Abstract
The SlyA protein of Salmonella enterica serovar Typhimurium is a member of the MarR family of transcription regulators and is required for virulence and survival in professional macrophages. Isolated SlyA protein was able to bind a specific DNA target without posttranslational modification. This suggested that SlyA might not be activated by directly sensing an external signal but rather that the intracellular concentration of SlyA is enhanced in appropriate environments through the action of other transcription factors. Analysis of slyA transcription reveals the presence of a promoter region located upstream of the previously recognized SlyA repressed promoter. The newly identified upstream promoter region did not respond to SlyA but was activated by Mg(II) starvation in a PhoP-dependent manner. We present here evidence for a direct link between two transcription factors (PhoP and SlyA) crucial for Salmonella virulence.
The Salmonella transcription factor SlyA has been shown to be required for virulence in mice and for survival within tissues of the reticuloendothelial system (4, 7, 12). Moreover, Salmonella slyA strains are sensitive to products of the respiratory burst, have a stationary-phase survival defect, and are attenuated in the presence of macrophages (4, 24). It has been suggested that SlyA is particularly important in systemic salmonellosis (25). Further investigation of the slyA phenotype indicated that SlyA acts as both an activator and a repressor of gene expression (4). Reduced levels of FliC and IroN, elevated levels of PagC, and altered patterns of Omp proteins were found to be associated with a slyA lesion (24). Moreover, a recent proteomic analysis revealed that SlyA affected the levels of 23 proteins with a variety of roles, including the response to oxidative stress (23). Thus, SlyA is a global transcription factor that is important for pathogenesis.
SlyA is a member of the MarR family of transcription factors. Members of this family are found in bacteria and archaea and play important roles in bacterial virulence. For example, the Escherichia coli MarR and EmrR proteins regulate genes involved in antibiotic resistance, PecS from Erwinia chrysanthemi controls pectinase and cellulase production, and RovA of Yersinia spp. regulates invasin expression (16, 17, 20). Recently, the crystal structure of MarR was determined (2). This revealed that the protein is a dimer with each subunit possessing a winged-helix-turn-helix DNA-binding motif (2). SlyA has also been shown to be a homodimer that negatively regulates its own expression (24). The Salmonella SlyA protein has been purified from an E. coli expression strain and the isolated protein was able to recognize and preferentially bind at a DNA sequence consisting of an inverted repeat (TTAGCAAGCTAA) within the slyA promoter (24). Because the isolated SlyA protein was able to specifically bind target DNA, it was suggested that SlyA itself might not sense an environmental signal but that another transcription factor, responding to a relevant signal, upregulates slyA expression (24). As a consequence the intracellular concentration of SlyA would increase to a level at which SlyA is able to bind DNA and regulate the transcription of target genes. Thus, the starting point for the work described here was to investigate factors affecting the regulation of slyA expression. A new slyA promoter region was identified that was positively regulated by PhoP. Thus, a link was established between two transcription factors (PhoP and SlyA) crucial for Salmonella virulence.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in the present study are listed in Table 1. DNA manipulations were performed by using standard protocols (21). The various slyA promoter regions were amplified by PCR with primers designed to introduce EcoRI and BamHI restriction sites to allow ligation into pRW50 (13). The authenticities of the PCR products were established by automated DNA sequencing. The Salmonella phoP strain was constructed by linear transformation of ST12/75 carrying pTP223. Plasmid pTP223 encodes the lam, bet, and exo genes of phage lambda, under the control of the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible lac promoter (19). The lambda genes promote targeted recombination with short regions of homology. Two PCR primers (Table 1) containing 40 bases of phoP sequence and 20 bases of the chloramphenicol acetyltransferase (cat) cassette from pACYC184 (5) were used to amplify the entire cat cassette flanked by phoP sequences. The linear PCR product was introduced into ST12/75 (pTP223) by electroporation, and chloramphenicol-resistant colonies were selected at 37°C. The amplified DNA promoted the replacement of 696 bp of the phoP gene, including amino acids 1 to 203 of PhoP, by the cat cassette. After the strain was cured of pTP223, the location of the phoP::cat lesion was confirmed by PCR from genomic DNA of a representative transformant (JRG4844).
