Abstract
This study focuses on the mechanisms for hydrogen peroxide detoxification in Synechococcus sp. strain PCC 7942. To gain better understanding of the role of different routes of hydrogen peroxide detoxification, we inactivated tplA (thioredoxin-peroxidase-like), which we recently identified. In addition, we inactivated the gene encoding catalase-peroxidase and examined the ability to detoxify H2O2 and to survive oxidative stress in both of the single mutants and in the double mutant. Surprisingly, we observed that the double mutant survived H2O2 concentrations that the single catalase-peroxidase mutant could not tolerate. This phenotype correlated with an increased ability of the double mutant to detoxify externally added H2O2 compared to the catalase-peroxidase mutant. Therefore, our studies suggested the existence of a hydrogen peroxide detoxification activity in addition to catalase-peroxidase and thioredoxin-peroxidase. The rate of detoxification of externally added H2O2 was similar in the wild-type and the TplA mutant cells, suggesting that, under these conditions, catalase-peroxidase activity was essential for this process and TplA was dispensable. However, during excessive radiation, conditions under which the cell might experience oxidative stress, TplA appears to be essential for growth, and cells lacking it cannot compete with the wild-type strain. Overall, these studies suggested different physiological roles for various cellular hydrogen peroxide detoxification mechanisms in Synechococcus sp. strain PCC 7942.
Cells of photosynthetic organisms possess substantial sources of reactive oxygen species (ROS) in addition to the ROS-producing processes common to all living cells. This stems from the need to harvest light energy for their phototrophic metabolism. Accordingly, these organisms face the challenge of capturing light energy efficiently while avoiding oxidative damage caused by a surplus of absorbed light. Excessive excitation stems from an imbalance between energy absorption and dissipation rates (11, 19, 22). Although high photon flux causes excess excitation, light intensity is not the only effective factor. Any environmental parameter that would slow down anabolism (nonoptimal temperature, nutrient limitation, etc.) would decrease photochemical dissipation and therefore may lead to oxidative stress caused by excess absorbed light.
Certain excited pigment molecules may produce ROS, i.e., singlet chlorophyll while decaying through triplet chlorophyll, causes the formation of singlet oxygen (11, 19). The photosynthetic electron transport chain may produce damaging oxygen species as well. For example, production of ROS may occur on the acceptor side of photosystem II through electron flow from phaeophytin or semiquinone. In addition, it is commonly accepted that under excess of absorbed light, photosystem I can reduce molecular oxygen to superoxide anion (Mehler reaction [1]), which can be converted by superoxide dismutase to H2O2. Apart from being potentially harmful by itself, in the presence of reduced metal ions H2O2 may be converted to hydroxyl radical, a highly reactive and damaging entity. A recent study (13) provided evidence for the involvement of A-type flavoproteins in photoreduction of O2 by electron transfer from photosystem I (the Mehler reaction) in Synechocystis sp. strain PCC 6803 (referred to here as Synechocystis strain 6803). One of the flavoproteins essential for photoreduction of O2 has been shown to reduce O2 directly to water in vitro (35). It has therefore been suggested that in contrast to eukaryotes, the Mehler reaction in cyanobacteria does not produce ROS.
To avoid the damaging consequences of hydrogen peroxide production, cells possess various enzymes that detoxify this compound (9, 11, 21, 24, 27, 32, 33). Hydrogen peroxide-detoxifying enzymes are traditionally classified as catalases or peroxidases (27). Enzymes of the former group convert H2O2 to water and molecular oxygen, whereas the peroxidases rely on electrons supplied by reductants of low molecular weight, such as ascorbate and glutathione, to reduce H2O2 or organic hydroperoxides. In general, catalases exhibit lower affinity for H2O2 and higher kcat compared to the peroxidases. A third type of hydrogen peroxide-detoxifying enzyme, one unique to prokaryotes, is designated catalase-peroxidase (38). As a typical catalase, this enzyme converts H2O2 to water and molecular oxygen but in vitro, it also shows a peroxidase activity with o-dianisidine or pyrogallol as substrates. As suggested by the substrate specificity (23), it is likely that, in vivo, the cyanobacterial catalase-peroxidase functions as a catalase rather than a peroxidase.
An additional antioxidant enzyme that drew attention recently is thioredoxin-peroxidase, a protein conserved from bacteria to mammals. The exact role and mode of regulation of these enzymes are not clear; however, it is established that they rely on reduced thioredoxin (6, 10, 15-18) which in a photosynthetic cell may be reduced by photosystem I through ferredoxin (5).
During studies on acclimation of Synechococcus sp. strain PCC 7942 (referred to here as Synechococcus) to nutrient limitation and high-light stress, we identified an open reading frame (ORF) that is highly homologous to thioredoxin-peroxidases (tplA, for thioredoxin-peroxidase-like). To obtain a comprehensive picture of the relevance of TplA, as well as of the catalase-peroxidase, for hydrogen peroxide detoxification and cell survival, we inactivated the genes encoding for these enzymes and characterized each of the single mutants, as well as the double mutant.
