Abstract
Temperature sensitivity of DNA polymerization and growth of a dnaX(Ts) mutant is suppressible at 39 to 40°C by mutations in the initiator gene, dnaA. These suppressor mutations concomitantly cause initiation inhibition at 20°C and have been designated Cs,Sx to indicate both phenotypic characteristics of cold-sensitive initiation and suppression of dnaX(Ts). One dnaA(Cs,Sx) mutant, A213D, has reduced affinity for ATP, and two mutants, R432L and T435K, have eliminated detectable DnaA box binding in vitro. Two models have explained dnaA(Cs,Sx) suppression of dnaX, which codes for both the τ and γ subunits of DNA polymerase III. The initiation deficiency model assumes that reducing initiation efficiency allows survival of the dnaX(Ts) mutant at the somewhat intermediate temperature of 39 to 40°C by reducing chromosome content per cell, thus allowing partially active DNA polymerase III to complete replication of enough chromosomes for the organism to survive. The stabilization model is based on the idea that DnaA interacts, directly or indirectly, with polymerization factors during replication. We present five lines of evidence consistent with the initiation deficiency model. First, a dnaA(Cs,Sx) mutation reduced initiation frequency and chromosome content (measured by flow cytometry) and origin/terminus ratios (measured by real-time PCR) in both wild-type and dnaX(Ts) strains growing at 39 and 34°C. These effects were shown to result specifically from the Cs,Sx mutations, because the dnaX(Ts) mutant is not defective in initiation. Second, reduction of the number of origins and chromosome content per cell was common to all three known suppressor mutations. Third, growing the dnaA(Cs,Sx) dnaX(Ts) strain on glycerol-containing medium reduced its chromosome content to one per cell and eliminated suppression at 39°C, as would be expected if the combination of poor carbon source, the Cs,Sx mutation, the Ts mutation, and the 39°C incubation reduced replication to the point that growth (and, therefore, suppression) was not possible. However, suppression was possible on glycerol medium at 38°C. Fourth, the dnaX(Ts) mutation can be suppressed also by introduction of oriC mutations, which reduced initiation efficiency and chromosome number per cell, and the degree of suppression was proportional to the level of initiation defect. Fifth, introducing a dnaA(Cos) allele, which causes overinitiation, into the dnaX(Ts) mutant exacerbated its temperature sensitivity.
DnaA protein, the principal chromosome replication initiation control factor (39), consists of an AAA+ core region, which has ATP binding and hydrolysis, DNA binding, and oligomerization activities, plus two less-well-characterized N-terminal domains (16, 20, 44, 51, 55, 60). DnaA initiates replication by protomer binding to DnaA boxes within the chromosome origin, oriC (21, 43), oligomerizing (49, 55) in the ATP-bound form (49) to unwind the AT-rich region and form an open complex, and recruiting the DnaB-DnaC complex (10, 22, 42, 50). Upon release of DnaC (48), DnaB helicases assemble on each strand, leading to assembly of two replisomes for polymerization in opposite directions (12, 17). Precision in activation timing is enhanced by integration host factor-facilitated distribution of DnaA to oriC binding sites (25). Initiation is negatively regulated to once and only once per cell cycle by the interplay of at least three mechanisms. First, the active ATP form is converted to the inactive ADP form after initiation by regulatory inactivation of DnaA (RIDA), a process effected by the DNA polymerase III processivity clamp (assembled onto DNA, the Hda protein, and ongoing DNA synthesis [31, 32]). A mutant DnaA with reduced intrinsic ATPase activity was insensitive to RIDA and overinitiated in vivo (45). Second, SeqA competes with DnaA in oriC binding and sequesters the newly replicated origin, thereby preventing premature initiations (9, 40, 58, 59). Third, DnaA protein is titrated by DnaA boxes outside oriC, including dat (13, 35).
Certain mutations within dnaA which reduce affinity for ATP or for DnaA boxes (J. R. Walker, K. A. Severson, M. J. Hermandson, K. M. Carr, J. M. Kaguni, and A. Blinkova, unpublished data) inhibit initiation at 20 or 44°C and also have the effect of suppressing temperature sensitivity of replication and growth of a DNA polymerization dnaX(Ts) mutant (23, 61). These mutations, which conferred changes A213D in the ATP binding region and R432L and T435K in the DNA binding specificity region, were designated Cs,Sx to indicate both cold-sensitive initiation at 20°C and suppression of dnaX(Ts) at 39 to 40°C (23, 61). One suppressor mutant has been shown not only to be initiation defective at 20 and 44°C, but also to initiate with reduced efficiency at the permissive 34°C and at 39°C, the temperature at which suppression was observed in dnaA(Cs,Sx) dnaX(Ts) double mutants (8). Wild-type cells in yeast extract-tryptone medium contained four or (mostly) eight origins at both 34 and 39°C, indicating synchronous initiation of all origins within individual cells. Similarly grown dnaA R432L mutant cells contained fewer origins per cell and initiated asynchronously, with many cells containing three, five, six, or seven origins (8). Asynchrony of initiation presumably resulted from reduced initiation frequency and consequent failure of some origins to function during the allowed initiation period (53, 54). Although the dnaA(Cs,Sx) mutation reduced initiation frequency, it efficiently suppressed a dnaX(Ts) mutation, and the dnaA(Cs,Sx) dnaX(Ts) double mutant grew at 39°C with the wild-type growth rate of about 25 min/generation (in yeast extract-tryptone medium) and plated at 39°C with an efficiency of about 1.0 (4).
The dnaX polymerization gene codes for both the τ and γ subunits of DNA polymerase III (7, 19, 57). As the replisome organizer, τ dimerizes the holoenzyme (34, 46) and stimulates fork progression by interaction with the DnaB helicase (14, 66) and primase (66). The γ subunit is a major component of the processivity clamp loading complex (24, 47, 64). The dnaX(Ts) mutant used in this study has residue 118, glycine, changed to aspartate, is altered in both τ and γ (5), and stops polymerization abruptly on a shift to 42°C (18). Two models have been proposed for the mechanism by which mutations in dnaA suppress the dnaX(Ts) growth defect. The initiation deficiency model assumes that reduced initiation efficiency allows dnaX(Ts) cells to survive the 39°C challenge, albeit with reduced chromosome content (4, 8; O. Skovgaard, J. Gregersen, C. Hubert, and K. Olesen, Abstr. EMBO Workshop Cell Cycle Nucleoid Organization Bacteria, abstr. 26, 2000). The stabilization model assumes direct or indirect contact of DnaA(Cs,Sx) with polymerization factors during replication (4, 8). Here we report five lines of evidence consistent with the initiation deficiency model.
MATERIALS AND METHODS
Bacterial strains and culture media.
The principal bacterial strains are listed in Table 1. Strains AB21, AB20 (8), EGC23 (4), SXC603, and SXC601 are isogenic, or nearly isogenic, derivatives of strain C600. Strains SXC603 and SXC601 were constructed by P1 transduction (63) of dnaA(Cs,Sx) A213D and T435K, respectively, along with rbs::Tn406 into strain C600. For simplicity, dnaA(Cs,Sx) alleles will be referred to as Sx or suppressor or indicated by the amino acid change. Strains AB2801, AB2803, AB2805, and AB2807 were constructed by P1 transduction of dnaX(Ts) G118D along with zbb::Tn10 into oriC+ strain WM2482, oriC160 strain WM2759, oriC17 strain WM2844, and oriC162 strain WM2845, respectively (62). A dnaA(Cos) dnaX(Ts) strain was made by P1 cotransduction of dnaA(Cos) and the closely linked tna::Tn10 from strain KA411 (30) into strain AB600, a dnaX(Ts) purE::Tn5 derivative of strain C600. Tetracycline-resistant transductants were selected by incubating for 3 days at 30°C and scored as Cos if they grew very slowly or dnaA+ if they grew at the wild-type growth rate at that temperature. Strains AB2852 and AB2853 are dnaA+ dnaX(Ts) and dnaA(Cos) dnaX(Ts), respectively. The defined C medium base (27) with MgSO4 reduced to 0.4 mM was supplemented with 0.1% glucose, 0.1% glycerol, and 0.2% Casamino Acids, as indicated, plus threonine and leucine (50 μg of each/ml) and thiamine HCl (5 μg/ml) for the derivatives of strain C600 and tryptophan and thymine (50 μg of each/ml) for strains AB2801 to AB2807. Tetracycline and ampicillin were added to 15 and 100 μg/ml, respectively, as needed for transduction to yeast extract (0.5%), Peptone (1%), NaCl (0.5%) medium supplemented with 50 μg of thymine/ml.
TABLE 1.