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides
| Strain, plasmid, or oligonucleotide | Relevant characteristics and/or sequencea | Source or reference |
|---|---|---|
| Strains | ||
| ST12/75 | Parental Salmonella strain | T. Wallis and S. J. Libby |
| ST12/75ΔslyA | slyA strain; Penr | T. Wallis and S. J. Libby |
| JRG4844 | ST12/75 phoP strain; Cmr | This work |
| Plasmids | ||
| pRW50 | A low-copy-number lac reporter vector; Tetr | 13 |
| pGS1459 (S1) | pRW50 containing the slyA promoter region (−593 to +165) | This work |
| pGS1384 (S2) | pRW50 containing the slyA promoter region (−237 to +165) | This work |
| pGS1474 (S3) | pRW50 containing the slyA promoter region (−152 to +165) | This work |
| pGS1475 (S4) | pRW50 containing the slyA promoter region (−108 to +165) | This work |
| pGS1476 (S5) | pRW50 containing the slyA promoter region (−68 to +165) | This work |
| pGS1477 (S6) | pRW50 containing the slyA promoter region (−46 to +165) | This work |
| pGS1534 (S7) | pRW50 containing the slyA promoter region (−593 to −136) | This work |
| pGS1536 (S8) | pRW50 containing the slyA promoter region (−360 to +165) | This work |
| pGS1535 (S9) | pRW50 containing the slyA promoter region (−292 to +165) | This work |
| pGS1557 (S10) | pRW50 containing the slyA promoter region (−238 to −136) | This work |
| pTP223 | lam, bet, and exo genes of phage lambda, under the control of the lac promoter | 19 |
| pACYC184 | Source of 3.6-kb cat cassette for the creation of the phoP mutant strain | 5 |
| Oligonucleotides | ||
| VN33 | Forward primer for the creation of phoP mutant; TTAAATAATGCCTGCCTCACCCTCTTTTCTTCAGAAAGAGAAGCCACTGGAGCACCTCAA | This work |
| VN34 | Reverse primer for the creation of phoP mutant; GCGTACGGTGGTAATGACATCGTGCGGATACTGGGCCTGTACGGGGAGAGCCTGAGCAAA | This work |
Penr, penicillin resistant; Cmr, chloramphenicol resistant; Tetr, tetracycline resistant. For oligonucleotides VN33 and VN34, the region of the phoP sequence is indicated in italics, and the 5′ end of the cat cassette sequence is shown in roman.
Growth conditions.
The growth media used in the present study are listed below. To measure the effects of progressive deletion of DNA from the slyA promoter region and for general culturing of bacteria, a rich medium (tryptone, 10 g liter−1; yeast extract, 5 g liter−1; NaCl, 5 g liter−1) was used. In other experiments a Tris minimal medium (100 mM Tris-HCl [pH 7.2], 11 mM glucose, 5 mM NH4Cl, 10 mM KH2PO4, 0.5 mM K2SO4, 0.1 mM CaCl2, 10 mM MgCl2) was used. For Mg(II)-starved cultures, the concentration of MgCl2 was reduced to 0.01 mM and the CaCl2 was omitted. For phosphate-starved cultures, KH2PO4 was reduced to 0.1 mM. For nitrogen-starved cultures NH4Cl was reduced to 0.5 mM. The chelator deferoxamine mesylate (0.3 mM) was used to sequester Fe(III) and, when indicated, FeCl3 was added to cultures at a final concentration of 0.1 mM. For investigation of the effects of pH on slyA expression a Bis-Tris minimal medium was used (8). Media were supplemented with antibiotics (tetracycline, 10 μg ml−1; chloramphenicol, 10 μg ml−1) when appropriate. The solid medium used for investigating the phenotype of the slyA mutant was Tris minimal medium (pH 7.2) containing 22 mM glucose, 0.5% agarose, and 10 or 0.25 mM MgCl2.
β-Galactosidase assay.
For β-galactosidase activity measurements (15), aerobic cultures were grown in shaking (250 rpm) 100-ml flasks containing 5 ml of the indicated medium at 37°C for either 6 h (exponential phase) or 16 h (stationary phase).
Transcript mapping by primer extension.
The transcription start point of the upstream slyA promoter(s) was determined by RNA extraction and primer extension. Total RNA was prepared from stationary-phase (24 h) strain ST12/75(pGS1534) grown aerobically in Mg(II)-starved Tris minimal medium by using a Qiagen RNeasy kit. For primer extension, the method outlined in the Qiagen Omniscript reverse transcription (RT) instruction manual was used with 10 μg of RNA and Omniscript reverse transcriptase (20 U; Qiagen). After ethanol precipitation the cDNA was fractionated on 6% urea-polyacrylamide gels for autoradiographic analysis. The gels were calibrated with a sequence ladder from the same DNA and primer.
RT-PCR.
For RT-PCR, total RNA was isolated from stationary-phase cultures grown in Bis-Tris minimal medium containing either 10 mM Mg(II) or 0.01 mM Mg(II). A specific oligonucleotide (CATCTCAGCGATCAGCGGCTC) designed to complement the 3′ end of the slyA gene was used to prime the Omniscript (Qiagen) reverse transcriptase (4 U). The resulting cDNA was used as the template for PCR amplification (using Taq polymerase) of fragments located within the slyA transcripts in the presence of [α-32P]dCTP (225 kBq). To prevent the amplification of any genomic DNA carried over during total RNA isolation, RNA preparations were digested with DNase I before cDNA production. The Mg(II)-responsive pagD transcript and the unresponsive dam transcript were used as positive and negative controls. The reactions were analyzed by polyacrylamide gel electrophoresis (6% Tris-borate-EDTA-buffered polyacrylamide gels) and autoradiography. Products were quantified by densitometry. The relevant primers were as follows: T1, TGGAATCGCCACTAGGTTCTG; T2, GCCAAGTGCGCACTATGTCTG; T3, GTAAGGGCAATCCTGTGGCGT; >T3, TTTTGAATTCGCAAAGCGTAAAGAGGGAGAGATC; pagD (reverse), CGTCATTGACTGGTGCGGACA; pagD (forward), CAGTTCAGGCCATTGTTCTGG; dam (reverse), CGACTCCTGGTTACAGA; and dam (forward), CGAGTGCCTTGTCGAACCTT.