MATERIALS AND METHODS
Culture conditions and competition experiments.
Synechococcus and its mutants were cultured as previously described (7). For growth experiments under high-light conditions the cultures were illuminated with 800 μmol of photons m−2 s−1 (provided by halogen lamps cooled with water jackets) and bubbled with air. The same conditions were applied for “competition tests” in which a mixture of equal amounts of wild type and a certain mutant (optical density at 750 nm [OD750] of 0.02) served to initiate the experiment. The mixed culture was diluted daily to the original OD; thus, the absorbed light remained fairly constant throughout the experiment. The generation time under these conditions was about 12 h. The number of wild-type and mutant cells in the culture at a given time was assessed by diluting and plating the cultures on solid medium, followed by restreaking of single colonies on plates containing the appropriate antibiotic for the selection of a specific mutant.
Construction of mutant strains, DNA manipulation, and isolation of RNA.
It is often observed that genes, the products of which are involved in cell growth and survival under certain environmental conditions, are clustered. Therefore, we sequenced further downstream of nblR, the gene encoding for a response regulator that is essential for cell survival during high-light illumination and nutrient starvation (30). This led to identification of an ORF highly homologous to thioredoxin-peroxidases located ca. 2 kb downstream of nblR. A 2-kb EcoRV/EcoRI subclone was used for interposon inactivation of tplA by insertion of a spectinomycin resistance cassette at an XcmI site.
The gene encoding for catalase-peroxidase of Synechococcus, katG, was amplified from genomic DNA by using primers designed according to the published sequences (23) (5′-CCAAACACCAACAGGAGA-3′ and 5′-GTTGCGATAGCATCGTGA-3′). The PCR product was cloned into a pGEM-T vector (Promega). Digestion by ClaI was used to delete a 609-bp fragment, which was replaced by a kanamycin resistance cassette. Each of the plasmids containing either the inactivated tplA or katG was used to transform the wild-type strain to yield the tplA mutant (TplΩ) and the katG mutant (KatΩ), respectively.
The double mutant, KatTplΩ, characterized throughout the present study was obtained by transforming the plasmid bearing the inactivated katG into TplΩ. Where indicated, a double mutant constructed by transforming KatΩ with inactivated tplA was analyzed. PCR on genomic DNA isolated from the transformants by using specific primers confirmed the complete segregation and replacement of the native gene with the inactivated one.
Molecular techniques were performed according to standard procedures (29). For transcript analyses, cultures were illuminated with 70 or 400 μmol of photons m−2 s−1 provided by fluorescent lamps. RNA isolation and Northern blot analyses were performed as described earlier (8). The 0.5-kbp EcoRI/XmnI fragment containing part of tplA was used as a probe.
Determination of viability and analysis of H2O2 detoxification by whole cells.
To determine viability after H2O2 treatment, cells at the exponential growth phase were adjusted to an OD750 of 0.5, and H2O2 was applied to a range of final concentrations (15 μM to 10 mM, depending on the specific strain). After incubation for 24 h under incandescent light (50 μmol of photons m−2 s−1), aliquots were placed onto solid medium plates to assess viability.
For analysis of H2O2 detoxification, cells harvested during exponential growth (5,000 × g, 5 min) were washed once with an equal volume of 20 mM NaNO3, 0.3 mM MgSO4 · 7H2O, and 0.2 mM CaCl2 · 2H2O (the concentrations of these salts in the growth medium) and resuspended in the same solution to an OD750 of 0.5. H2O2 was then added to the desired final concentrations. Aliquots were drawn at various times and diluted when required, and the amount of H2O2 was determined by oxidation of Fe2+ in the presence of xylenol orange (36) using a FOX assay (36) by measuring the absorbance with a Multiskan RC (Labsystems) with a 560-nm band-pass filter. The initial slope of the absorbance as a function of time served to quantify the amount of H2O2. Qualitative determination of H2O2 was obtained by carrying out color development to its maximal level. In these assays, yellow color represents the absence and purple represents the presence of H2O2.
Where indicated, 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) was added to a final concentration of 10 μM. Green light (50 μmol of photons m−2 s−1) was provided with a 520-nm filter.
As previously shown for Synechocystis strain 6803 (34), we noticed that the ability to decompose H2O2 was strongly dependent on cell density. Therefore, all of the analyses presented in the present study were performed at a fixed cell density. Changing the cell density, however, affected the absolute H2O2 concentrations that the strains could tolerate and detoxify but not the relative differences between the mutants.
The nucleotide sequence of the genomic region containing tplA was deposited in GenBank under accession no. AF492495.