Principal bacterial strains
| Strain | Genotype
|
|||||
|---|---|---|---|---|---|---|
| oriC | oriC mutation | dnaA | DnaA change | Tn | dnaX | |
| AB21a | + | + | zib::Tn10 | + | ||
| AB20a | + | (Sx)721 | R432L | zib::Tn10 | + | |
| AB27b | + | + | zbb::Tn10 | (Ts)c | ||
| EGC23d | + | (Sx)721 | R432L | zib::Tn10 | (Ts) | |
| SXC603b | + | (Sx)73 | A213D | zib::Tn10 | + | |
| SXC601b | + | (Sx)71 | T435K | zib::Tn10 | + | |
| AB2801b | + | + | zbb::Tn10 | (Ts) | ||
| AB2803b | 160 | Δ nte-275-352 | + | zbb::Tn10 | (Ts) | |
| AB2805b | 17 | Scrambled box M | + | zbb::Tn10 | (Ts) | |
| AB2807b | 162 | + 14 nt R3/R4 | + | zbb::Tn10 | (Ts) | |
| AB2851b | + | (Cos) | A184Vf | tna::Tn10 | + | |
| AB2853b | + | (Cos) | A184Vf | tna::Tn10g | (Ts) | |
Flow cytometry analysis of DNA content per cell.
Cultures were grown for at least nine generations to approximately 0.2 absorbance (595 nm) and fixed without further treatment or incubated in the presence of 300 μg of rifampin and 15 μg of cephalexin/ml for 240 to 300 min to inhibit new initiation events and cell divisions before fixing (39). The cells were fixed by adding 1 ml of culture to 9 ml of 95% ethanol and stored at 4°C. For DNA staining, the cells were centrifuged and resuspended in Tris-EDTA (10 mM [pH 8.0] and 1 mM, respectively) at an absorbance of 0.4 and mixed with the specific dye PicoGreen (Molecular Probes Inc., Eugene, Oreg.), as described by Marie et al. (41). The PicoGreen concentrated solution provided by the supplier was diluted 1:100 in 25% dimethyl sulfoxide and added to cells at a ratio of 10 μl of diluted dye to 50 μl of cells. The cells were stained for 3 h at room temperature and diluted with 1 ml of Tris-EDTA containing a 1:1,000 dilution of PicoGreen. DNA content per cell was analyzed by using a Becton Dickinson FACSCalibur flow cytometer using a 488-nm laser. Data were processed with CellQuest software.
Nucleoid staining and light microscopy.
Cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) (1) and examined for nucleoid content and cell lengths by both fluorescence and phase-contrast microscopy of the same microscope field. DAPI was added at 2.5 μg/ml to growing cultures at an absorbance of approximately 0.1 and incubation continued for 30 min.
Real-time PCR measurement of marker frequency.
The number of copies of oriC and proA, relative to an xasA(gadC) terminus marker (28), were estimated by real-time PCR in an Applied Biosystems ABI Prism 7700 SDS with Primer Express (PE) software following instructions of the manufacturer. The assay for amplification used Taq polymerase to hydrolyze specific probes labeled with a fluorescent 6-carboxyfluorescein reporter (6FAM) or VIC (a trademark name of Applied Biosystems) at the 5′ end and a major groove binding nonfluorescent binding quencher on the 3′ end. Quantitation was achieved using the standard curve method in which the threshold cycle (Ct) was plotted versus the log of DNA concentration. A linear relationship was observed for each amplicon between Ct and the log of DNA concentration over the 30-fold range of 0.3 to 10 ng of stationary-phase chromosomal DNA extracted from the wild-type strain AB21 grown in glucose-Casamino Acids medium. Primers, with melting temperatures of 58 to 60°C and with 50 to 80% GC content, were chosen within oriC, proA, and xasA by PE software to amplify regions of about 60 bp. The forward and reverse primers, respectively, from the 5′ ends were as follows: for oriC, GCACTGCCCTGTGGATAACAA (2889 to 2909) and ACAGTTAATGATCCTTTCCAGGTTG (2955 to 2931) (numbering system from GenBank AE accession number 000451); for proA, AATGGCGGAAAGCGGC (828 to 843) and CCTGCAACTGCGCCAGT (886 to 870) (numbering system started with 1 as the first nucleotide of the coding sequence [GenBank accession number NC_000913]); and for xasA, TGGGTGTTCTGGCGGAA (896 to 912) and TCCCGCGAGAAGGACCA (946 to 930) (numbering system started with 1 as the first nucleotide of the coding sequence [GenBank NC_000913]). Primers were obtained from Integrated DNA Technologies, Inc. (Coralville, Iowa). Probes chosen with the aid of PE software and obtained from Applied Biosystems (Foster City, Calif.) were as follows: for oriC, 5′ VIC CGGCTTTTAAGATC (2914 to 2928); for proA, 5′ VIC CACGCAGATGCAGC (853 to 866); and for xasA, 5′ 6FAM TCGCTCCTGGATTA (914 to 928).
The 25-μl assay volume, optimized for each amplicon, contained, for oriC and for xasA, 50 nM forward primers and 900 nM reverse primers; for proA, the forward and reverse primers were each 300 nM. All the probes were 250 nM.
Marker frequencies of oriC and proA relative to the xasA gene for exponentially growing cells were determined using chromosomal DNA extracted from cultures which had been growing exponentially for about nine generations to absorbances at 595 nm of 0.2.
Chromosomal DNA extraction.
Stationary- and exponential-phase chromosomal DNA was extracted by using a Puregene DNA isolation kit (Gentra Systems, Minneapolis, Minn.), following the manufacturer's instructions.
RESULTS
A dnaA suppressor mutation reduces initiation efficiency in dnaX+ and Ts strains at 34°C in glucose-Casamino Acids medium as measured by flow cytometry.
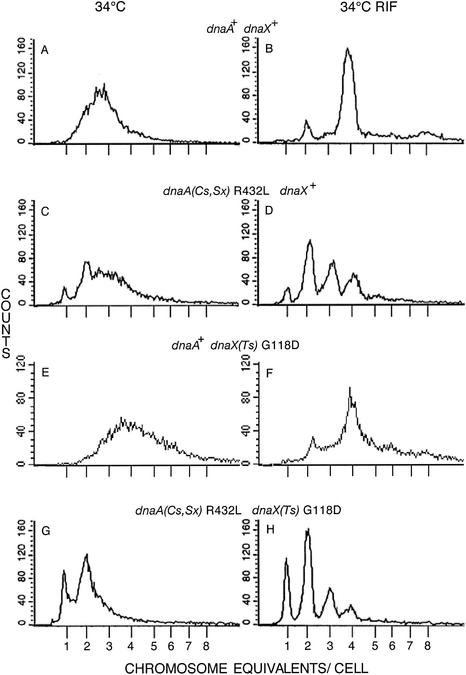
Growth rate, chromosome content, and synchrony of initiation were examined in the wild-type, dnaA(Sx), dnaX(Ts), and dnaA(Sx) dnaX(Ts) strains growing exponentially in glucose-Casamino Acids medium at the permissive 34°C. The cells were stained directly with PicoGreen and also after incubation with rifampin and cephalexin (to allow replication rounds to be completed while initiation and cell division were inhibited), and both stained preparations were examined for chromosome content by flow cytometry (39). The number of completed chromosomes after rifampin-cephalexin treatment reflected the number of origins present at the time the inhibitors were added. Exponentially growing wild-type cultures initiated synchronously and consisted mostly of cells with two replicating chromosomes (Fig. 1A) and four origins per cell (Fig. 1B). Although some cells contained 2 or 8 origins, the average was 4.26/cell (Table 2). The dnaA R432L suppressor mutant grew at 34°C with a doubling time of 39 min, slightly slower than the wild-type 36 min, and contained fewer chromosomes per cell. The most numerous classes contained 2, 3, or 4 origins (Fig. 1D), and a significant fraction of exponentially growing cells contained only one completed chromosome (Fig. 1C). The average number of origins was 3.01 (Table 2). Asynchronous initiation was a consequence also; about 28% of the cells contained three origins (Table 2).
FIG. 1.
A dnaA(Sx) mutation reduces chromosome and origin content of dnaX+ and Ts strains in glucose-Casamino Acids medium at 34°C. Cells growing exponentially were stained directly (A, C, E, and G) or after incubation in the presence of rifampin and cephalexin to allow replication completion (B, D, F, and H) and analyzed for DNA content by flow cytometry (39). Counts refer to the number of cells. Wild-type (dnaA+ dnaX+) strain AB21 (A and B), dnaA R432L dnaX+ strain AB20 (C and D), dnaA+ dnaX(Ts) strain AB27 (E and F), and the double mutant dnaA R432L dnaX(Ts) strain EGC23 (G and H) were analyzed.
TABLE 2.
