seven genes corresponded exactly with that of their homologues in E. coli K12. The seventh gene appeared to be a duplication of the ecpD gene. This apparent fimbrial operon was flanked by two truncated transposase genes with greatest similarity to transposases in E. herbicola and Shigella sonnei. The 129-bp sequence of serovar Derby immediately following selC was more similar to LEE of enteropathogenic E. coli (75% identity over 128 bp) and the SHI-2 pathogenicity island of Shigella flexneri (68% identity over 96 bp) than any other Salmonella sequence. The right-hand end of the insertion (bases 8779 to 9216) had a high level of similarity to IS630 of S. sonnei and included the 5′ end of a transposase gene, albeit with a nonsense mutation (codon 17) and a 14-base deletion (at base 9041) that caused a frame shift. Similarity between the insertion in serovar Derby and the sequence found in SPI-3 in serovar Typhimurium occurred 80 bases 3′ to the similarity between serovar Typhimurium and serovars Ratchaburi, Bovismorbificans, Infantis, Virchow, and Zanzibar (Fig. 2). The insertion of 263 bases (relative to serovar Typhimurium) found in serovars Ratchaburi, Bovismorbificans, Infantis, Virchow, and Zanzibar within the SPI-3 sequences preceding rmbA was also found in serovar Derby (Fig. 2). Like serovars Ratchaburi, Bovismorbificans, Infantis, and Zanzibar, serovar Derby did not contain the 10-base tandem repeat seen in serovar Virchow. Limited sequencing confirmed that serovar Hessarek was very similar to serovar Derby between base 21 and base 808.
TABLE 4.
ORFs predicted within the insertion in S. Derby SPI3
| ORF | Locationa | Size (bp) | G+C (%) | E valueb | Identity (%) | Description of homologous gene product (accession no.) | Species |
|---|---|---|---|---|---|---|---|
| Tpase1 | 438-774 | >337 | 47.6 | 4.30 × 10−21 | 47 | Putative transposase A (AAG53985) | E. herbicola |
| yadC | 1193-2419 | 1,227 | 39.4 | 2.50 × 10−37 | 33.5 | Putative fimbrial-like protein (AAC73246) | E. coli 0157:H7 |
| yadK | 2446-3036 | 591 | 46.4 | 1.20 × 10−56 | 46.2 | Putative fimbrial-like protein (AAG54440) | E. coli K12 |
| yadL | 3055-3636 | 582 | 49.1 | 2.70 × 10−34 | 38.8 | Putative fimbrial-like protein (AAC73248) | E. coli K12 |
| yadM | 3678-4259 | 582 | 45.5 | 1.60 × 10−49 | 46 | Putative fimbrial-like protein (AAG54442) | E. coli 0157:H7 |
| htrE | 4276-6750 | 2,475 | 46.9 | 0.0 | 64 | Probable porin/fimbrial assembly protein (AAC73250) | E. coli K12 |
| ecpD1 | 6897-7643 | 747 | 45.6 | 6.60 × 10−91 | 55 | Probable pilin chaperone similar to PapD (AAC73251) | E. coli K12 |
| ecpD2 | 7784-8422 | 639 | 49.3 | 4.80 × 10−172 | 100 | Probable pilin chaperone similar to PapD (AAC73251) | E. coli K12 |
| Tpase2c | 8779-9110 | >245 | 52.8 | 7.50 × 10−46 | 88 | Transposase (BAA85092) | S. sonnei |
Nucleotide position in the sequenced DNA.
Expected value obtained using BlastP and FastA (ORF only) to search the GenBank nonredundant bacterial database.
ORF contains one nonsense codon and a deletion of 14 bases compared to homologue. Analysis based on longest ORF in serovar Derby sequence.
Variation between Salmonella serovars within SPI-4.