RESULTS
Identification of a thioredoxin-peroxidase-like gene.
During studies of the acclimation of Synechococcus to nutrient limitation and high-light stress, we have identified an ORF highly homologous to thioredoxin-peroxidases (also termed peroxiredoxins), enzymes conserved from microorganisms to mammals. For example, the Synechococcus TplA exhibits 88% similarity to sll0755 of Synechocystis strain 6803 (http://www.kazusa.or.jp/cyanobase/), 82% similarity to BAS1 from barley (2), 75% similarity to human thioredoxin-peroxidase (31), and 55% similarity to AhpC from Escherichia coli (4). The highest homology was observed between TplA and a subfamily of thioredoxin-peroxidases designated 2-Cys peroxiredoxins. These enzymes function as homodimers in which a disulfide bridge between two conserved cysteins is formed during catalysis (6). Sequence alignment indicated that TplA possesses the two conserved cystein residues essential for catalysis (not shown). Although the physiological role of these enzymes is not completely understood, it has been established that peroxiredoxins reduce hydrogen peroxide or alkyl hydroperoxides by using electrons from thioredoxin (10, 12, 14, 25).
Viability of wild-type and mutant strains after H2O2 treatment.
To study the physiological role of TplA in Synechococcus, we inactivated its gene and characterized the mutant with respect to its ability to survive externally added H2O2, to detoxify H2O2, and to grow during high-light illumination, conditions under which the cells might experience oxidative stress. In order to gain better understanding of the ability of the cell to cope with oxidative stress, we also inactivated the gene encoding for catalase-peroxidase and characterized this mutant, as well as the double mutant lacking both genes.
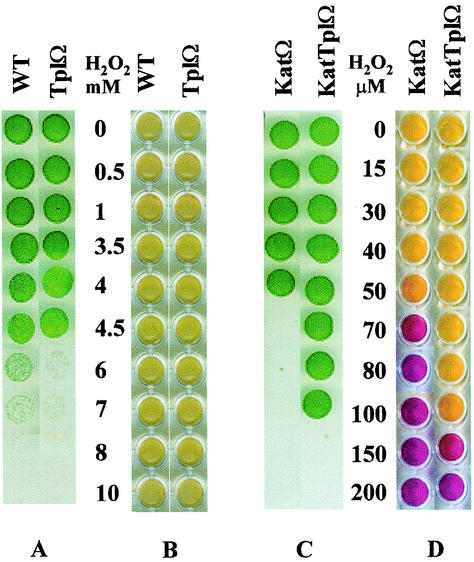
The various strains were exposed to H2O2 in liquid cultures for 24 h, followed by transfer of aliquots onto solid growth medium (Fig. 1). Strains possessing catalase-peroxidase (wild type and TplΩ) survived exposure to 50- to 100-fold-higher concentrations of H2O2 than strains lacking this activity (KatΩ and KatTplΩ) (Fig. 1A and C, respectively). Apparently, catalase-peroxidase is required for detoxification of high concentrations of externally added H2O2.
FIG. 1.
Cells spotted on solid growth medium (A and C) and qualitative assay for remaining H2O2 (B and D) after incubation for 24 h with the indicated concentrations of H2O2. Yellow represents the absence and purple represents the presence of H2O2. WT, wild type; TplΩ, tplA mutant; KatΩ, catalase-peroxidase mutant; KatTplΩ, double mutant. In the specific experiment shown, viability and residual H2O2 were determined simultaneously, and thus cells were prepared as described for the determination of H2O2 content (see Materials and Methods).
The ability to detoxify H2O2 likely contributes to cell survival during oxidative stress and, therefore, we expected the double mutant to be the most sensitive to the application of H2O2. Surprisingly, the double mutant survived higher H2O2 concentrations than did the catalase-peroxidase mutant (Fig. 1C). Since the mutants were fully segregated (see Materials and Methods), the data suggested that inactivation of both katG and tplA resulted in the induction of H2O2 detoxification activity that is supplementary to catalase-peroxidase and thioredoxin-peroxidase.
Detoxification of H2O2 by wild-type and mutant cultures.
Qualitative determination of the residual H2O2 remaining after 24 h of incubation was performed on the cultures that served to assess the viability (Fig. 1). These analyses showed that the higher ability of the double mutant to survive H2O2 (compared to the catalase-peroxidase mutant) coincided with its increased capacity to detoxify these levels of H2O2 (Fig. 1C and D). These experiments also indicated that catalase-possessing strains were able to detoxify 100- to 200-fold-higher concentrations of H2O2 than catalase-lacking strains (Fig. 1B and D). It is interesting that in catalase-lacking strains (KatΩ and KatTplΩ), cell survival under a certain concentration of H2O2 correlated with the ability to decompose it (Fig. 1C and D). On the other hand, catalase-possessing strains (wild type and TplΩ) could detoxify H2O2 concentrations that caused cell death (Fig. 1A and B). Presumably, KatG remains active in H2O2-damaged cells, whereas peroxidases can no longer function if their electron source is exhausted.