Growth rate and origin content at 34°C in media with different carbon sources
| Medium and strain | dnaA | dnaX | Gena | Origins/cell (ave) | % Cells with indicated no. of originsb
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5-7 | 8 | >8 | |||||
| Glucose-Casamino Acids | |||||||||||
| AB21 | + | + | 36 | 4.26 | 2 | 7 | 4 | 66 | 13 | 8 | |
| SXC603 | A213D | + | 43 | 3.07 | 5 | 29 | 25 | 26 | 13 | 1 | 1 |
| AB20 | R432L | + | 39 | 3.01 | 5 | 33 | 28 | 22 | 9 | 1 | 2 |
| SXC601 | T435K | + | 48 | 2.59 | 16 | 40 | 15 | 11 | 11 | 2 | 5 |
| EGC23 | R432L | Ts | 40 | 2.21 | 19 | 45 | 20 | 11 | 4 | 1 | |
| Glucose | |||||||||||
| AB21 | + | + | 69 | 2.40 | 1 | 77 | 3 | 19 | |||
| AB20 | R432L | + | 68 | 1.97 | 21 | 64 | 8 | 5 | |||
| EGC23 | R432L | Ts | 77 | 1.90 | 31 | 52 | 10 | 6 | |||
| Glycerol | |||||||||||
| AB21 | + | + | 88 | 1.97 | 10 | 85 | 3 | 2 | |||
| AB20 | R432L | + | 96 | 1.79 | 38 | 50 | 8 | 3 | |||
| EGC23 | R432L | Ts | 110 | 1.77 | 44 | 41 | 10 | 4 | |||
Generation time in minutes.
Blank spaces indicate less than 1%.
The dnaX(Ts) mutant grew exponentially at 34°C with a doubling time of 39 min, and most of the cells contained four origins (Fig. 1F), similar to the wild type. Although there was no apparent defect in initiation and run-out polymerization completed most chromosomes, polymerization failed in some of the chromosomes even at this permissive temperature. (The average number of origins per cell was not calculated because of the failure of some chromosomes to be completed [Fig. 1F].) Inhibited polymerization of some chromosomes did not interfere with colony-forming ability of individual cells, because this mutant formed colonies at 34°C with an efficiency of approximately 1.0.
The dnaX(Ts) effect on polymerization was proportional to temperature. At 30°C, chromosome replication was complete after rifampin-cephalexin addition in both wild-type and Ts mutant cells, and the chromosome contents were essentially identical. The average number of origins per cell was 2.80 and 2.82, respectively. At 36°C, the Ts mutant grew and plated with wild-type efficiency, but the fraction of chromosomes which could not be completed (after rifampin-cephalexin addition) was greater than that observed at 34°C (data not shown).
The dnaA suppressor dnaX(Ts) double mutant grew with a doubling time of 40 min, initiated asynchronously, and contained fewer chromosomes than the dnaA(Sx) strain. Many of these cells growing exponentially (before adding inhibitors) contained one or two apparently completed, or nearly completed, chromosomes (Fig. 1G), and adding rifampin-cephalexin demonstrated that 64% of them contained only one or two origins (Fig. 1H; Table 2). The average number of origins per cell was 2.21, compared to 3.01 for the dnaA(Sx) strain (Table 2).
Initiation deficiency is a common characteristic of dnaA(Sx) mutants.
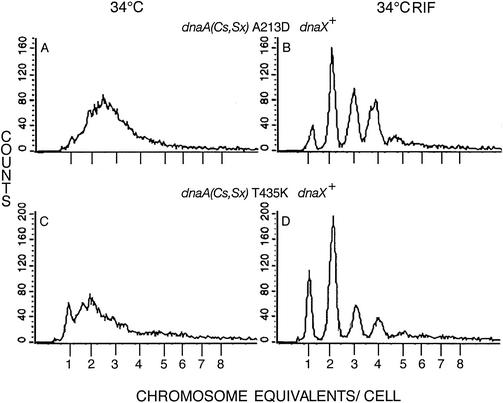
Two additional suppressor mutations also reduced the number of replicating chromosomes per cell and caused asynchronous initiation even at permissive temperature. The dnaA A213D mutant was similar to the R432L mutant; the average cell contained 3.07 origins (Table 2), and the most numerous classes contained 2, 3, or 4 origins (Fig. 2A and B). The T435K mutant culture was more severely affected. The growth rate was slowed, increasing the generation time from the wild-type 36 min to 48 min, and about 16% of the exponentially growing cells contained a fully replicated chromosome before adding inhibitors (Fig. 2C; Table 2). After allowing replication run-out, 56% of the cells contained 1 or 2 completed chromosomes, indicating 1 or 2 origins/cell at the time of rifampin addition (Fig. 2D). The average number of origins per cell was 2.59, compared with 4.26 for the wild-type cells. Asynchronous initiation was a consequence of this mutation also; about 15% of the cells contained 3 origins.
FIG. 2.
Chromosome number and origin reduction are characteristic of additional dnaA suppressor mutants growing in glucose-Casamino Acids medium at 34°C. Cells growing exponentially (A and C) or after replication run-out (B and D) were analyzed as described in the Fig. 1 legend. dnaX+ strains with dnaA suppressor A213D (strain SXC603) (A and B) and T435K (strain SXC601) (C and D) were analyzed.
Chromosome content at the suppressive 39°C in glucose-Casamino Acids medium.
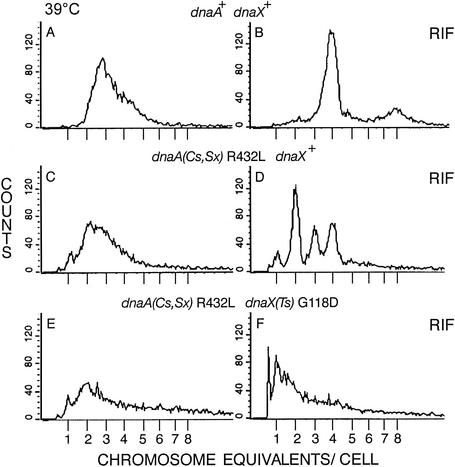
Wild-type cells growing at 39°C were similar to those grown at 34°C; they contained a heterogeneous distribution of mostly two replicating chromosomes (Fig. 3A) with four origins (Fig. 3B). The dnaA R432L mutant DNA content was distributed more broadly (Fig. 3C) and, after rifampin-cephalexin addition, the cells completed replication of two, three, or four chromosomes (Fig. 3D). This pattern was similar to those of cells grown at 34°C, except that a greater fraction of the 39°C-grown cells contained four origins (c.f., Fig. 1D). The dnaA(Sx) dnaX(Ts) double mutant cell DNA content was even more broadly distributed; some cells contained one nonreplicating chromosome, whereas most of them contained one or more replicating chromosomes (Fig. 3E and F). However, many replication forks were permanently inhibited at 39°C, because polymerization did not go to completion in all the chromosomes, evident after rifampin-cephalexin addition (Fig. 3F), even during 8 h of incubation (data not shown).
FIG. 3.
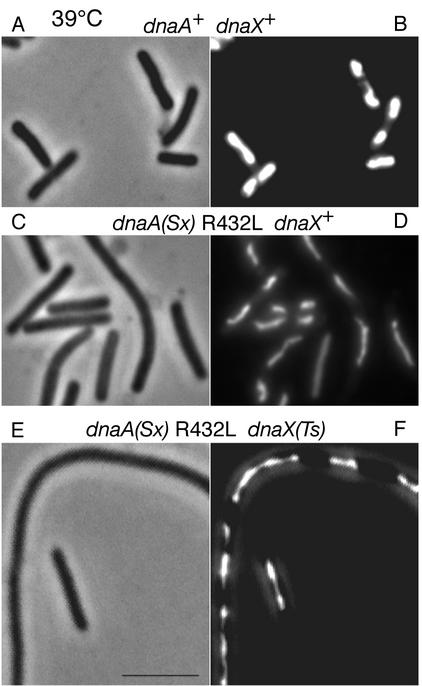
A dnaA(Sx) mutation reduces chromosome content of dnaX+ and Ts strains in glucose-Casamino Acids medium at 39°C. Cells growing exponentially (A, C, and E) or after replication run-out (B, D, and F) were analyzed as described in the Fig. 1 legend. Wild-type (dnaA+ dnaX+) strain AB21 (A and B), dnaA (R432L) dnaX+ strain AB20 (C and D), and dnaA R432L dnaX(Ts) strain EGC23 (E and F) cells were analyzed. In panel F, the peak at less than 1 chromosome/cell was not characterized but might represent cell fragments with trapped DNA.
Cell length and nucleoid content.