Southern blots of chromosomal DNA digested with EcoRV were found to be the most suitable for analysis of SPI-4. Digestion with HindIII produced patterns that were too variable, and BglII yielded a number of fragments of similar size that could not be clearly differentiated. The hybridization patterns obtained using EcoRV-digested chromosomes showed that SPI-4 genes A, B, C, D, E, and F were stable for all serovars examined, whereas the regions containing genes G, H, I, J, K, L, M, N, O, P, Q, and R showed variation (ftp://jb:jb@ftp.vet.unimelb.edu.au/). However, all differences were attributable to loss or gain of restriction endonuclease cleavage sites, rather than insertions or deletions.
Variation between Salmonella serovars within SPI-5.
The EcoRV SPI-5 hybridization patterns of serovars Dublin, Bovismorbificans, Enteritidis, Hessarek, Infantis, and Virchow chromosomal DNA were the same and differed from those of all the serovar Typhimurium and serovar Choleraesuis strains, which had the same pattern as each other. Serovars Derby, Heidelberg, Ohio, Ratchaburi, and Zanzibar had patterns different from those of the other serovars (ftp://jb:jb@ftp.vet.unimelb.edu.au/).
The primers 5/F and 5/R were used in a PCR to amplify SPI-5 from all serovars tested. The PCR products from serovars Bovismorbificans, Choleraesuis, Enteritidis, Heidelberg, Hessarek, Infantis, Ratchaburi, Virchow, Typhimurium, Zanzibar, and Dublin were approximately 6.5 kb, and digestion with EcoRV yielded patterns that either were identical to those of serovar Dublin or differed only by the loss or gain of cleavage sites. Serovars Derby and Ohio yielded PCR products of approximately 7 kb.
Digestion with DraI and double digestion with EcoRV and DraI revealed that the variation in the restriction patterns of serovars Derby and Ohio might occur in sopB, pipC, or pipD but that the region containing pipD, orfX, and pipA was conserved.
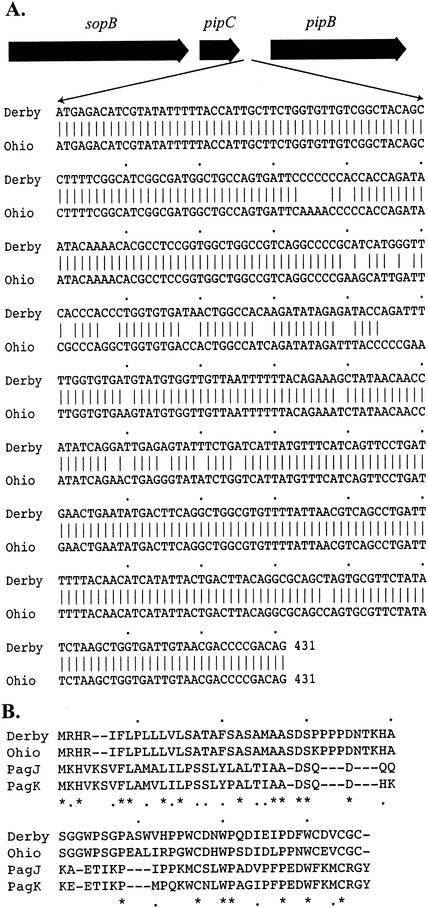
Further study using the primers SopB/F and SopB/R to amplify sopB from different serovars showed that sopB was the same size as in serovar Dublin but that there was an additional EcoRV cleavage site within sopB of serovars Derby, Ohio, Ratchaburi, and Zanzibar. Primers pipC/F and pipB/R were used to amplify chromosomal DNA of these serovars. The PCR yielded a 1.5-kb product in both serovar Derby and serovar Ohio, whereas serovar Dublin and the other serovars examined yielded only a 1-kb product. The DNA sequences of the 1.5-kb PCR products were determined, and it was found that serovar Derby and serovar Ohio had pipC and pipB nucleotide sequences similar to those of serovar Dublin, but an extra 431 bp was inserted within the intergenic region between pipC and pipD (Fig. 4). The 431-bp insertions in serovars Derby and Ohio were similar to each other and might encode a small ORF. Searches of the databases showed that this ORF had 24% amino acid identity and 42% amino acid similarity to the PagK or PagJ protein of serovar Typhimurium (Fig. 4).