RESULTS
Dissection of the slyA promoter region.
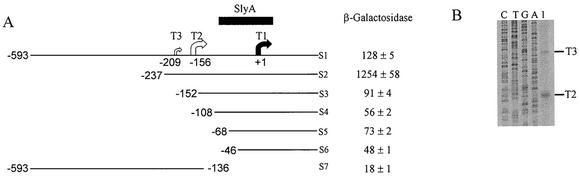
The starting point for this work was the construction of a series of slyA::lacZ promoter fusions in the low-copy-number plasmid pRW50 (13). In this series (S1 to S6, Fig. 1A), the DNA upstream of the previously mapped slyA transcription start (T1 in Fig. 1A) varied between 46 and 593 bp. Thus, measurement of β-galactosidase activity was used to identify promoter elements that influence expression of the Salmonella slyA gene. During aerobic growth deletion of the section between positions −593 and −237 enhanced slyA expression by ∼10-fold compared to the full-length promoter (compare S1 with S2) (Fig. 1A). This enhancement was abolished by further deletion to position −152 (S3). More extensive deletions reduced transcription still further, with the exception of the section extending from position −68 (S5), which had slightly greater activity than promoter sequences beginning at position −108 (S4) or position −46 (S6) (Fig. 1A). These data suggested that an activatory element was located between positions −237 and −152. Thus, a further slyA::lacZ fusion was created that encompassed the region from positions −593 to −136 (S7). Low levels of β-galactosidase activity were observed with this latter lacZ fusion, suggesting the presence of a weak slyA promoter located between positions −593 and −136. Therefore, primer extension was used to investigate whether any slyA transcripts initiate in this region. Two new transcripts, a minor product beginning at position −209 (T3) and a major product beginning at position −156 (T2), (numbering relative to T1) were detected (Fig. 1B). However, the sum of the activities of the downstream (represented by slyA::lacZ fusion S7) and upstream (represented by slyA::lacZ fusion S3) promoters did not approach the activity associated with S2, suggesting that there must be some synergistic interaction between the promoters that is partially dampened by the action of an as-yet-unidentified factor acting between positions −593 and −237.
FIG. 1.
Identification of a second slyA promoter region. (A) β-Galactosidase activities of aerobically grown cultures of ST12/75 each containing one of a series (S1 to S6) of slyA::lacZ fusions in which DNA upstream of the previously established transcript start (T1, solid arrow) (24), was progressively deleted. The slyA DNA ends at position +165 in this series of fusions. A further slyA::lacZ fusion (S7) in which the previously recognized slyA promoter (T1) was deleted was also analyzed. The solid box indicates the region of the promoter protected by SlyA in footprinting studies. Two newly identified slyA transcript starts (T2 and T3, open arrows; see panel B) are also indicated. β-Galactosidase activities were measured in duplicate from at least two independent cultures; the means ± the standard errors are shown. Units of β-galactosidase activity are as defined by Miller (15). (B) Primer extension analysis of RNA isolated form ST12/75(pGS1534). Lanes C, T, G, and A are the sequence ladders for this region of slyA; lane 1 shows the primer extension products from the slyA gene.
Effect of growth conditions on slyA expression.
To investigate whether conditions could be identified in which expression from the slyA::lacZ fusion S1 (Fig. 1A) could approach that observed with the S2 fusion, cultures were grown under a variety of conditions and β-galactosidase activities were measured (Table 2). Although expression levels did not reach those estimated for the S2 fusion, the experiments showed that the expression of slyA was reduced in nitrogen-starved cultures, slightly (∼50%) higher in phosphate-starved cultures, and significantly enhanced in Mg(II)-starved cultures. Moreover, the ∼4-fold enhancement in slyA expression under conditions of Mg(II) limitation suggests that the Mg(II)-responsive PhoP-PhoQ two-component system (9) might be involved in regulating slyA expression.
TABLE 2.
Expression of slyA::lacZ (S1) under different growth conditions
| Growth medium | Mean β-galactosidasea activity ± SE in:
|
|
|---|---|---|
| Exponential phase | Stationary phase | |
| Base medium | 99 ± 8 | 91 ± 5 |
| N starved | ND | 20 ± 1 |
| PO4 starved | ND | 137 ± 3 |
| Mg(II) starved | 134 ± 4 | 398 ± 12 |
The activities were measured in duplicate on at least two independent cultures. Cultures were grown under aerobic conditions in Tris-minimal medium at pH 7.2 containing 5 mM NH4Cl, 10 mM KH2PO4, 0.5 mM K2SO4, and 10 mM MgCl2 (base medium) at 37°C for 6 h (exponential phase) or 16 h (stationary phase). This medium was amended as follows: 0.5 mM NH4Cl (N starved), 0.1 mM KH2PO4, (PO4 starved), or 0.01 mM MgCl2 [Mg(II) starved]. ND, not determined.