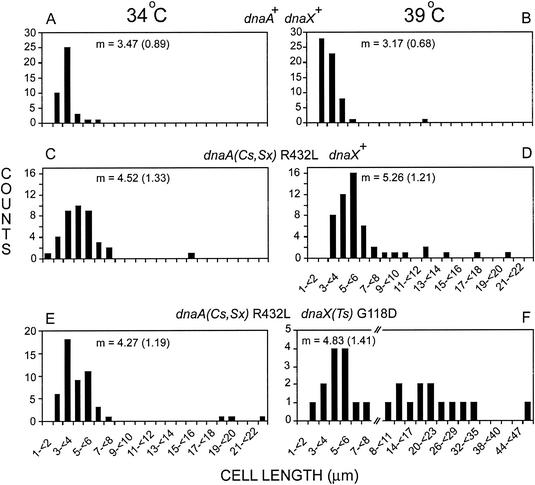
Cells growing at 34 and 39°C in glucose-Casamino Acids medium were stained with DAPI (1), and the same cells were examined both by fluorescence microscopy for nucleoid content and phase-contrast microscopy for length measurements. At 34°C, wild-type cell lengths were distributed around a mean of 3.47 μm, dnaA suppressor mutant cells were elongated with a mean of 4.52 μm, and most of the dnaA suppressor dnaX(Ts) cells were also elongated (mean of 4.27 μm), but this culture also contained a few filaments that were up to about 5 times the normal length (Fig. 4). At 39°C, the wild-type cells were approximately the same length as at 34°C, and the dnaA suppressor culture contained mostly elongated cells and a significant proportion of short filaments 2 to 5 times the normal length. The dnaA suppressor dnaX(Ts) double mutant culture consisted of two populations: about half the cells were elongated (mean, 4.83 μm), and about half were filaments with 2 to 10 times the normal length (Fig. 4). DAPI staining of cells growing at 39°C revealed that those cells within the length range of 1 to 8 μm contained about 2 nucleoids per cell, regardless of the genotype (Fig. 5). Filaments among the dnaA(Sx) and dnaA(Sx) dnaX(Ts) cultures contained nucleoids which appeared less compact than those of the wild-type cells (Fig. 5).
FIG. 4.
Cell lengths in glucose-Casamino Acids medium at 34 and 39°C. Wild-type strain AB21 (A and B), dnaA (R432L) dnaX+ strain AB20 (C and D), and dnaA R432L dnaX(Ts) strain EGC23 (E and F) cells growing exponentially at 34°C (A, C, and E) and 39°C (B, D, and F) were observed by phase-contrast microscopy. The abscissa scale changes in panel F. The average cell lengths (in micrometers) of cells within the 1- to 8-μm range are indicated in each panel; parentheses contain 1 standard deviation.
FIG. 5.
Phase contrast (A, C, and E) and DAPI-stained (B, D, and F) wild-type strain AB21 (A and B), dnaA (R432L) dnaX+ strain AB20 (C and D), and dnaA R432L dnaX(Ts) strain EGC23 (E and F) cells growing at 39°C. The same microscope fields were examined by both fluorescence and phase microscopy. Bar, 5 μm.
Growth of a dnaA(Sx) dnaX(Ts) mutant on media with different carbon sources.
If the initiation deficiency model is correct, it is possible that suppression would fail if cultures were plated (at 39°C) on media with poorer carbon sources. On a poor medium which limited the chromosome content to about one per cell, a dnaA(Sx) dnaX(Ts) double mutant might not survive the decreased chromosome content caused by the poor medium (26) combined with reduced initiation frequency and the inhibitory effect of Ts DnaX products at 39°C. These factors might be expected to reduce replication efficiency to the point that growth would cease (and suppression could not be observed). This possibility was tested by comparing growth on media containing only glucose or glycerol carbon sources to that on glucose-Casamino Acids medium.
The dnaA(Sx) mutant retained cold sensitivity but grew at 39°C on all three media (Table 3). The dnaX(Ts) mutant did not plate efficiently at 39°C on any of those media. The dnaA(Sx) dnaX(Ts) double mutant also retained cold sensitivity and was suppressed at 39°C on the two richer media, the glucose-Casamino Acids and glucose media. However, the double mutant did not plate efficiently on the glycerol medium at 39°C (Table 3). That is, growth and suppression failed as chromosome content was reduced. However, glycerol medium did support growth of the double mutant at 38°C (Table 3). [The dnaA(Sx) dnaX(Ts) double mutant was able to grow for several generations at 39°C in liquid glycerol medium (see below), albeit with a generation time of about 3.5 h.]
TABLE 3.
Efficiency of plating on media with different carbon sources
| Strain | dnaA | dnaX | Efficiency of platinga
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Glucose-Casamino Acids
|
Glucose
|
Glycerol
|
||||||||
| 20°C | 39°C | 20°C | 39°C | 20°C | 37°C | 38°C | 39°C | |||
| AB21 | + | + | 1.04 | 1.01 | 0.86 | 0.78 | 1.11 | 0.91 | ||
| AB20 | Sx | + | 1.04 × 10−5 | 1.01 | <4.4 × 10−6 | 1.09 | <7 × 10−5 | 1.2 | ||
| AB27 | + | Ts | 1.07 | 7.8 × 10−5 | 0.94 | <6 × 10−5 | 0.92 | 0.93 | 8.7 × 10−6 | 2.1 × 10−7 |
| EGC23 | Sx | Ts | 1.3 × 10−5 | 1.03 | 7.3 × 10−6 | 1.0 | 5.2 × 10−6 | 0.89 | 0.66 | 5.7 × 10−3 |
Relative to 34°C or, in the case of strain AB27, 30°C. Blank spaces indicate that no plating was done.
Chromosome content in glucose medium at 34 and 39°C.
Growing exponentially at 34°C in glucose medium, wild-type cells contained mostly one (77%) or two (19%) replicating chromosomes (Table 2). The dnaA R432L mutant cells contained fewer chromosomes: one nonreplicating (21% of the cells) and one replicating (64% of the cells) and initiated asynchronously (Table 2). The dnaA R432L dnaX(Ts) double mutant cells had still fewer chromosomes: 31% had one nonreplicating chromosome and 52% had one replicating chromosome (Table 2). At the suppressive 39°C, the wild-type cells contained mostly one replicating chromosome, the dnaA R432L mutant contained fewer chromosomes and some of them were nonreplicating, and the dnaA R432L dnaX(Ts) double mutant cells had mostly one or two completely or partially replicated chromosomes, some of which could not be completed during prolonged incubation in rifampin-cephalexin (data not shown).
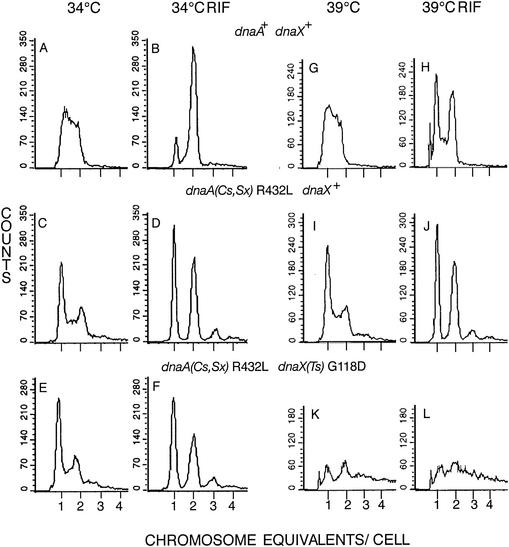
Chromosome content in glycerol medium at 34 and 39°C.
Wild-type cells growing exponentially at 34°C in glycerol medium had a generation time of 88 min, contained mostly one replicating chromosome, and had an average of 1.97 origins/cell (Table 2; Fig. 6A and B). The dnaX(Ts) mutant cells were not significantly different at 34°C, with a generation time of 89 min and an average of 1.93 origins/cell (data not shown). It is significant that, at 34°C in this medium with one replicating chromosome per cell, the Ts mutant chromosomes replicated fully (after rifampin-cephalexin addition), in contrast to the failure of some chromosomes of the Ts mutant to be fully replicated at 34°C in glucose-Casamino Acids medium, in which cells contained mostly two replicating chromosomes (Fig. 1F). Most of the dnaA R432L mutant cells, doubling every 96 min, contained one completed, one replicating, or two completed chromosomes (Table 2; Fig. 6C and D). The dnaA R432L dnaX(Ts) double mutant cells had a generation time of 110 min and contained mostly one completed or one replicating chromosome (Table 2; Fig. 6E and F). The suppressor mutation reduced the average number of origins per cell from the wild-type and dnaX(Ts) levels of 1.97 and 1.93 to 1.79 in the dnaA suppressor dnaX+ mutant and to 1.77 in the dnaA suppressor dnaX(Ts) double mutant (Table 2). The suppressor mutation also caused asynchronous initiation, as indicated by the presence of some three-chromosome cells (Fig. 6D and F).
FIG. 6.
Chromosome content of cells growing exponentially at 34°C (A, C, and E) and at 39°C (G, I, and K) and after incubation in rifampin and cephalexin at 34°C (B, D, and F) and at 39°C (H, J, and L) in glycerol medium. Wild-type strain AB21 (A, B, G, and H), dnaA R432L strain AB20 (C, D, I, and J), and dnaA R432L dnaX(Ts) strain EGC23 (E, F, K, and L) cells were analyzed as described in the Fig. 1 legend.