FIG. 4.
(A) The 431-bp insertion in serovars Derby and Ohio, between pipC and pipB of SPI-5. (B) Predicted amino acid sequence of this insertion compared to PagJ and PagK, showing 24% identity and 42% similarity. Asterisks and dots indicate identical and similar amino acid residues, respectively.
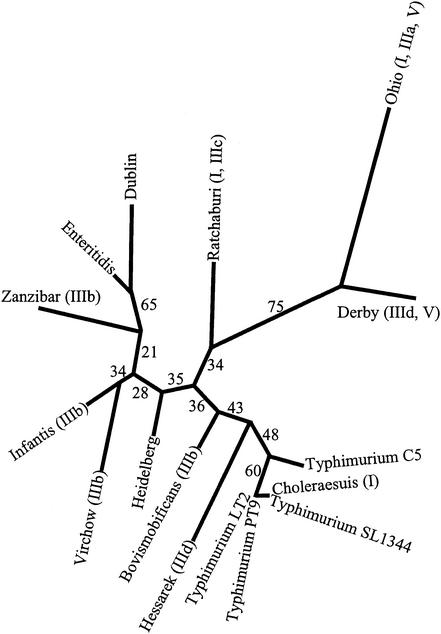
Phylogenetic relationships between serovars.
The phylogenetic inference using SPI restriction sites suggested that serovar Derby and serovar Ohio had a close genetic relationship to each other, and similarly, serovar Dublin and serovar Enteritidis were closely related (Fig. 5). The statistical analysis of the tree topology supported these inferences, as did the distribution of some of the insertions and deletions in the SPIs. Some serovars were grouped as closely related, but the distribution of the deletions and insertions in the SPIs did not concur with the inferred relationships. For example, serovars Choleraesuis, Hessarek, and Typhimurium strains LT2, SL1344, C5, and PT9 had a close relationship in the phylogenetic tree, but only serovar Choleraesuis had a deletion in SPI-1 and only serovar Hessarek had an insertion within SPI-3. However, the tree topology in these regions was not strongly supported by statistical analysis, implying that further data may be needed to confirm the recency of these events.
FIG. 5.
The phylogenetic tree for the pathogenicity islands of 13 Salmonella serovars inferred using restriction site data and Restml of PHYLIP 3.6. Major genetic events identified in the pathogenicity islands (insertions and deletions compared to serovar Typhimurium) are indicated as follows: (I), deletion of avrA and replacement with a 0.2-kb sequence; (IIIa), deletion within sugR; (IIIb), deletion of sugR and rhuM and replacement with a 0.7-kb sequence; (IIIc), deletion of sugR and rhuM and replacement with a 3-kb sequence; (IIId), deletion of sugR and rhuM and replacement with a 10-kb sequence; and (V), insertion of a 0.5-kb sequence within SPI-5. The number next to each branch indicates the confidence that can be placed on that branch as determined by bootstrap resampling of the restriction site data 100 times, inferring the best tree for each set of resampled data and then inferring a consensus tree using Consensus (PHYLIP 3.6). Each number indicates the percentage of the trees inferred from the resampled data that contained that branch.
DISCUSSION
The serovars chosen for this study included the host-specific serovars Choleraesuis (pigs) and Dublin (cattle), which, in nonendemic herds, cause invasive disease, with localization in the meninges, lungs, joints, and reproductive tract (21, 37). Both these serovars also cause invasive disease in other species, including humans. Serovar Virchow and serovar Enteritidis can cause invasive disease in humans but are not common causes of disease in chickens, the food source implicated in many human cases. Serovars Typhimurium and Heidelberg, and to a lesser extent serovars Derby and Infantis, are common causes of enteric salmonellosis in humans and other animals. Serovars Ohio, Hessarek, Zanzibar, Bovismorbificans, and Ratchaburi are relatively rare causes of disease and are principally associated with enteritis (13). With the exception of serovar Typhimurium, the variation observed in the SPIs was conserved within a serovar. Most of the isolates used in this study were independent isolates obtained from sick animals, humans, or the environment of outbreaks. For example, one isolate of serovar Virchow was of human origin, isolated in Queensland, Australia, and another isolate originated from poultry in Victoria, Australia. The serovar Enteritidis strains were isolated from two different countries but had identical RFLP patterns in the SPI regions.