It has previously been observed that under certain conditions slyA expression is maximal in stationary-phase cultures (4). Therefore, the response of the S1 slyA::lacZ fusion to growth under conditions of Mg(II) starvation was monitored by measuring β-galactosidase activities in exponential- and stationary-phase cultures grown in Mg(II)-replete (10 mM) and Mg(II)-starved (0.01 mM) conditions. The data obtained suggested that the response to Mg(II) starvation was more marked in stationary-phase cultures (∼4.4-fold enhancement) than in exponential-phase cultures (∼1.4-fold enhancement) (Table 2). Moreover, in the Mg(II)-replete cultures slyA expression was not enhanced in the stationary phase compared to the exponential-phase cultures (Table 2). This suggests that Mg(II) starvation is the trigger that promotes slyA expression in stationary-phase cultures.
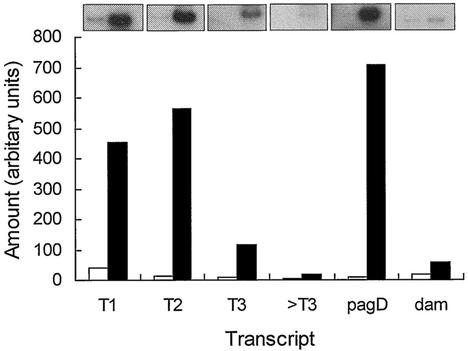
To test whether the plasmid-based transcription studies accurately reflected the normal regulation of the slyA gene on the chromosome total RNA was prepared from cultures grown under Mg(II)-starved and Mg(II)-replete conditions. RT-PCR showed that slyA transcripts extended as far as T3 (Fig. 2). The T2 transcript was highly induced under Mg(II)-starved conditions compared to Mg(II)-replete conditions (Fig. 2). Note that the T1 transcript was increased by a similar amount because it is embedded within the T2 transcript. A known Mg(II)-responsive transcript, pagD (10), behaved as expected, as did the PhoP-unresponsive (11) dam transcript. A forward primer located upstream of T3 (>T3) failed to yield significant levels of product under both growth conditions, suggesting that there are no further promoters upstream of T3. Therefore, it was concluded that the plasmid-based reporters were representative of chromosomal slyA regulation.
FIG. 2.
Effect of Mg(II) on transcription of chromosomal slyA. Expression of slyA was estimated by RT-PCR with total RNA (2 μg) isolated from Mg(II)-replete (open bars) and Mg(II)-starved (solid bars) stationary-phase cultures as the template. Oligonucleotide primers complementary to the T1, T2, and T3 transcript starts, as well as a primer complementary to a region upstream of T3 (>T3), were used to define the upstream limit of the slyA message. The Mg(II)-responsive pagD and unresponsive dam transcripts served as controls. Radiolabeled PCR products were separated on polyacrylamide gels, and the amount of product formed was estimated by quantitative densitometry of the corresponding autoradiographs. Representative autoradiographs are shown above the bar chart.
The effect of PhoP on slyA expression.
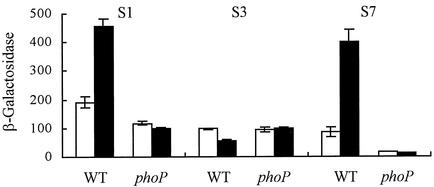
To test whether PhoP-mediated the enhanced expression of slyA under conditions of Mg(II) starvation, β-galactosidase activities were measured for parental and phoP strains containing the S1 slyA::lacZ promoter fusion. The data obtained show that slyA expression was enhanced in the Mg(II)-starved parental culture but that this was not the case for the phoP strain (Fig. 3). This suggests that PhoP either directly or indirectly activates slyA expression during exposure of bacteria to low-Mg(II) conditions.
FIG. 3.
Transcription from the upstream region of the slyA promoter is regulated by PhoP in response to Mg(II) starvation. β-Galactosidase activities were measured for cultures of ST12/75 and ST12/75ΔphoP containing the indicated slyA::lacZ fusion (S1, S3, or S7; see Fig. 1A) in Mg(II)-replete (open bars) and Mg(II)-starved (solid bars) minimal medium. β-Galactosidase activities were measured in triplicate from at least two independent cultures; means ± the standard errors are shown. Units of β-galactosidase activity are as defined by Miller (15).
To determine which region of the slyA promoter was responding to PhoP, expression from the slyA::lacZ promoter fusions S3 (T1 promoter only) and S7 (T2 and T3 promoters) was compared to that obtained with S1 (T1, T2, and T3 promoters). The activity of the S3 fusion was similar in both parental and phoP strains (Fig. 3). Moreover, in contrast to the S1 fusion, expression from the S3 fusion was lower in the parental Mg(II)-starved cultures (Fig. 3). This suggests that the promoter associated with transcript 1 (T1) is unresponsive to PhoP and Mg(II) availability. However, the activity of the S7 fusion did respond to Mg(II) starvation in a PhoP-dependent manner (Fig. 3).