At 39°C in glycerol medium, wild-type cells doubled in about 61 min and contained one chromosome that was completed or in various stages of replication (Fig. 6G and H). The dnaA R432L mutant cells grew more slowly, doubling in about 86 min, and usually contained one completed, one replicating, or two chromosomes and initiated asynchronously (Fig. 6I and J). dnaA R432L dnaX(Ts) double mutant cells had a generation time of about 3.5 h and contained one or two apparently fully replicated chromosomes or one or more chromosomes in various stages of completion. Many of those replication forks apparently were irreversibly stalled, because the distribution did not change markedly during rifampin and cephalexin incubation (Fig. 6K and L). Although the double mutant grew in liquid glycerol medium at 39°C, growth was limited to several generations after shifting to 39°C, and this strain did grow sufficiently at 39°C to form colonies on solid glycerol medium (Table 3).
A dnaA suppressor mutation reduces the origin/terminus ratio at 34 and 39°C as measured by real-time PCR.
The origin-to-terminus ratio was reduced by a dnaA suppressor mutation at both the permissive 34°C and the 39°C temperature used for suppression. The number of copies of the origin (min 84.5) relative to the copies of xasA(gadC) (28), a gene located at min 33.7 near the terminus (min 34.6), was determined by real-time PCR as a measurement of the origin/terminus ratio. An additional marker located about 40% of the distance from oriC to the terminus was proA, located at min 5.6 (2). Amplicons of less than 60 bp within the minimal oriC, proA, and xasA were amplified by the TaqMan hydrolysis assay. Wild-type stationary-phase cells contained fully replicated chromosomes, as indicated by oriC/terminus and proA/terminus ratios each of 1.01 with standard deviations of 0.01 and 0.03, respectively, and this DNA was chosen as the standard.
oriC/terminus and proA/terminus ratios were determined for wild-type, dnaX(Ts), dnaA suppressor, and dnaA(Sx) dnaX(Ts) double mutants growing in glucose-Casamino Acids medium at both the permissive and suppressive temperatures (Table 4). At permissive temperatures, multifork replication occurred in both the wild-type and dnaX(Ts) strains, as indicated by origin/terminus and proA/terminus ratios of 3.00 and 2.04 for the wild-type strain at 34°C and 3.49 and 2.09 for the Ts strain at 30°C. The reason for the greater origin/terminus ratio in the Ts mutant than in the wild type is not known, but it is clear that the Ts mutation had no inhibitory effect on initiation. (The Ts mutant was grown at the fully permissive 30°C, to avoid the high origin/terminus ratio expected from stalled replication forks already shown at 34°C [Fig. 1F]). On the other hand, the dnaA(Sx) mutation reduced origin/terminus ratios at 34°C to less than 2.0 (1.77 and 1.84) for dnaX+ and Ts strains (Table 4). The proA/terminus ratios were similarly reduced by the suppressor mutation. At 39°C, the wild-type origin/terminus ratio was 3.47, but the dnaA suppressor reduced it to 1.83 in an otherwise wild-type strain and to 1.5 in a dnaX(Ts) strain (Table 4).
TABLE 4.
A dnaA suppressor mutation reduces the origin-to-terminusa ratio, as measured by real-time PCR analysis
| Medium and strain | dnaA | dnaX | Temp (°C) | Marker frequency
|
|
|---|---|---|---|---|---|
| oriC/xasA (SD)b | proA/xasA (SD) | ||||
| Glucose-Casamino Acids | |||||
| AB21 | + | + | 34 | 3.00 (0.40) | 2.04 (0.14) |
| 39 | 3.47 (0.49) | ||||
| AB20 | Sx | + | 34 | 1.77 (0.10) | 1.36 (0.12) |
| 39 | 1.83 (0.22) | ||||
| AB27 | + | Ts | 30 | 3.49 (0.32) | 2.09 (0.34) |
| EGC23 | Sx | Ts | 34 | 1.84 (0.16) | 1.32 (0.05) |
| 39 | 1.50 (0.10) | ||||
| Glycerol | |||||
| AB21 | + | + | 34 | 2.00 (0.19) | 1.45 (0.07) |
| AB20 | Sx | + | 34 | 1.48 (0.07) | 1.22 (0.05) |
| AB27 | + | Ts | 30 | 1.79 (0.13) | 1.43 (0.10) |
| EGC23 | Sx | Ts | 34 | 1.16 (0.10) | 1.16 (0.10) |
The xasA marker was used as an indicator of the terminus.
Standard deviations for six to nine determinations.
In glycerol medium at 34°C, the wild-type strain doubled with a generation time of about 88 min and the origin and proA marker frequencies were 2.00 and 1.45 (Table 4), compared to the terminus, as expected if replication occurs once each cell cycle. Yoshikawa and Sueoka (65) determined that the relative frequency of markers in cells growing exponentially with one chromosome replicating once per cell cycle is 21−x, where x is the position along the chromosome with 0 and 1 representing the origin and terminus, respectively. The relationship is applicable also to markers located on arms of bidirectionally replicating chromosomes. The proA/terminus ratio for wild-type cells growing exponentially in glycerol medium was calculated to be 1.52, which compares favorably with the observed 1.45. The dnaX(Ts) mutant origin frequency was 1.79, slightly lower than the wild-type level of 2.00. However, the dnaA suppressor mutation had a major effect on origin/terminus ratios, reducing the wild-type level from 2.00 to 1.48 and the dnaX(Ts) level from 1.79 to 1.16 (Table 4).
Suppression of dnaX(Ts) by oriC mutations.
The dnaX(Ts) temperature sensitivity was suppressible also by oriC mutations which reduced initiation efficiency. Weigel et al. (62) isolated a series of chromosomal oriC mutants, some of which had a reduced initiation frequency as evidenced by reduced numbers of origins per cell and initiated asynchronously. dnaX(Ts) was introduced into the wild-type strain used by Weigel et al. (62) and into three oriC mutant strains differing in the degree of asynchrony. The oriC+ dnaX(Ts) strain contained an average 2.76 origins/cell, initiated synchronously, and was temperature sensitive at both 38 and 39°C (Table 5). The oriC160 mutation, which deleted nucleotides 275 to 352, caused only small changes in a dnaX+ strain: it slowed the growth rate and increased the number of origins per cell slightly but had no significant effect on slowing initiation or asynchrony. In a dnaX(Ts) background, oriC160 increased the average number of origins to 4.09/cell and caused a slight degree of asynchrony (from the wild-type level of 5% three-chromosome cells to 10%) but had no detectable effect on temperature sensitivity. Scrambling the DnaA box M sequence in oriC17 had no significant effect on the origin copy number, increased asynchrony, reduced growth rate from the wild type 46 to 53 min/doubling, and restored growth of the dnaX(Ts) mutant at 38°C to a plating efficiency of 0.5, but did not suppress at 39°C. The most deleterious mutation, oriC162, which inserted 14 bp between the DnaA boxes R3 and R4, reduced the origin content, caused significant asynchrony, reduced growth rate to 65 min/doubling, and suppressed dnaX(Ts) at both 38 and 39°C (Table 5). The suppression by oriC162 at 38°C was “efficient” in the sense that the colonies had the wild-type appearance and growth rate, but suppression caused by oriC17 at 38°C and oriC162 at 39°C was not efficient. Those colonies grew more slowly than did the oriC+ control strain, were more variable in diameter, and were flattened and irregularly shaped. (The oriC mutations had no detectable effect on dnaX+ or Ts strain growth at 20°C [data not shown].)
TABLE 5.
oriC mutations which reduce initiation efficiency suppress dnaX(Ts)
| Strain | oriC | dnaX | Gena | % Cells with indicated no. of originsb
|
Origins/cell (ave)c | Efficiency of platingd at:
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | >4 | 38°C | 39°C | |||||
| AB2801 | + | Ts | 46 | 1 | 60 | 5 | 32 | 2 | 2.76 | <7 × 10−5 | <4 × 10−6 |
| AB2803 | 160 | Ts | 49 | 14 | 10 | 53 | 23 | 4.03 | 5 × 10−5 | 5.6 × 10−7 | |
| AB2805 | 17 | Ts | 53 | 3 | 49 | 13 | 33 | 3 | 2.80 | 0.5 | 5.8 × 10−5 |
| AB2807 | 162 | Ts | 65 | 12 | 54 | 20 | 11 | 3 | 2.42 | 0.94 | 0.83 |
Generation time in minutes at 30°C.
At 30°C, the blank space indicates less than 1%.
At 30°C.
Relative to efficiency at 30°C.
Exacerbation of dnaX(Ts) by the dnaA(Cos) mutation.