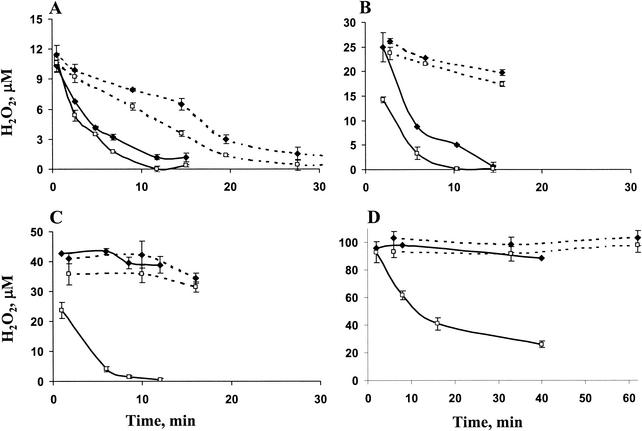
The contribution of the different enzymes to H2O2 detoxification was assessed by monitoring the rate of H2O2 decomposition by the various strains. Comparisons between the rates of detoxification by illuminated and darkened cultures were performed since photosynthetic electron transport may be essential for replenishment of the reducing equivalents required for peroxidase activity.
The KatΩ and the double mutant decomposed relatively low concentrations of H2O2 (15 or 30 μM) at a similar rate under either illumination or darkness (Fig. 2A and B). Challenging these strains with higher H2O2 levels (50 and 100 μM) revealed a substantial difference between these mutants (Fig. 2C and D). The KatTplΩ mutant completely eliminated 50 μM within 12 min of incubation in the light and was capable of partially reducing even 100 μM H2O2 within 40 min of illumination, unlike KatΩ, which did not reduce such concentrations (Fig. 2C and D). These experiments confirmed our finding (Fig. 1) that the ability of the double mutant to survive high H2O2 levels originated from its ability to detoxify them.
FIG. 2.
Time courses of decomposition of H2O2 by illuminated (solid lines) or darkened (dashed lines) cultures of catalase-peroxidase mutant (♦) or the double mutant KatTplΩ (□). At time zero, 15 μM (A), 30 μM (B), 50 μM (C), or 100 μM (D) H2O2 was added to the cultures.
In the dark, incubation with H2O2 did not reveal a difference between KatΩ and the double mutant; both strains were unable to reduce either 50 or 100 μM H2O2 (Fig. 2C and D). These two mutants, however, detoxified lower H2O2 concentrations similarly, although at a slower rate compared to cultures incubated in the light. The dependence of H2O2 detoxification in these strains on light suggests a requirement for reductants produced by the photosynthetic electron transport chain. This was further supported by the lack of light-dependent H2O2 detoxification upon addition of DCMU or illumination with a nonphotosynthetic (green) light (not shown). Therefore, reduction of relatively low concentrations of H2O2 in the dark (Fig. 2A and B) may depend on the availability of cellular pools of reduced compounds.
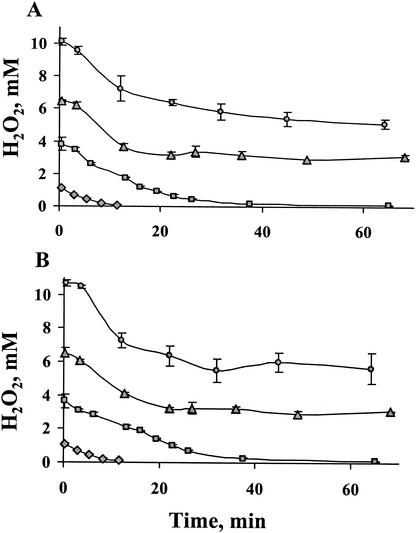
In accordance with their ability to survive high levels of H2O2, wild type and TplΩ were capable of detoxifying relatively high concentrations of H2O2 compared to KatΩ and the double mutant; 1 and 4 mM H2O2 were completely decomposed within 15 and 65 min, respectively. High concentrations of H2O2, such as 7 and 10 mM, were only partially reduced within 65 min (Fig. 3), presumably due to accumulating damage to enzyme activities. In addition, the rate of detoxification of H2O2 in the wild type and the tplA mutant were essentially identical (Fig. 3), suggesting that catalase activity was not affected by the inactivation of tplA.
FIG. 3.
Time courses of decomposition of H2O2 by illuminated cultures of wild type (A) and tplA mutant (B). At zero time, 1 mM ( ), 4 mM (
), 4 mM ( ), 7 mM (
), 7 mM ( ), and 10 mM (
), and 10 mM ( ) H2O2 was added to the cultures. Identical curves were obtained with darkened cultures.