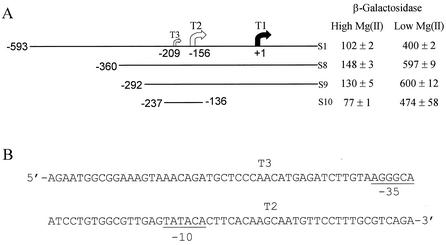
To further delimit the site of PhoP action, three more slyA::lacZ fusions (S8, S9, and S10; Fig. 4A) were created with deletions upstream of T2 and T3. The activities of these promoters did not approach that of the S2 fusion in rich medium (Fig. 1A), suggesting that the negatively acting element is located between positions −292 and −237 and that T1 is the probable source of the enhanced activity of the S2 fusion. The response to Mg(II) starvation of these new fusions was then tested. These experiments revealed that β-galactosidase activities from all three strains were elevated in Mg(II)-starved cultures. Consequently, it was concluded that the site of PhoP action was located between positions −237 and −136. When transcription from the S7 fusion (Fig. 1) was tested in a slyA strain, the β-galactosidase activities were similar [124 ± 4 Miller units in Mg(II)-replete cultures and 324 ± 31 Miller units in Mg(II)-starved cultures] to those obtained for the parental strain [87 ± 16 Miller units in Mg(II)-replete cultures and 402 ± 44 Miller units in Mg(II)-starved cultures], and thus it was concluded that SlyA does not regulate expression from the newly identified upstream slyA promoter.
FIG. 4.
The slyA T2 transcript is Mg(II) responsive. (A) β-Galactosidase activities of aerobic cultures of ST12/75 containing the indicated slyA::lacZ fusion (S1, S8, S9, or S10) in Mg(II)-replete and Mg(II)-starved minimal medium. β-Galactosidase activities were measured in duplicate from at least two independent cultures; means ± the standard errors are shown. Units of β-galactosidase activity are as defined by Miller (15). (B) Sequence of the PhoP-responsive slyA promoter. The locations of the 5′ ends of the major T2 and the minor T3 upstream slyA transcripts are indicated. Potential −10 and −35 elements associated with T2 are indicated.
A consensus PhoP box has been defined as a direct repeat of two heptanucleotide consisting of (T)G(T)TT(AA) by analysis of promoter regions of PhoP-regulated genes and by footprinting of the mgtA-treR intergenic region (9, 27). Inspection of the DNA sequence of the S10 slyA::lacZ fusion (positions −237 to −136) revealed no close matches to a PhoP box (Fig. 4B), suggesting that PhoP acts indirectly.
Among the genes upregulated by PhoP is pmrD (9). PmrD mediates transcriptional activation of genes that are regulated by the PmrA-PmrB two-component sensor regulator during growth in low concentrations of Mg(II). Alternatively, PmrA-PmrB can be activated by extracellular Fe(III) (26). Thus, if PhoP acts through PmrA to enhance slyA expression, the activity of the S10 slyA::lacZ fusion might be expected to respond to extracellular Fe(III) in both Mg(II)-replete and Mg(II)-starved cultures. However, experiments in which Tris-minimal medium was supplemented with Fe(III) or with the Fe(III) chelator deferoxamine revealed that expression from the S10 slyA::lacZ fusion was not influenced by extracellular Fe(III) (not shown). Accordingly, inspection of the S10 slyA DNA sequence did not reveal any good matches to the PmrA consensus, which consists of YTTAAKNNNNYTTAAK (1).
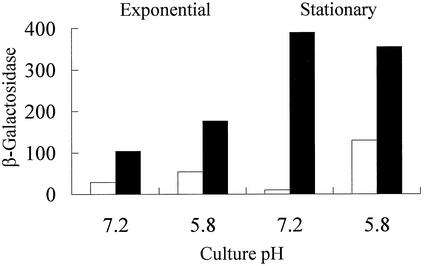
It has been shown that expression of a slyA::lacZ fusion possessing all of the transcript starts identified here is enhanced in the stationary phase (4). Experiments with the S1 slyA::lacZ fusion, which encodes all three slyA transcripts, suggested that Mg(II) starvation triggered stationary-phase induction of slyA expression (see above). To test whether the T2 and T3 transcripts responded to growth phase, the activity of the S7 fusion was measured in exponential- and stationary-phase cultures grown in Mg(II)-replete and Mg(II)-starved Tris-minimal medium with glucose as the carbon source. This showed that the upstream promoter(s) were responsive to growth phase under these conditions, with an ∼5-fold enhancement in stationary-phase cultures compared to exponential-phase cultures (79 Miller units compared to 15 Miller units) grown in Mg(II)-replete medium. A similar enhancement (∼6-fold) was observed for cultures grown in Mg(II)-starved medium (326 Miller units compared to 52 Miller units). These observations suggest that the upstream promoter region is growth phase responsive irrespective of the Mg(II) status of the growth medium. However, it was noted that the pH of these cultures became more acidic as growth proceeded, and it is known that a subset of PhoP-regulated genes respond to mild acid pH. Although it is thought that PhoP-PhoQ are not directly involved in this acid response (9), it was of interest to investigate the activity of the PhoP-regulated slyA promoter during growth under acidic conditions. The activity of the S7 slyA::lacZ fusion was monitored in aerobic exponential- and stationary-phase cultures grown in Bis-Tris minimal medium buffered at pH 7.2 and pH 5.8. When the pH of Mg(II)-replete cultures was maintained at pH 7.2, β-galactosidase activities were not enhanced in stationary phase (Fig. 5). However, at pH 5.8 expression from the S7 slyA::lacZ fusion was enhanced in stationary-phase cultures versus exponential-phase cultures. Moreover, compared to values obtained at pH 7.2, slyA::lacZ expression was higher at pH 5.8 under Mg(II)-replete conditions (Fig. 5). This suggests that transcription from the upstream slyA promoter during Mg(II)-replete growth (open bars in Fig. 5) is enhanced under acidic growth conditions, and this is most readily apparent in the stationary phase.