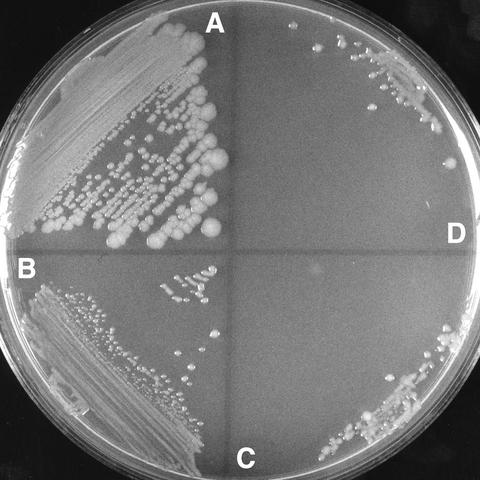
If reducing initiation efficiency was the mechanism of suppression, one would predict that overinitiation should make a dnaX(Ts) strain more sensitive to incubation at higher temperatures. This prediction was tested by construction and testing dnaA(Cos) dnaX(Ts) mutants. The Cos mutations suppress the dnaA(Ts)46 mutant phenotype and restore growth at 39 and 42°C by causing overinitiation (29, 33, 52). However, this overinitiation also limits growth at low temperature, and Cos mutants do not form colonies below 39°C during 24 h of incubation. Cos mutants do grow at 20 and 30°C and form small colonies on enriched medium with an efficiency of 1, provided that incubation is continued for 3 to 5 days. We took advantage of the fact that dnaA(Cos) strains grow at 30°C, although slowly, to construct a double cold-sensitive dnaA(Cos), temperature-sensitive dnaX mutant. dnaA(Cos) and the nearby tna::Tn10 were transduced into dnaX+ and Ts recipients, tetracycline-resistant transductants were selected at 30°C, and dnaA(Cos) transductants were identified by colony growth and morphology at 30°C. The dnaA(Cos) dnaX(Ts) transductants were slightly more temperature sensitive than the Cos dnaX+ control strain, but only over the narrow temperature range of 34 to 37°C (Table 6). Streaked at 37°C on glucose-Casamino Acids medium, the wild-type strain grew normally in 24 h, the dnaX(Ts) strain formed smaller colonies with an efficiency of about 0.24, and the dnaA(Cos) dnaX(Ts) strain did not plate efficiently (Table 6; Fig. 7). The dnaA+ and Cos dnaX(Ts) strains were similar at 30 and 39°C. Therefore, the hyperinitiation dnaA mutation Cos exacerbates the temperature sensitivity of dnaX(Ts) at the somewhat intermediate range of 34 to 37°C.
TABLE 6.
dnaA(Cos) exacerbates dnaX(Ts) temperature sensitivity
| Strain | dnaA | dnaX | Efficiency of platinga at:
|
||
|---|---|---|---|---|---|
| 34°C | 37°C | 39°C | |||
| C600 | + | + | 1.0 | 1.03 | 1.01 |
| AB600 | + | Ts | 1.02 | 0.24 | 2.7 × 10−3 |
| AB2851 | Cos | + | 0.96 | 1.01 | 1.03 |
| AB2853 | Cos | Ts | 0.27 | 1.8 × 10−3 | 1.7 × 10−4 |
Relative to efficiency at 30°C.
FIG. 7.
Growth of dnaA(Cos) dnaX(Ts) mutant strains is inhibited at 37°C. Wild-type strain AB21 (A), dnaX(Ts) strain AB600 (B), and dnaA(Cos) dnaX(Ts) strain AB2853 (streaked twice) (C and D) were incubated on glucose-Casamino Acids medium at 37°C for 24 h.
DISCUSSION
The principal characteristics of dnaA(Sx) suppression of dnaX(Ts) temperature sensitivity include the following. First, suppression allows growth of the Ts mutant at the somewhat intermediate 39 to 40°C but does not restore growth at 42°C, suggesting that DnaA(Sx) permits growth of the Ts mutant with partial DnaX activity but cannot substitute for completely inactive DnaX products. Second, the DnaA suppressor proteins are defective in different, specific DnaA activities in vitro. The DnaA A213D mutant protein is defective in ATP binding, and the R432L and T435K mutants are defective in DnaA box binding (J. R. Walker, K. A. Severson, M. J. Hermandson, K. M. Carr, J. M. Kaguni, and A. Blinkova, unpublished data). These defects are not so severe as to prevent DnaA(Sx) function in vivo—all the suppressor mutants grew readily at 34°C. Third, dnaA(Sx) mutations are recessive to dnaA+, and dnaX(Ts) cells which contain both wild-type and suppressor dnaA alleles do not grow at 39°C (4). Restoration of wild-type initiator activity abolished the suppression phenotype. Fourth, suppression requires functional oriC (4), the target on which DnaA acts. Elimination of the target also eliminated suppression (4). (“Suppression” refers to restoration of growth at 39 to 40°C to the wild-type rate and plating with an efficiency of about 1 at the fast growth rate provided by yeast extract-tryptone medium. “Inefficient” suppression does occur in the absence of oriC, but colony formation requires prolonged incubation and the plating efficiency is about 0.1 to 0.2 [4].) Fifth, one suppressor mutation has been shown to reduce initiation efficiency and cause asynchrony at both 34 and 39°C in a dnaX+ background (8).
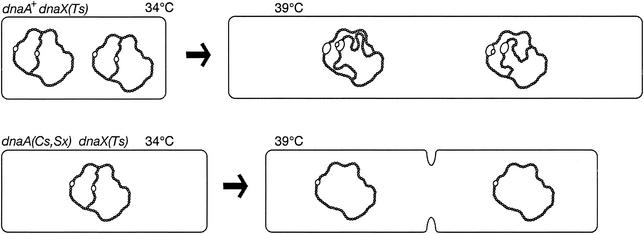
These basic characteristics are consistent with the model (Fig. 8) in which reduced initiation frequency per se results in suppression (4, 8). Lethality of the Ts dnaX mutant at 39 to 40°C could result from gradual failure of polymerization, exacerbated perhaps by failure of the RIDA system (31), which is known to require functional DnaX products (36). Premature initiations might be expected to titrate limiting amounts of partially active DNA polymerase III, thereby contributing to polymerization and growth arrest. dnaA(Sx) mutations would reduce initiation frequency in the Ts dnaX mutant, resulting in reduced chromosome content and allowing partially active DNA polymerase III to complete enough chromosomes at 39 to 40°C to sustain growth, but with fewer chromosomes per cell. Cell division was inhibited in some double mutant cells at 39°C and filaments, which would be expected eventually to lose viability, accounted for about half the culture. However, cell divisions also continued and produced slightly elongated cells which would be expected to sustain continuing growth (Fig. 4 and 5). This mechanism of suppressing dnaX(Ts) has also been proposed by Skovgaard et al., who isolated additional dnaA suppressor mutants and showed that cold-sensitivity is not obligatory for suppression (Skovgaard et al., Abstr. EMBO Workshop Cell Cycle Nucleoid Organization Bacteria).
FIG. 8.
Initiation deficiency model for dnaA(Sx) suppression of dnaX temperature sensitivity. The dnaX(Ts) cells growing at 34°C contain two replicating chromosomes and four origins (open bubbles). On shifting to 39°C, replication forks slow and gradually stall. If the RIDA process is inactivated by the elevated temperature (36), multiple initiations might titrate the limiting, partially active DNA polymerase III. Replication cannot keep pace with growth, and cells lose viability. The dnaA(Sx) mutation reduces the chromosome content even at 34°C and, on shifting to 39°C, the partially active polymerase provides enough activity to replicate a reduced number of chromosomes per cell. Growth and cell division continue (although some filamentous cells form also).
The initiation deficiency model is supported here by five lines of evidence. First, reduction of chromosome content in a dnaA(Sx) dnaX(Ts) double mutant growing at the suppressing 39°C has been demonstrated directly. A dnaA R432L mutation reduced the chromosome content per cell (measured by flow cytometry) and the origin-to-terminus marker frequency (measured by real-time PCR) in both dnaX+ and dnaX(Ts) strains not only at the suppressive 39°C but also at the permissive 34°C in glucose-Casamino Acids medium. Moreover, these reductions were more pronounced at 39 than at 34°C (Fig. 3; Table 4). Second, reduction of the number of origins and chromosome content per cell was a common property of all three known dnaA suppressor mutants (Fig. 1 and 2). Third, suppression was less efficient as the growth rate decreased. It was very efficient at 39°C in glucose-Casamino Acids medium (about 4 origins/wild-type cell) but did not occur at 39°C in glycerol medium (about 1.5 origins/wild-type cell). This is explained as the cumulative effect of the poor carbon source, the dnaA(Sx) mutation reducing initiation efficiency, the temperature-sensitive dnaX gene products, and the 39°C incubation reducing replication to the point that growth (and, therefore, suppression) was not possible. Importantly, glycerol medium did support suppression at 38°C (Table 3). Fourth, some mutations in oriC which caused reduced initiation frequency (62) also suppressed the dnaX(Ts) mutation (Table 5), and the degree of suppression was proportional to the degree of initiation deficiency. oriC160, which had no significant effect on initiation, did not suppress. oriC17, which caused an intermediate effect on initiation frequency, partially suppressed dnaX(Ts), but only at the lower 38°C. The most severely defective mutation, oriC162, also caused the highest degree of suppression; at 38°C, oriC162 restored growth to approximately the wild-type rate and at 39°C, oriC162 also partially suppressed dnaX(Ts) (Table 5). Fifth, an oriC mutation which increased initiation frequency exacerbated the temperature sensitivity of a Ts dnaX strain at the intermediate 34 to 37°C range (Table 6; Fig. 7).