) H2O2 was added to the cultures. Identical curves were obtained with darkened cultures.
Identical rates of H2O2 detoxification were observed in illuminated (Fig. 3) or darkened cultures (not shown) of the wild-type and TplΩ cells. Since peroxidases but not catalases rely on reduced constituents originating from photosynthetic electron transfer, this result indicates that the catalase function of the catalase-peroxidase provides the dominant activity required for detoxification of the high H2O2 levels. Catalase also appears to be the principal means for decomposition of lower concentration of H2O2; TplΩ detoxified 5 μM H2O2 much faster than did KatΩ (not shown).
Response of mutant strains to high-light conditions during growth.
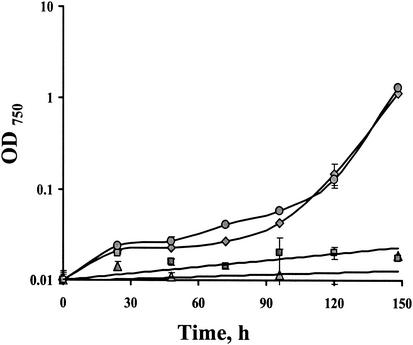
In addition to detoxification of externally supplied H2O2 (Fig. 1 to 3), it was important to assess the ability of the various strains to cope with environmental conditions that might result in oxidative stress, such as growth under intensive radiation. Mutants where tplA was inactivated (TplΩ and KatΤplΩ) grew very poorly under these conditions, whereas KatΩ grew similarly to the wild-type cells (Fig. 4). Furthermore, growth experiments performed under high-light irradiance demonstrated that TplΩ and the double mutant were outcompeted by the wild type, specifically under these conditions (Fig. 5), but not when the light intensity was 100 μmol of photons m−2 s−1 (not shown). Taken together, these data may indicate a crucial role for TplA under a high-light regime. Furthermore, the novel H2O2 detoxification activity observed in the double mutant cannot compensate for the lack of TplA under these conditions. An additional support for the importance of TplA for high-light growth was provided by the fivefold rise in the abundance of the tplA transcript (Fig. 6) after the high-light treatment.
FIG. 4.
Growth as measured by the change in OD750 of wild type ( ), KatΩ (
), KatΩ ( ), TplΩ (
), TplΩ ( ), and the double mutant KatTplΩ (
), and the double mutant KatTplΩ ( ) during illumination with high-light intensity.
) during illumination with high-light intensity.
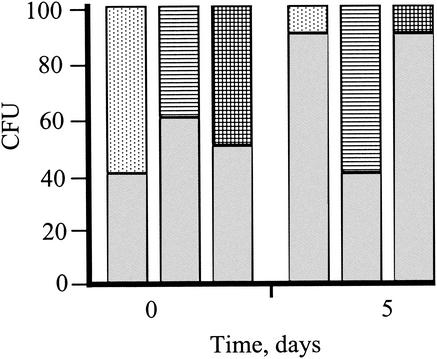
FIG. 5.
Percentage of CFU of wild type (shaded), KatΩ (hatched), TplΩ (dotted), and the double mutant KatTplΩ (check pattern) at time zero and after 5 days of growth in a mixed culture illuminated with 800 μmol of photons m−2 s−1 (see Materials and Methods for details of the competition tests).
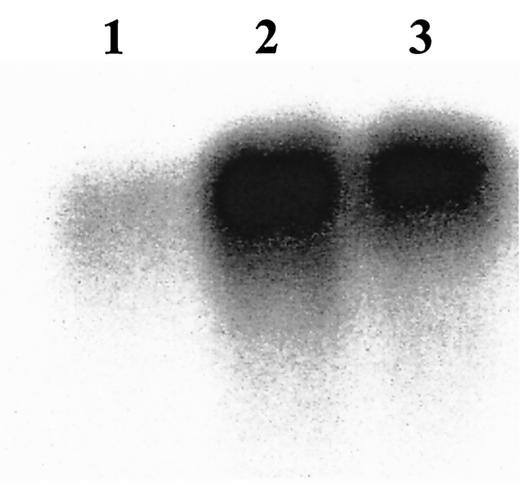
FIG. 6.
Northern blot hybridization with a tplA specific probe to RNA isolated from Synechococcus grown at 70 μmol of photons m−2 s−1 (lane 1) or exposed to 400 μmol of photons m−2 s−1 for 45 min (lane 2) or 2 h (lane 3). The probe hybridized to a single band of ca. 800 bp. Each lane was loaded with 5 μg of RNA.
DISCUSSION
A novel H2O2 detoxification activity in Synechococcus.