FIG. 5.
Effect of culture pH and Mg(II) availability on slyA expression. β-Galactosidase activities were measured for cultures of ST12/75 containing the S7 slyA::lacZ fusion (see Fig. 1A) in Mg(II)-replete (10 mM, open bars) and Mg(II)-starved (0.01 mM, solid bars) Bis-Tris minimal medium buffered at either pH 7.2 or pH 5.8. β-Galactosidase activities were measured in triplicate, and the mean values that varied by <10% are presented. Units of β-galactosidase activity are as defined by Miller (15).
In contrast to the Mg(II)-replete cultures, when the pH of Mg(II)-starved cultures was maintained at pH 7.2 slyA expression was significantly enhanced in the stationary phase (solid bars in Fig. 5). Moreover, exponential-phase expression was also greater than for the corresponding Mg(II)-replete cultures. In Mg(II)-starved cultures maintained at pH 5.8, slyA::lacZ, expression in the exponential phase was enhanced compared to cultures held at pH 7.2, and expression was further enhanced in the stationary phase (Fig. 5). The response to Mg(II) starvation was most marked in stationary-phase cultures maintained at pH 7.2 (Fig. 5). Thus, at both pH values tested the stationary-phase induction of expression from the upstream slyA promoter region was enhanced in response to Mg(II) starvation.
Phenotypic effects of a slyA lesion.
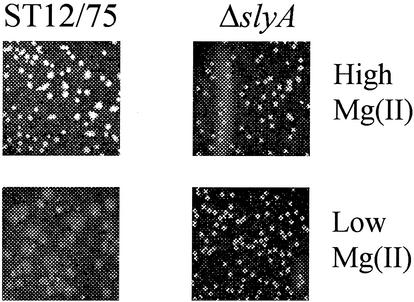
A number of slyA-associated phenotypes have been determined, including sensitivity to oxidative stress, attenuation for virulence in mice, reduced survival in the stationary phase, and the inability to survive within tissues of the reticuloendothelial system (4, 7, 12, 24). The regulatory studies described above suggested that slyA expression is upregulated in response to Mg(II) starvation, stationary phase, and acidity, which is consistent with elevated expression of SlyA during infection (9). Consequently, it might be expected that a slyA mutant would display a phenotype under conditions of Mg(II) starvation. It has been shown that phoP strains have altered growth kinetics in medium containing low levels of Mg(II) (3, 22). However, growth of the parent and slyA strains was similar in low-Mg(II) liquid cultures (not shown). Because growth in Mg(II)-starved broth cultures is unaffected by lesions in some genes of the PhoP regulon, growth of the slyA strain on solid medium containing either 10 or 0.25 mM Mg(II) was investigated. Both parent and mutant strains were able to grow, but there was a clear morphological phenotype associated with the slyA lesion (Fig. 6). Although parent and slyA strains had a similar appearance when grown on the Mg(II)-replete medium, the colonies of the parental strain responded to the low-Mg(II) medium by becoming mucoidal, whereas the colonies of the slyA strain retained the same morphology as that observed when grown on the high-Mg(II) medium. This result suggests that SlyA mediates the adaptation of the cell envelope properties of Salmonella in response to Mg(II) when cultured on a solid surface.
FIG. 6.
Growth of ST12/75 and ST12/75 (ΔslyA) on solid medium containing high or low levels of Mg(II). The indicated strains were grown for 48 h at 37°C on agarose medium containing either 10 mM MgCl2 (upper row) or 0.25 mM MgCl2 (lower row).
DISCUSSION
Previous studies have shown that SlyA negatively regulates its own expression by occlusion of a promoter with a transcription start located 41 bp upstream of the translational start site (24). We have now identified two further transcripts originating in the far-upstream region of the slyA promoter. Transcription from this upstream region is unaffected by SlyA but is enhanced in Mg(II)-starved cultures. This latter response is dependent on the PhoP-PhoQ two-component system. Thus, we provide evidence for an intimate link between two Salmonella transcription factors, namely, PhoP and SlyA, that are required for virulence.