Although the bootstrap values obtained for some of the branches on the tree inferred from the restriction site data were relatively low, the phylogenetic relationships between seven of the serovars used in our study (serovars Enteritidis, Dublin, Infantis, Heidelberg, Typhimurium, Choleraesuis, and Derby) have been studied previously using multilocus enzyme electrophoresis (2, 5, 30), and for these serovars, our phylogenetic analysis based on the restriction endonuclease maps in the five pathogenicity islands yielded a tree with a topology very similar to those obtained previously using multilocus enzyme electrophoresis. The other six serovars in our study have not been included in multilocus enzyme electrophoresis studies, so the inference of phylogenetic relationships is dependent on the restriction endonuclease cleavage data. The benefit of using restriction map data across all five islands, rather than restricted sequence data on one or a few genes, is that these data are less likely to be influenced by horizontal transfer of individual segments within the islands, a process that has been suggested by recent work (6). Indeed, such horizontal transfer between serovars might be expected to result in low bootstrap values, no matter what data were used, since the consensus tree would represent a compromise between multiple regions within the same genome with differing evolutionary histories.
Major genetic variation was detected in SPI-1, SPI-3, and SPI-5 for some serovars examined. SPI-2 and SPI-4 were found to be relatively conserved, with only minor variation in the presence or absence of restriction endonuclease sites. The only significant variation within SPI-1 was found in the avrA gene. The detection of the deletion of avrA within serovars Choleraesuis, Ohio, and Ratchaburi confirmed and extended the previous studies (15) that found the deletion of this gene in serovars Typhi, Choleraesuis, Ohio, Montevideo, Othmarschen, Nienstaedten, and Arizona and examined the sequence of this region in serovar Typhi and serovar Choleraesuis. While AvrA is secreted by the SPI-1 type III secretion system (15), its expression is not regulated by HilA or InvF (8). Although it is similar to YopJ of Yersinia pseudotuberculosis, expression of AvrA in either serovar Dublin or Y. pseudotuberculosis showed that it does not exert a YopJ-like activity on cytokine expression or in killing macrophages (29).
While avrA is deleted in the host-adapted serovars Typhi and Choleraesuis, it is present in serovar Dublin, another host-specific serovar, and it is present in serovars from a range of hosts and with a range of pathogenicities. There is no clear phylogenetic relationship between the serovars containing a deletion in avrA (Fig. 5), suggesting that an essentially identical deletion may have occurred several times during the evolution of different Salmonella serovars. This, and the similarity of the sequences that have replaced the deleted region in the different serovars, suggests that the deletion has been generated by insertion and imperfect excision of a site-specific, transmissible genetic element, such as a phage or insertion element, or alternatively by horizontal transfer of a discrete region of the SPI. The characterization of the full significance and genesis of this deletion will depend on the determination of a function for AvrA and on detection of the sequence that has been substituted for avrA in some other location.
The general conservation within the remainder of the SPI-1 region confirmed the proposition (26) that the genes within SPI-1 were an early acquisition. It has been suggested (22) that SPI-1 was acquired before the divergence of S. bongori and S. enterica, whereas SPI-2 was acquired by S. enterica after its split from S. bongori. Our studies found that all the genes within SPI-2 were conserved and that the structure of SPI-4 was also conserved in all the serovars examined, with the only variation identified being loss or gain of restriction sites, presumably as a result of single base changes.