The dnaA R432L dnaX(Ts) double mutant cells responded to 39°C incubation in glycerol medium somewhat differently in liquid and on agar-containing media. In glycerol-containing liquid medium at 39°C, the cells survived and grew at least nine generations from about 105 to about 108 cells/ml, although the generation time was extended to about 3.5 h. On glycerol-containing agar medium at 39°C, they formed colonies with an efficiency of only about 6 × 10−3 (Table 3). Apparently, they survived for a limited number of generations in glycerol-containing liquid medium at 39°C, but most single cells did not grow enough generations at 39°C to form visible colonies on glycerol-containing agar plates.
Prokaryotic DnaA protein structural studies (16, 51) indicate that the A213D mutation, known to decrease affinity for ATP in vitro (J. R. Walker, K. A. Severson, M. J. Hermandson, K. M. Carr, J. M. Kaguni, and A. Blinkova, unpublished data), lies within one of the alpha helices which sandwich the five-stranded beta sheet of domain IIIa (16). The R432L and T435K mutations are located within the DNA binding domain IV DnaA signature sequence, and specifically in the turn region of the helix-turn-helix motif, which binds within the DNA major groove (16) and is critical for DnaA box binding specificity (3, 56). Both these mutations eliminated specific DnaA box binding in vitro (J. R. Walker, K. A. Severson, M. J. Hermandson, K. M. Carr, J. M. Kaguni, and A. Blinkova, unpublished data).
Although the evidence presented here is consistent with the initiation deficiency model for dnaA(Sx) suppression of the Ts dnaX mutant, there are indications that wild-type DnaA might normally associate with polymerization factors. First, DnaA activity is inhibited by polymerization proteins in the RIDA mechanism, suggesting that DnaA must be present at replication forks, or as least those near oriC (31). Second, replication factor τ is the replisome organizer (24, 66), interacting with several components, including binding DnaA directly in vitro (unpublished data cited by Datta et al. [15]). Third, a dnaA(Sx) mutation has been shown to be synthetically lethal, with several dna mutations other than dnaX, including dnaB, -C, -E, and -G (6). Localization of replication proteins within stationary replication factories (reviewed by Lemon and Grossman [37]) could permit multiple interactions of initiation, polymerization, and partition (38) proteins.
Acknowledgments
We thank Julia Grimwade for many helpful discussions about flow cytometry and the suggestion to use PicoGreen, Laura Runyon-Janecky for advice about flow cytometry, Makkuni Jayaram and Sundarapandian Velmurugan for advice about DAPI staining, Clarence Chan for use of a microscope, and Cecil W. Harkey and Allyson Mangum of the University of Texas Institute for Cellular and Molecular Biology Core Facility for advice on use of a flow cytometer.
This work was supported, in part, by Welch Foundation grant F-1379.
REFERENCES
- 1.Akerlund, T., R. Bernander, and K. Nordström. 1992. Cell division in Escherichia coli minB mutants. Mol. Microbiol. 6:2073-2083. [DOI] [PubMed] [Google Scholar]
- 2.Berlyn, M. K. B., K. B. Low, and K. E. Rudd. 1996. Linkage map of Escherichia coli K-12, p. 1715-1902. In F. C. Neidhardt, R. Curtiss III, H. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C.
- 3.Blaesing, F., C. Weigel, M. Welzeck, and W. Messer. 2000. Analysis of the DNA-binding domain of Escherichia coli DnaA protein. Mol. Microbiol. 36:557-569. [DOI] [PubMed] [Google Scholar]
- 4.Blinkova, A., E. Ginés-Candelaria, J. D. Ross, and J. R. Walker. 2000. Suppression of a DnaX temperature-sensitive polymerization defect by mutation in the initiation gene, dnaA, requires functional oriC. Mol. Microbiol. 36:913-925. [DOI] [PubMed] [Google Scholar]
- 5.Blinkova, A., C. Hervas, P. T. Stukenberg, R. Onrust, M. E. O'Donnell, and J. R. Walker. 1993. The Escherichia coli DNA polymerase III holoenzyme contains both products of the dnaX gene, τ and γ, but only τ is essential. J. Bacteriol. 175:6018-6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blinkova, A., and J. R. Walker. 1983. Interactions of DNA replication factors in vivo as detected by introduction of suppressor alleles of dnaA into other temperature-sensitive dna mutants. J. Bacteriol. 153:535-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blinkova, A., and J. R. Walker. 1990. Programmed ribosome frameshift generates the Escherichia coli DNA polymerase III γ subunit from within the τ subunit reading frame. Nucleic Acids Res. 18:1725-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boye, E., A. Blinkova, and J. R. Walker. 2001. Defective initiation in an Escherichia coli dnaA(Cs,Sx) mutant. Biochimie 83:25-32. [DOI] [PubMed] [Google Scholar]
- 9.Boye, E., T. Stokke, N. Kleckner, and K. Skarstad. 1996. Coordinating DNA replication initiation with cell growth: differential roles for DnaA and SeqA proteins. Proc. Natl. Acad. Sci. USA 93:12206-12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bramhill, D., and A. Kornberg. 1988. Duplex opening by DnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell 52:743-755. [DOI] [PubMed] [Google Scholar]
- 11.Braun, R. E., K. O'Day, and A. Wright. 1987. Cloning and characterization of dnaA(Cs), a mutation which leads to overinitiation of DNA replication in Escherichia coli K-12. J. Bacteriol. 169:3898-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carr, K. M., and J. M. Kaguni. 2001. Stoichiometry of DnaA and DnaB protein in initiation at the Escherichia coli chromosomal origin. J. Biol. Chem. 276:44919-44925. [DOI] [PubMed] [Google Scholar]
- 13.Christensen, B. B., T. Atlung, and F. G. Hansen. 1999. DnaA boxes are important elements in setting the initiation mass of Escherichia coli. J. Bacteriol. 181:2683-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dallmann, H. G., S. Kim, A. E. Pritchard, K. J. Marians, and C. S. McHenry. 2000. Characterization of the unique C terminus of the Escherichia coli τ DnaX protein. Monomeric C-τ binds α and DnaB and can partially replace τ in reconstituted replication forks. J. Biol. Chem. 275:15512-15519. [DOI] [PubMed] [Google Scholar]
- 15.Datta, H. J., G. S. Khatri, and D. Bastia. 1999. Mechanism of recruitment of DnaB helicase to the replication origin of the plasmid pSC101. Proc. Natl. Acad. Sci. USA 96:73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erzberger, J. P., M. M. Pirruccello, and J. M. Berger. 2002. The structure of bacterial DnaA: implications for general mechanisms underlying DNA replication initiation. EMBO J. 221:4763-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang, L., M. J. Davey, and M. O'Donnell. 1999. Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol. Cell 4:541-553. [DOI] [PubMed] [Google Scholar]
- 18.Filip, C. C., J. S. Allen, R. A. Gustafson, R. C. Allen, and J. R. Walker. 1974. Bacterial cell division regulation: characterization of the dnaH locus of Escherichia coli. J. Bacteriol. 119:443-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flower, A. M., and C. S. McHenry. 1990. The γ subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshift. Proc. Natl. Acad. Sci. USA 87:3713-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujita, M. Q., H. Yoshikawa, and N. Ogasawara. 1990. Structure of the dnaA region of Micrococcus luteus: conservation and variations among eubacteria. Gene 3:73-78. [DOI] [PubMed] [Google Scholar]
- 21.Fuller, R. S., B. E. Funnell, and A. Kornberg. 1984. The DnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell 38:889-900. [DOI] [PubMed] [Google Scholar]
- 22.Funnell, B. E., T. A. Baker, and A. Kornberg. 1987. In vitro assembly of a prepriming complex at the origin of the Escherichia coli chromosome. J. Biol. Chem. 262:10327-10334. [PubMed] [Google Scholar]
- 23.Ginés-Candelaria, E., A. Blinkova, and J. R. Walker. 1995. Mutations in dnaA which suppress a dnaX(Ts) polymerization mutation are dominant when located in the chromosomal allele and recessive on plasmids. J. Bacteriol. 177:705-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glover, B. P., and C. S. McHenry. 2000. The DnaX-binding subunits δ′ and ψ are bound to γ and not τ in the DNA polymerase III holoenzyme. J. Biol. Chem. 275:3017-3020. [DOI] [PubMed] [Google Scholar]
- 25.Grimwade, J. E., V. T. Ryan, and A. C. Leonard. 2000. IHF redistributes bound initiator protein, DnaA, on supercoiled oriC of Escherichia coli. Mol. Microbiol. 35:835-844. [DOI] [PubMed] [Google Scholar]
- 26.Helmstetter, C. E. 1996. Timing of synthetic activities in the cell cycle, p. 1627-1639. In F. C. Neidhardt, R. Curtiss III, H. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C.