The PhoP-PhoQ system is required for Salmonella virulence in mice, for survival within macrophages, and to resist killing by some antimicrobial peptides (for reviews, see references 6 and 9). PhoP-PhoQ also controls the modification of the cell envelope components of Salmonella. The signal sensed by PhoP-PhoQ is extracellular Ca(II) and Mg(II) and, whereas the levels of these cations are relatively high in extracellular spaces, they are low in the phagosomal vacuoles of host cells. Thus, it appears that the PhoP-PhoQ system can sense whether the bacteria are inside or outside a host cell and adapt the surface properties of the bacterium accordingly (6). Like PhoP-PhoQ, SlyA is required for survival in the macrophage environment and influences the cell envelope of Salmonella (4, 7, 12, 24). Therefore, both PhoP-PhoQ and SlyA are active in the same environment, and it is likely that the relationship between PhoP and SlyA identified here has evolved to integrate some of the responses to the plethora of signals [for example, Mg(II) starvation and acidity] received while within a host. Such a coordinated transcriptional response is probably crucial for Salmonella strains to adapt to and survive within such a potentially hostile environment. Consequently, it is not surprising to find that Salmonella strains lacking either of these key regulators are attenuated.
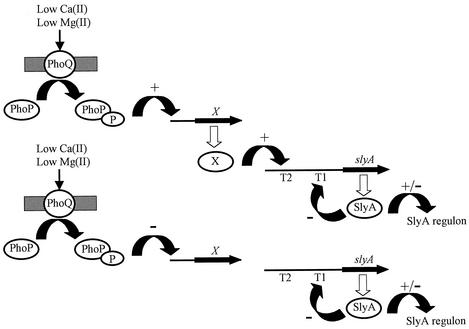
The section of the slyA promoter that responds to Mg(II) availability via PhoP contains two possible transcript starts: a minor transcript (T3) and a major transcript (T2). We believe that it is more likely that the T2 region responds to Mg(II), rather than the T3 region, for three reasons. First, the slyA::lacZ fusion S10 has only 29 bp of slyA sequence upstream of the T3 transcript, and yet this fusion is fully Mg(II) responsive. Second, there is no obvious −10 or −35 element associated with the T3 transcript, whereas such features are identifiable for the T2 transcript. Third, the induction of the T2 transcript in low- compared to high-Mg(II) cultures, as estimated by RT-PCR, is much greater than that observed for the T3 transcript (Fig. 2). Analysis of the DNA sequence upstream of T2 revealed no close matches to consensus sites of the virulence-associated transcription factors: PhoP-PhoQ (27), PmrA-PmrB (1), HilA (14), HilC, or HilD (18). Thus, the factor through which PhoP-PhoQ exerts its effects (factor X in Fig. 7) is unknown. Clearly, future studies should concentrate on identifying the transcription factors involved and on characterizing their cognate binding sites and their relationships to PhoP-PhoQ.
FIG. 7.
Model for PhoP-mediated regulation of slyA expression. Intracellular Salmonella are exposed to a low-Mg(II) environment. This is sensed by the periplasmic domain of PhoQ protein and, consequently, the phospho-PhoP dephosphorylase activity of PhoQ is inhibited. Phospho-PhoP regulates the expression of >40 genes. We suggest that there is an as-yet-unidentified transcription factor (X), which activates transcription from the slyA promoter T2. Alternatively, if factor X acts as a repressor of the slyA T2 promoter, PhoP-PhoQ might repress the expression of X and thereby relieve repression of slyA expression. Consequently, the intracellular concentration of SlyA is increased to a level at which it can regulate transcription from its target promoters, including repression of the slyA T1 promoter.
In conclusion, the work described here has established an intimate link between two transcription factors that are required for the survival of Salmonella in the macrophage environment. By linking the expression of transcription factors in this way a variety of signals can be integrated into a transcriptional cascade to produce an optimal pattern of gene expression for a particular niche. Such relationships between transcription factors are likely to be crucial in coordinating an appropriate transcriptional response to the host environment.
Acknowledgments
We thank A. J. G. Moir for DNA sequencing. We thank Stephen Libby (North Carolina State University, Raleigh) and Tim Wallis (IAH, Compton, United Kingdom) for bacterial strains.
The BBSRC UK supported this work with a research studentship (M.R.S.) and through project grant BFP11284.