The double mutant, KatTplΩ, survived and detoxified H2O2 concentrations that were not tolerated or reduced by the KatΩ (Fig. 1 and 2). These observations indicated a novel H2O2 detoxifying activity, in addition to that of TplA and catalase-peroxidase. Presumably, the supplementary H2O2 detoxification activity is expressed in the absence of both KatG and TplA. This novel activity relies on reductants produced by the photosynthetic electron transport chain, as suggested by its dependence on photosynthetic light and inhibition by DCMU. It is plausible that inactivation of both tplA and katG caused the induction of the novel H2O2 detoxification activity. An alternative possibility is that a spontaneous mutation present in TplΩ contributed to the phenotype of the double mutant since the latter was raised by inactivation of katG in TplΩ. However, a TplKatΩ mutant that was raised by inactivation of tplA in KatΩ also exhibited higher resistance to H2O2 compared to KatΩ (not shown), thus supporting the hypothesis that the lack of both KatG and TplA causes the induction of a supplementary H2O2-decomposing activity.
Interestingly, the activity observed in the katG mutant (most likely originating from TplA) is more susceptible to H2O2 treatment than is the activity exhibited by the double mutant. For example, the lack of detoxification of 50 or 100 μM H2O2 by the katG mutant implies rapid oxidation of the reduced substrates required for H2O2 detoxification and/or damage to the enzyme itself. On the other hand, the activity present in the double mutant was not impaired under these conditions (Fig. 2), although the addition of 200 μM of H2O2 eliminated this activity as well (not shown).
Although originally classified as lacking ascorbate peroxidase (21), Synechococcus was shown to possess an ascorbate peroxidase-like activity (28). This activity, which appeared to be cytosolic, copurified with a small non-heme iron-containing compound and was heat stable. This ascorbate-peroxidase-like activity probably does not account for the H2O2-detoxifying activity observed in the double mutant since initial characterization of this activity indicated that it is membrane associated and heat inactivated, unlike the ascorbate-dependent activity. Currently, we are characterizing the novel H2O2-detoxifying activity observed in the double mutant to clarify its nature and physiological significance.
Differential role for catalase-peroxidase and TplA.
Analyses of viability (Fig. 1) and of H2O2 detoxification (Fig. 1 to 3) indicated that catalase-peroxidase is essential for survival and the elimination of relatively high concentrations of externally added H2O2. Studies of Synechocystis strain 6803 and its katG mutant also suggested such a role (34). Despite its importance for the elimination of relatively high concentrations of H2O2, catalase-peroxidase seems to be dispensable for growth under high-light illumination (Fig. 4 and 5), conditions under which the cell might experience severe oxidative stress (26). Further, although it reduced externally added H2O2 as efficiently as the wild type (Fig. 3), a mutant lacking TplA grew very poorly and was outcompeted by wild-type cells (Fig. 4 and 5, respectively) during excessive radiation. Under the latter conditions, the double mutant exhibited a phenotype similar to that of TplΩ (Fig. 4 and 5). This indicated that TplA was crucial for growth under high-light conditions and the supplementary H2O2-detoxifying activity observed in the double mutant could not replace it. Evidence for the importance of thioredoxin-peroxidase for high-light growth is also provided by the effect of high-light treatment on the quantum yield in Synechocystis strain 6803 and its thioredoxin-peroxidase mutant; the latter exhibited a lower quantum yield compared to wild-type cells (18). In addition to the high-light-sensitive phenotype of the TplA-lacking mutants (Fig. 4 and 5), transcription induction of tplA observed in wild-type cells, following high-light illumination (Fig. 6), also supports the physiological relevance of this gene product under high-light conditions. The tplA transcript appears to be monocistronic (the probe hybridized to a 800-bp transcript), and therefore it is likely that the phenotype is not a result of a polar effect.
As is apparent from our study, neither catalase-peroxidase nor the additional peroxidase activity present in the double mutant could substitute for TplA function during high-light growth. Given that the photosynthetic electron transport chain reduces thioredoxin, one may speculate that a thioredoxin-dependent peroxidase provides an energy dissipation route, which might be critical under excessive excitation. Support for this hypothesis comes from studies on Synechococcus by Miller et al. (20), which showed photosystem II-dependent oxygen evolution and fluorescence quenching consequent on photoreduction of H2O2. Further support is provided by experiments on Synechocystis strain 6803 showing that when linear electron flow is blocked, the addition of t-butyl-hydroperoxide enabled oxygen evolution by wild type but not by a thioredoxin-peroxidase mutant (37). Production of organic hydroperoxides during excessive radiation and their specific reduction by TplA may provide an additional explanation for the indispensable role of this enzyme.