An insertion encoding 58 amino acids with similarity to PagJ and PagK between PipC and PipB in SPI-5 was found in serovars Derby and Ohio (Fig. 4). The genes pagJ and pagK are PhoP-activated genes and are nearly identical to each other (14). The PhoPQ regulatory system is necessary for activation of invasion genes in response to environmental signals. The distribution of pagJ and pagK is limited; Southern blot analysis identified hybridizing sequences only in serovar Typhimurium and serovar Enteritidis and not in serovar Typhi or serovar Paratyphi or a range of other Enterobacteriaceae (14). However, there has not been any investigation of the distribution of these genes in other serovars. The amino acid sequences within and around pagJ and pagK are similar to those of proteins associated with mobile or extrachromosomal elements, including transposases, phage proteins, and proteins encoded on plasmids (10, 14, 28, 35). Deletion of either pagK or pagJ does not attenuate virulence in mice, but this may be because they have similar or identical functions. Thus far, no single mutation of a PhoP-activated gene has resulted in significant changes in the phenotype (14). It is possible that the insertion in SPI-5 is a remnant of pagJ and pagK from a previous recombination event or that it is a functional homologue of these genes. The phylogenetic analysis suggested that the event that led to the insertion in SPI-5 was likely to have been a single event, since the serovars carrying this insertion were clustered on the tree, and the topology of the tree in this area was strongly supported by the statistical analysis (Fig. 5).
The position of the genetic variation seen in SPI-3 in our work was similar to that seen for different subspecies (3), using specific regions of each gene as hybridization probes. However, the serovars examined in our studies were all pathogenic serovars within subspecies I. The high prevalence of genetic variation within SPI-3 in subspecies I and the similarity of some of the inserted sequences to other virulence-associated genes provides more information on Salmonella diversity, since information to date is derived mainly from studies of serovars Typhimurium and Typhi.
Most genetic variation in SPI-3 occurred at the left-hand end next to selC in the region containing genes sugR and rhuM. This region is the integration site for many pathogenicity islands of enteric bacteria, including E. coli and Shigella, and also the retron phage of E. coli. In serovar Ohio there was a deletion of 0.5 kb in sugR. The roles of SugR and RhuM have not yet been reported, but their position suggests that they may have been acquired independently of the remainder of SPI-3 and, even though the intergenic region between them is 580 bp, they are likely to form an operon because a lacZ fusion to this intergenic region resulted in some β-galactosidase activity (3). The link between these two genes is further supported by the deletion of both of them in many serovars.
The similarity of the variation at the left end of SPI-3 in serovars Bovismorbificans, Infantis, Virchow, and Zanzibar suggests a close relationship between these serovars, and this was supported by phylogenetic analysis (Fig. 5).
The insertion that replaced sugR and rhuM in serovar Ratchaburi and that lay immediately adjacent to selC had a high similarity to retron phage ΦR73, which is located in a similar position on the chromosome of E. coli K-12. The P4-like ΦR73 prophage is 12.6 kb and is flanked by 29-bp direct repeats and integrated 3′ to selC (33). Even though part of the prophage is similar to bacteriophage P4, it has a unique region at the right end that differs from P4. The retron-Ec73 region encodes the gene for RT and the genes msr, msd, and orf316. The low similarity of RT-Ec73 to other bacterial RT genes, for example, 27% identity to RT-Ec67 of E. coli (more than 47 amino acid residues), 25% identity to RT-Ec86 of E. coli (more than 43 amino acid residues), and 26% identity to RT-Mx65 of Myxococcus xanthus (more than 174 amino acid residues), indicates that it is equally distant from the other bacterial RTs (33). The high level of DNA sequence similarity between RT-Ec73 and the insertion in serovar Ratchaburi implies a much closer relationship than that between RT-Ec73 and other bacterial RTs. However, the insertion in serovar Ratchaburi contains a significant deletion compared to the full-length phage, with more than 80% of the genome missing and the integrase gene disrupted. This suggests that the phage genome in serovar Ratchaburi is no longer fully competent, although at least two ORFs from the phage, including the RT gene, appear to be translationally competent. The presence of retron phage sequences within an SPI suggests that such phages may also have played a role in the evolution of SPIs. It is well established that retroviruses of eukaryotes can acquire host sequences. It may be that retron phages similarly acquire and transfer sequences from one host to another.