REFERENCES
- 1.Aguirre, A., S. Lejona, E. G. Vescovi, and F. C. Soncini. 2000. Phosphorylated PmrA interacts with the promoter region of ugd in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:3874-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alekshun, M. N., S. B. Levy, T. R. Mealy, B. A. Seaton, and J. F. Head. 2001. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 Å resolution. Nat. Struct. Biol. 8:710-714. [DOI] [PubMed] [Google Scholar]
- 3.Blanc-Potard, A.-B., and E. Groisman. 1997. The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J. 16:5376-5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchmeier, N., S. Bossie, C. Chen, F. C. Fang, D. G. Guiney, and S. Libby. 1997. SlyA, a transcriptional regulator of Salmonella typhimurium, is required for resistance to oxidative stress and is expressed in the intracellular environment of macrophages. Infect. Immun. 65:3725-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cotter, P. A., and V. J. DiRita. 2000. Bacterial virulence gene regulation: an evolutionary perspective. Annu. Rev. Microbiol. 54:519-565. [DOI] [PubMed] [Google Scholar]
- 7.Daniels, J. J. D., I. B. Autenrieth, A. Ludwig, and W. Goebel. 1996. The gene slyA of Salmonella typhimurium is required for destruction of M cells and intracellular survival but not for invasion or colonization of the murine small intestine. Infect. Immun. 64:5075-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deiwick, J., T. Nikolaus, S. Erdogan, and M. Hensel. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31:1759-1773. [DOI] [PubMed] [Google Scholar]
- 9.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gunn, J. S., C. M. Alpuche-Aranda, W. P. Loomis, W. J. Belden, and S. I. Miller. 1995. Characterization of the Salmonella pagC/pagD chromosomal region. J. Bacteriol. 177:5040-5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heithoff, D. M., R. L. Sinsheimer, D. A. Low, and M. J. Mahan. 1999. An essential role for DNA methylation in bacterial virulence. Science 284:967-970. [DOI] [PubMed] [Google Scholar]
- 12.Libby, S. J., W. Goebel, A. Ludwig, N. Buchmeier, F. Bowe, F. C. Fang, D. G. Guiney, J. G. Songer, and F. Heffron. 1994. A cytolysin encoded by Salmonella is required for survival within macrophages. Proc. Natl. Acad. Sci. USA 91:489-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lodge, J., R. Williams, A. Bell, B. Chan, and S. Busby. 1990. Comparison of promoter activities in Escherichia coli and Pseudomonas aeruginosa: use of a new broad host range promoter probe plasmid. FEMS Microbiol. Lett. 67:221-225. [DOI] [PubMed] [Google Scholar]
- 14.Lostroh, C. P., and C. A. Lee. 2001. The HilA box and sequences outside it determine the magnitude of HilA-dependent activation of PprgH from Salmonella pathogenicity island 1. J. Bacteriol. 183:4876-4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller, J. H. 1972. Assay of β-galactosidase, p. 352-355. In Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Miller, P. F., and M. C. Sulavik. 1996. Overlaps and parallels in the regulation of intrinsic multiple-antibiotic resistance in Escherichia coli. Mol. Microbiol. 21:441-448. [DOI] [PubMed] [Google Scholar]
- 17.Nagel, G., A. Lahrz, and P. Dersch. 2001. Environmental control of invasin expression in Yersinia pseudotuberculosis is mediated by regulation of RovA, a transcriptional activator of the SlyA/Hor family. Mol. Microbiol. 41:1249-1269. [DOI] [PubMed] [Google Scholar]
- 18.Olekhnovich, I. N., and R. J. Kadner. 2002. DNA-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:4186-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poteete, A., and A. Fenton. 1984. Lambda rec-dependent growth and recombination of phage P22. Virology 134:161-167. [DOI] [PubMed] [Google Scholar]
- 20.Revell, P. A., and V. L. Miller. 2000. A chromosomally encoded regulator is required for expression of the Yersinia enterocolitica inv gene and for virulence. Mol. Microbiol. 35:677-685. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Soncini, F. C., E. G. Vescovi, F. Soloman, and E. Groisman. 1996. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J. Bacteriol. 178:5092-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sporey, A., A. Bosserhoff, C. van Rhein, W. Goebel, and A. Ludwig. 2002. Differential regulation of multiple proteins of Escherichia coli and Salmonella enterica serovar Typhimurium by the transcriptional regulator SlyA. J. Bacteriol. 184:3549-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stapleton, M. R., V. A. Norte, R. C. Read, and J. Green. 2002. Interaction of the Salmonella typhimurium transcription and virulence factor SlyA with target DNA and identification of members of the SlyA regulon. J. Biol. Chem. 277:17630-17637. [DOI] [PubMed] [Google Scholar]
- 25.Watson, P. R., S. M. Paulin, A. P. Bland, S. J. Libby, P. W. Jones, and T. S. Wallis. 1999. Differential regulation of enteric and systemic salmonellosis by slyA. Infect. Immun. 67:4950-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wosten, M. M. S., L. F. F. Kox, S. Chamnongpol, F. C. Soncini, and E. Groisman. 2000. A signal transduction system that responds to extracellular iron. Cell 103:113-125. [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto, K., H. Ogasawara, N. Fujita, R. Utsumi, and A. Ishihama. 2002. Novel mode of transcription regulation of divergently overlapping promoters by PhoP, the regulator of two-component system sensing external magnesium availability. Mol. Microbiol. 45:423-438. [DOI] [PubMed] [Google Scholar]