It is important to point out that the Arabidopsis genome contains 10 ORFs showing homology to members of the peroxiredoxins (thioredoxin-peroxidases) family (10). Use of Arabidopsis plants expressing antisense to the chloroplast-located thioredoxin-peroxidase suggested a protective function for the enzyme (3). However, the exact role and physiological significance of these enzymes and that of peroxiredoxins identified in other plants is yet to be established. Sequences showing homology to catalase-peroxidase were not identified in the complete genomes of Anabaena sp. strain PCC 7120, Nostoc punctiforme ATCC 29133, Synechococcus strain WH 8102, and two Prochlorococcus marinus strains (MI9313 and MED4). In fact, the latter three marine organisms do not possess sequences that show homology to any catalase (mono-functional heme or manganese enzyme). On the other hand, each of the above-mentioned organisms possesses at least one sequence that is highly homologous to thioredoxin-peroxidase. These observations further emphasize the physiological significance of thioredoxin-peroxidases.
Acknowledgments
A.P. and A.U. contributed equally to this study.
This work was supported by United States-Israel Binational Science Foundation (grant 9800146) and by The Israel Science Foundation (grant 210/99).
REFERENCES
- 1.Asada, K. 1999. The water-water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50:601-639. [DOI] [PubMed] [Google Scholar]
- 2.Baier, M., and K. J. Dietz. 1996. Primary structure and expression of plant homologues of animal and fungal thioredoxin-dependent peroxide reductases and bacterial alkyl hydroperoxide reductases. Plant Mol. Biol. 31:553-564. [DOI] [PubMed] [Google Scholar]
- 3.Baier, M., and K. J. Dietz. 1999. Protective function of chloroplast 2-cysteine peroxiredoxin in photosynthesis: evidence from transgenic Arabidopsis. Plant Physiol. 119:1407-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Buchanan, B. B. 1984. The ferredoxin/thioredoxin system: a key element in the regulatory function of light in photosynthesis. Bioscience 34:378-383. [PubMed] [Google Scholar]
- 6.Chae, H. Z., S. J. Chung, and S. G. Rhee. 1994. Thioredoxin-dependent peroxide reductase from yeast. J. Biol. Chem. 269:27670-27678. [PubMed] [Google Scholar]
- 7.Collier, J. L., and A. R. Grossman. 1992. Chlorosis induced by nutrient deprivation in Synechococcus sp. strain PCC 7942: not all bleaching is the same. J. Bacteriol. 174:4718-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collier, J. L., and A. R. Grossman. 1994. A small polypeptide triggers complete degradation of light-harvesting phycobiliproteins in nutrient-deprived cyanobacteria. EMBO J. 13:1039-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dat, J. F., D. Inze, and F. Van Breusegem. 2001. Catalase-deficient tobacco plants: tools for in planta studies on the role of hydrogen peroxide. Redox Rep. 6:37-42. [DOI] [PubMed] [Google Scholar]
- 10.Dietz, K. J., F. Horling, J. Konig, and M. Baier. 2002. The function of the chloroplast 2-cysteine peroxiredoxin in peroxide detoxification and its regulation. J. Exp. Bot. 53:1321-1329. [PubMed] [Google Scholar]
- 11.Foyer, C. H., and G. Noctor. 2000. Oxygen processing in photosynthesis: regulation and signalling. New Phytol. 146:359-388. [Google Scholar]
- 12.Grant, C. 2001. Microreview: role of the glutathione/glutaredoxin and thioredoxin systems in yeast growth and response to stress conditions. Mol. Microbiol. 39:533-541. [DOI] [PubMed] [Google Scholar]
- 13.Helman, Y., D. Tchernov, L. Reinhold, M. Shibata, T. Ogawa, R. Schwarz, I. Ohad, and A. Kaplan. 2003. Genes encoding A-type flavoproteins are essential for photoreduction of O2 in cyanobacteria. Curr. Biol. 13:230-235. [DOI] [PubMed] [Google Scholar]
- 14.Hofmann, B., H. J. Hecht, and L. Flohe. 2002. Peroxiredoxins. Biol. Chem. 383:347-364. [DOI] [PubMed] [Google Scholar]
- 15.Ishii, T., K. Itoh, H. Sato, and S. Bannai. 1999. Oxidative stress-inducible proteins in macrophages. Free Radic. Res. 31:351-355. [DOI] [PubMed] [Google Scholar]
- 16.Ishii, T., M. Yamada, H. Sato, M. Matsue, S. Taketani, K. Nakayama, Y. Sugita, and S. Bannai. 1993. Cloning and characterization of a 23-kDa stress-induced mouse peritoneal macrophage protein. J. Biol. Chem. 268:18633-18636. [PubMed] [Google Scholar]