The 10-kb insertions adjacent to selC in serovars Derby and Hessarek were similar to each other but not to any SPI-3 genes. The majority of the insertion appeared to be composed of an operon for synthesis of fimbriae, although, surprisingly, this operon was most similar to an E. coli fimbrial operon. The inserted operon was bracketed by the remnants of two distinct transposases, suggesting the involvement of at least two different insertion elements in the evolution of this sequence. It is notable that both of the major insertions observed in SPI-3 were likely to have been derived from E. coli, implying that the evolution of pathogenicity islands is an ongoing process in pathogenic salmonellae and that the sources of the elements are likely to be other enteric bacteria. A surprising finding was that although serovars Hessarek and Derby do not appear to be closely related phylogenetically (Fig. 5), they shared the same insertion, suggesting that this element has inserted independently into different serovars or that horizontal transfer of this region has occurred.
The possibility that a single event led to the deletion of sugR and rhuM in serovars Bovismorbificans, Infantis, Virchow, and Zanzibar is refuted by the observation that exactly the same apparent deletion was observed in serovar Ratchaburi, although in this case the deletion was replaced by an inserted sequence derived from a retron phage. Furthermore, a very similar deletion appears to have occurred during the insertion of the fimbrial operon into serovar Hessarek and serovar Derby. For serovar Typhi, sugR and rhuM have also been substantially deleted, with pseudogene remnants, pseudogenes of transposase genes, and two short ORFS with no significant similarity to other sequences in the databases. These findings suggest that there is a region within SPI-3, immediately adjacent to selC and 5′ to rmbA, that is particularly prone to deletion and/or insertion of transposable elements from a variety of sources and that the acquisition of the sugR/rhuM region in serovar Typhimurium is likely to have been a relatively recent event. It seems most probable that the ancestral SPI-3 sequence in this region was most similar to the sequence for serovars Bovismorbificans, Infantis, and Zanzibar, with multiple insertions, deletions, and short sequence duplications giving rise to the variations seen with the other serovars. It is clear that the region immediately following selC has been the target of a variety of insertion sequences, including at least two distinct transposable elements, similar to IS3 of E. herbicola and IS630 of S. sonnei, and a retron phage most similar to ΦR73 of E. coli. A fourth element is likely to have introduced the sugR/rhuM region into some serovars.
The high level of variability of DNA sequences adjacent to selC and the similarity of selC-related DNA sequences of other enteric bacteria to those in the serovars investigated here imply the occurrence of genetic transfer of sequences in this region between enteric bacteria and imply that in contrast to the other SPIs, SPI-3 may be still evolving through major sequence acquisitions.
Although the Salmonella serovars used in these studies cause a range of syndromes in both humans and animals, the major genetic variations identified were not significantly correlated with prominent differences in clinical features, host range, or levels of virulence. It may be that the variant genes are not important in pathogenesis or that there are other Salmonella genes that have the same action. Alternatively, the effect of these deletions and insertions may be reflected in less obvious benefits, for instance, in maintenance hosts that rarely suffer from disease or in survival in the environment. Understanding such issues will require a broader focus on the molecular biology of the ecology of these organisms.
This study has shown that even in relatively recent evolutionary history there have been a number of major genetic events shaping SPI-3, all of which appear to be focused on one specific region. Further studies of this region with a range of other serovars, using the techniques developed in this work, may reveal the range of transmissible elements that can contribute to the ongoing evolution of pathogenicity islands and thus provide a better understanding of the history of the more stable islands in both Salmonella and other enteric bacteria.
Supplemental information.
Additional information on this study, including oligonucleotide primers used, PCR conditions, maps of restriction endonuclease sites in different serovars, and aligned sequences, can be obtained from our website at ftp://jb:jb@ftp.vet.unimelb.edu.au/.