- 17.Kang, S. W., H. Z. Chae, M. S. Seo, K. Kim, I. C. Baines, and S. G. Rhee. 1998. Mammalian peroxiredoxin isoforms can reduce hydrogen peroxide generated in response to growth factors and tumor necrosis factor-alpha. J. Biol. Chem. 273:6297-6302. [DOI] [PubMed] [Google Scholar]
- 18.Klughammer, B., M. Baier, and K. J. Dietz. 1998. Inactivation by gene disruption of 2-cysteine-peroxiredoxin in Synechocystis sp. PCC 6803 leads to increased stress sensitivity. Physiol. Plant 104:699-706. [Google Scholar]
- 19.Krishna, K., and Niyogi. 1999. Photoprotection revisited: genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50:333-359. [DOI] [PubMed] [Google Scholar]
- 20.Miller, A. G., K. J. Hunter, S. J. O'Leary, and L. J. Hart. 2000. The photoreduction of H2O2 by Synechococcus sp. PCC 7942 and UTEX 625. Plant Physiol. 123:625-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyake, C., F. Michihata, and K. Asada. 1991. Scavenging of hydrogen-peroxide in prokaryotic and eukaryotic algae: acquisition of ascorbate peroxidase during the evolution of cyanobacteria. Plant Cell Physiol. 32:33-43. [Google Scholar]
- 22.Muller, P., X. P. Li, and K. K. Niyogi. 2001. Non-photochemical quenching: a response to excess light energy. Plant Physiol. 125:1558-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mutsuda, M., T. Ishikawa, T. Takeda, and S. Shigeoka. 1996. The catalase-peroxidase of Synechococcus PCC 7942: purification, nucleotide sequence analysis and expression in Escherichia coli. Biochem. J. 316:251-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noctor, G., and C. H. Foyer. 1998. Ascorbate and glutathione: keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49:249-279. [DOI] [PubMed] [Google Scholar]
- 25.Nordberg, J., and E. S. Arner. 2001. Reactive oxygen species, antioxidants, and the mammalian thioredoxin system. Free Radic. Biol. Med. 31:1287-1312. [DOI] [PubMed] [Google Scholar]
- 26.Patterson, C. O. P., and J. Myers. 1973. Photosynthetic production of hydrogen peroxide by Anacystis nidulans. Plant Physiol. 51:104-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Regelsberger, G., C. Jakopitsch, L. Plasser, H. Schwaiger, P. G. Furtmuller, G. A. Peschek, M. Zamocky, and C. Obinger. 2002. Occurrence and biochemistry of hydroperoxidases in oxygenic phototrophic prokaryotes (cyanobacteria). Plant Physiol. Biochem. 40:479-490. [Google Scholar]
- 28.Rozen, A., R. Mittler, Y. Burstein, and E. Tel-Or. 1992. A unique ascorbate peroxidase active component in the cyanobacterium Synechococcus PCC 7942 (R2). Free Radic. Res. Commun. 17:1-8. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., and W. D. Russell. 2001. Molecular Cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, New York, N.Y.
- 30.Schwarz, R., and A. R. Grossman. 1998. A response regulator of cyanobacteria integrates diverse environmental signals and is critical for survival under extreme conditions. Proc. Natl. Acad. Sci. USA 95:11008-11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shau, H., L. H. Butterfield, R. Chiu, and A. Kim. 1994. Cloning and sequence analysis of candidate human natural killer-enhancing factor genes. Immunogenetics 40:129-134. [DOI] [PubMed] [Google Scholar]
- 32.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2:188-194. [DOI] [PubMed] [Google Scholar]
- 33.Tel-Or, E., M. E. Huflejt, and L. Packer. 1986. Hydroperoxide metabolism in cyanobacteria. Arch. Biochem. Biophys. 246:396-402. [DOI] [PubMed] [Google Scholar]
- 34.Tichy, M., and W. Vermaas. 1999. In vivo role of catalase-peroxidase in Synechocystis sp. strain PCC 6803. J. Bacteriol. 181:1875-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vicente, J. B., C. M. Gomes, A. Wasserfallen, and M. Teixeira. 2002. Module fusion in an A-type flavoprotein from the cyanobacterium Synechocystis condenses a multiple-component pathway in a single polypeptide chain. Biochem. Biophys. Res. Commun. 294:82-87. [DOI] [PubMed] [Google Scholar]
- 36.Wolff, S. P. 1994. Ferrous ion oxidation in presence of ferric ion indicator xylenol orange for measurement of hydroperoxides. Methods Enzymol. 233:182-189. [Google Scholar]
- 37.Yamamoto, H., C. Miyake, K. J. Dietz, K. Tomizawa, N. Murata, and A. Yokota. 1999. Thioredoxin peroxidase in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 447:269-273. [DOI] [PubMed] [Google Scholar]
- 38.Zamocky, M., G. Regelsberger, C. Jakopitsch, and C. Obinger. 2001. The molecular peculiarities of catalase-peroxidases. FEBS Lett. 492:177-182. [DOI] [PubMed] [Google Scholar]