Acknowledgments
P.A. was supported by an AusAid Scholarship, a Melbourne University Faculty Research Scholarship, and a scholarship from Kasetsart University.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Beltran, P., J. M. Musser, R. Helmuth, J. J. Farmer III, W. M. Frerichs, I. K. Wachsmuth, K. Ferris, A. C. McWhorter, J. G. Wells, A. Cravioto, et al. 1988. Toward a population genetic analysis of Salmonella: genetic diversity and relationships among strains of serotypes S. choleraesuis, S. derby, S. dublin, S. enteritidis, S. heidelberg, S. infantis, S. newport, and S. typhimurium. Proc. Natl. Acad. Sci. USA 85:7753-7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanc-Potard, A. B., F. Solomon, J. Kayser, and E. Groisman. 1999. The SPI-3 pathogenicity island of Salmonella enterica. J. Bacteriol. 181:998-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd, E. F., F.-S. Wang, T. S. Whittam, and R. K. Selander. 1996. Molecular genetic relationships of the salmonellae. Appl. Environ. Microbiol. 62:804-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, E. F., F. S. Wang, P. Beltran, S. A. Plock, K. Nelson, and R. K. Selander. 1993. Salmonella reference collection B (SARB): strains of 37 serovars of subspecies I. J. Gen. Microbiol. 139:1125-1132. [DOI] [PubMed] [Google Scholar]
- 6.Brown, E. W., M. L. Kotewicz, and T. A. Cebula. 2002. Detection of recombination among Salmonella enterica strains using the incongruence length difference test. Mol. Phylogenet. Evol. 24:102-120. [DOI] [PubMed] [Google Scholar]
- 7.Collazo, C. M., and J. E. Galan. 1997. The invasion-associated type-III protein secretion system in Salmonella—a review. Gene 192:51-59. [DOI] [PubMed] [Google Scholar]
- 8.Eichelberg, K., and J. E. Galan. 1999. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and HilA. Infect. Immun. 67:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 10.Figueroa-Bossi, N., S. Uzzau, D. Maloriol, and L. Bossi. 2001. Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol. Microbiol. 39:260-272. [DOI] [PubMed] [Google Scholar]
- 11.Galan, J. E. 1996. Molecular genetic bases of Salmonella entry into host cells. Mol. Microbiol. 20:263-271. [DOI] [PubMed] [Google Scholar]
- 12.Galan, J. E., and R. Curtiss. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. USA 86:6383-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilbert, R. J., and D. Roberts. 1990. Foodborne gastroenteritis, p. 490-512. In L. H. Collier (ed.), Topley and Wilson's principles of bacteriology, virology and immunity, 8th ed., vol. 3. Edward Arnold, London, United Kingdom.
- 14.Gunn, J. S., W. J. Belden, and S. I. Miller. 1998. Identification of PhoP-PhoQ activated genes within a duplicated region of the Salmonella typhimurium chromosome. Microb. Pathog. 25:77-90. [DOI] [PubMed] [Google Scholar]
- 15.Hardt, W.-D., and J. E. Galan. 1997. A secreted Salmonella protein with homology to an avirulence determinant of plant pathogenic bacteria. Proc. Natl. Acad. Sci. USA 94:9887-9892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hensel, M., T. Nikolaus, and C. Egelseer. 1999. Molecular and functional analysis indicates a mosaic structure of Salmonella pathogenicity island 2. Mol. Microbiol. 31:489-498. [DOI] [PubMed] [Google Scholar]
- 17.Hoiseth, S. K., and B. A. D. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as vaccines. Nature 291:238-239. [DOI] [PubMed] [Google Scholar]
- 18.Hone, D., R. Morona, S. Attridge, and J. Hackett. 1987. Construction of defined galE mutants of Salmonella for use as vaccines. J. Infect. Dis. 156:167-174. [DOI] [PubMed] [Google Scholar]
- 19.Lilleengen, K. 1948. Typing Salmonella typhimurium by means of bacteriophage. Acta Pathol. Microbiol. Scand. Suppl. 77:11-125. [Google Scholar]
- 20.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 21.McDonough, P. L., D. Fogelman, S. J. Shin, M. A. Brunner, and D. H. Lein. 1999. Salmonella enterica serotype Dublin infection: an emerging infectious disease for the northeastern United States. J. Clin. Microbiol. 37:2418-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ochman, H., and E. A. Groisman. 1996. Distribution of pathogenicity islands in Salmonella spp. Infect. Immun. 64:5410-5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 24.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence analysis. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rakeman, J. L., H. R. Bonifield, and S. I. Miller. 1999. A HilA-independent pathway to Salmonella typhimurium invasion gene transcription. J. Bacteriol. 181:3096-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salender, R. K., J. Li, and K. Nelson. 1996. Evolutionary genetics of Salmonella enterica, p. 2691-2707. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Maganasik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), E. coli and Salmonella: cellular and molecular biology, vol. 2. ASM Press, Washington, D.C.
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., vol. 1, 2, and 3. Cold Spring Harbour Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Sanger, F., A. R. Coulson, G. F. Hong, D. F. Hill, and G. B. Petersen. 1982. Nucleotide sequence of bacteriophage lambda DNA. Mol. Biol. 162:729-773. [DOI] [PubMed] [Google Scholar]
- 29.Schesser, K., J.-M. Dukuzumuremyi, C. Cilio, S. Borg, T. S. Wallis, S. Pettersson, and E. E. Galyov. 2000. The Salmonella YopJ-homologue AvrA does not possess YopJ-like activity. Microb. Pathog. 28:59-70. [DOI] [PubMed] [Google Scholar]
- 30.Selander, R. K., P. Beltran, N. H. Smith, R. Helmuth, F. A. Rubin, D. J. Kopecko, K. Ferris, B. D. Tall, A. Cravioto, and J. M. Musser. 1990. Evolutionary genetic relationships of clones of Salmonella serovars that cause human typhoid and other enteric fevers. Infect. Immun. 58:2262-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selander, R. K., J. Li, and K. Nelson. 1996. Evolutionary genetics of Salmonella enterica, p. 2691-2707. In H. E. Umbarger (ed.), Escherichia coli and Salmonella. Cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C.
- 32.Shea, J. E., M. Hensel, C. Gleeson, and D. W. Holden. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 93:2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun, J., M. Inouye, and S. Inouye. 1991. Association of a retroelement with a P4-like cryptic prophage (retronphage FR73) intregrated into the selenocystyl tRNA gene of Escherichia coli. J. Bacteriol. 173:4171-4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tominaga, A., S. Ikemizu, and M. Enomoto. 1991. Site-specific recombinase genes in three Shigella subgroups and nucleotide sequences of a pinB gene and an invertible B segment from Shigella boydii. J. Bacteriol. 173:4079-4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warren, G., and D. J. States. 1993. Identification of protein coding regions by database similarity search. Nat. Genet. 3:266-272. [DOI] [PubMed] [Google Scholar]
- 37.Wilcock, B. P., C. H. Armstrong, and H. J. Olander. 1976. The significance of the serotype in the clinical and pathological features of naturally occurring porcine salmonellosis. Can. J. Comp. Med. 40:80-88. [PMC free article] [PubMed] [Google Scholar]
- 38.Wong, K. K., M. McClelland, L. C. Stillwell, E. C. Sisk, S. J. Thurston, and J. D. Saffer. 1998. Identification and sequence analysis of a 27-kilobase chromosomal fragment containing a Salmonella pathogenicity island located at 92 minutes on the chromosome map of Salmonella enterica serovar Typhimurium LT2. Infect. Immun. 66:3365-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wood, M. W., M. A. Jones, P. R. Watson, S. Hedges, T. S. Wallis, and E. E. Galyov. 1998. Identification of a pathogenicity island required for Salmonella enteropathogenicity. Mol. Microbiol. 29:883-891. [DOI] [PubMed] [Google Scholar]