- 27.Helmstetter, C. E., and C. A. Krajewski. 1982. Initiation of chromosome replication in dnaA and dnaC mutants of Escherichia coli B/r F. J. Bacteriol. 149:685-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hersch, B. M., F. T. Farooq, D. N. Barstad, D. L. Blankenhorn, and J. L. Slonczewski. 1996. A glutamate-dependent acid resistance gene in Escherichia coli. J. Bacteriol. 178:3978-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katayama, T., and A. Kornberg. 1994. Hyperactive initiation of chromosomal replication in vivo and in vitro by a mutant initiator protein, DnaAcos, of Escherichia coli. J. Biol. Chem. 269:12698-12703. [PubMed] [Google Scholar]
- 30.Katayama, T., and T. Nagata. 1991. Initiation of chromosomal DNA replication which is stimulated without oversupply of DnaA protein in Escherichia coli. Mol. Gen. Genet. 226:491-502. [DOI] [PubMed] [Google Scholar]
- 31.Katayama, T., T. Kubota, K. Kurokawa, E. Crooke, and K. Sekimizu. 1998. The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosomal replicase. Cell 94:61-71. [DOI] [PubMed] [Google Scholar]
- 32.Kato, J.-I., and T. Katayama. 2001. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 20:4253-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kellenberger-Gujer, G., A. J. Podhajska, and L. Caro. 1978. A cold sensitive dnaA mutant of E. coli which overinitiates chromosome replication at low temperature. Mol. Gen. Genet. 162:9-16. [DOI] [PubMed] [Google Scholar]
- 34.Kim, S., H. G. Dallmann, C. S. McHenry, and K. J. Marians. 1996. τ couples the leading- and lagging-strand polymerases at the E. coli DNA replication fork. J. Biol. Chem. 271:21406-21412. [DOI] [PubMed] [Google Scholar]
- 35.Kitagawa, R., T. Ozaki, S. Moriya, and T. Ogawa. 1998. Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 12:3032-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurowaka, W., S. Nishida, A. Emoto, K. Sekimizu, and T. Katayama. 1999. Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J. 18:6642-6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemon, K. P., and A. D. Grossman. 2001. The extrusion-capture model for chromosome partitioning in bacteria. Genes Dev. 15:2031-2041. [DOI] [PubMed] [Google Scholar]
- 38.Levine, C., and K. J. Marians. 1998. Identification of dnaX as a high-copy suppressor of the conditional lethal and partition phenotypes of the parE10 allele. J. Bacteriol. 180:1232-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lobner-Olesen, A., and E. Boye. 1992. Different effects of mioC transcription on initiation of chromosomal and minichromosomal replication in Escherichia coli. Nucleic Acids Res. 20:3029-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu, M., J. L. Campbell, E. Boye, and N. Kleckner. 1994. SeqA: a negative modulator of replication initiation in E. coli. Cell 77:413-426. [DOI] [PubMed] [Google Scholar]
- 41.Marie, D., D. Vaulot, and F. Partensky. 1996. Application of the novel nucleic acid dyes YOYO-1, YO-PRO-1, and PicoGreen for flow cytometric analysis of marine prokaryotes. Appl. Environ. Microbiol. 62:1649-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marszalek, J., and J. M. Kaguni. 1994. DnaA protein directs the binding of DnaB protein in initiation of DNA replication in Escherichia coli. J. Biol. Chem. 269:4883-4890. [PubMed] [Google Scholar]
- 43.Masui, M., A. Oka, M. Takanami, S. Yasuda, and Y. Hirota. 1985. Sites of DnaA protein-binding in the replication origin of the E. coli K-12 chromosome. J. Mol. Biol. 184:529-533. [DOI] [PubMed] [Google Scholar]
- 44.Messer, W., F. Blaesing, J. Majka, J. Nardmann, S. Schaper, A. Schmidt, H. Seitz, C. Speck, D. Tuengler, G. Wegrzyn, C. Weigel, M. Welzeck, and J. Zakrzewska-Czerwinska. 1999. Functional domains of DnaA protein. Biochimie 81:819-825. [DOI] [PubMed] [Google Scholar]
- 45.Nishida, S., K. Fujimitsu, K. Sekimizu, T. Ohmura, T. Ueda, and T. Katayama. 2002. A nucleotide switch in the Escherichia coli DnaA protein initiates chromosomal replication. Evidence from a mutant DnaA protein defective in regulatory ATP hydrolysis in vitro and in vivo. J. Biol. Chem. 277:14986-14995. [DOI] [PubMed] [Google Scholar]
- 46.Onrust, R., J. Finkelstein, J. Turner, V. Naktinis, and M. O'Donnell. 1995. Assembly of a chromosomal replication machine: two DNA polymerases, a clamp loader, and sliding clamp in one holoenzyme particle. III. Interface between two polymerases and the clamp loader. J. Biol. Chem. 270:13366-13377. [DOI] [PubMed] [Google Scholar]
- 47.Onrust, R., P. T. Stukenberg, and M. O'Donnell. 1991. Analysis of the ATPase subassembly which initiates processive DNA synthesis by DNA polymerase III holoenzyme. J. Biol. Chem. 266:21681-21686. [PubMed] [Google Scholar]
- 48.Seitz, H., C. Weigel, and W. Messer. 2000. The interaction domains of the DnaA and DnaB replication proteins of Escherichia coli. Mol. Microbiol. 37:1270-1279. [DOI] [PubMed] [Google Scholar]
- 49.Sekimizu, K., D. Bramhill, and A. Kornberg. 1987. ATP activates DnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell 50:259-265. [DOI] [PubMed] [Google Scholar]
- 50.Sekimizu, K., D. Bramhill, and A. Kornberg. 1988. Sequential early stages in the in vitro initiation of replication at the origin of the Escherichia coli chromosome. J. Biol. Chem. 263:7124-7130. [PubMed] [Google Scholar]
- 51.Shaper, S., and W. Messer. 1997. Prediction of the structure of the replication initiator protein DnaA. Proteins Struct. Funct. Genet. 28:1-9. [PubMed] [Google Scholar]
- 52.Simmons, L. A., and J. M. Kaguni. 2003. The dnaAcos allele of Escherichia coli: hyperactive initiation is caused by substitution of A184V and Y271H, resulting in defective ATP binding and aberrant DNA replication control. Mol. Microbiol. 47:755-765. [DOI] [PubMed] [Google Scholar]
- 53.Skarstad, K., E. Boye, and H. B. Steen. 1986. Timing of initiation of chromosome replication in individual Escherichia coli cells. EMBO J. 5:1711-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skarstad, K., K. von Meyenburg, F. Hansen, and E. Boye. 1988. Coordination of chromosome replication initiation in Escherichia coli: effects of different dnaA alleles. J. Bacteriol. 170:852-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sutton, M. D., and J. M. Kaguni. 1997. The Escherichia coli dnaA gene: four functional domains. J. Mol. Biol. 274:546-561. [DOI] [PubMed] [Google Scholar]
- 56.Sutton, M. D., and J. M. Kaguni. 1997. Threonine 435 of Escherichia coli DnaA protein confers sequence-specific DNA binding activity. J. Biol. Chem. 272:23017-23024. [DOI] [PubMed] [Google Scholar]
- 57.Tsuchihashi, Z., and A. Kornberg. 1990. Translational frameshifting generates the γ subunit of DNA polymerase III holoenzyme. Proc. Natl. Acad. Sci. USA 87:2516-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.von Freiesleben, U., M. A. Krekling, F. G. Hansen, and A. Lobner-Olesen. 2000. The eclipse period of Escherichia coli. EMBO J. 19:6240-6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.von Freiesleben, U., K. V. Rasmussen, and M. Schaechter. 1994. SeqA limits DnaA activity in replication from oriC in Escherichia coli. Mol. Microbiol. 14:763-772. [DOI] [PubMed] [Google Scholar]
- 60.Walker, J. R., and A. Blinkova. 2002. Mutational analysis of Escherichia coli DnaA protein activities in initiation of chromosome replication, p. 310-317. In V. A. Lanzov (ed.), Bresler memorial lectures: molecular genetics, biophysics and medicine today. Russian Academy of Sciences, St. Petersburg, Russia.
- 61.Walker, J. R., J. A. Ramsey, and W. E. Haldenwang. 1982. Interaction of the Escherichia coli DnaA initiation protein with the DnaZ polymerization protein in vivo. Proc. Natl. Acad. Sci. USA 79:3340-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weigel, C., W. Messer, S. Preiss, M. Welzeck, Morigen, and E. Boye. 2001. The sequence requirements for a functional Escherichia coli replication origin are different for the chromosome and a minichromosome. Mol. Microbiol. 40:498-507. [DOI] [PubMed] [Google Scholar]
- 63.Willets, N. S., A. J. Clark, and B. Low. 1969. Genetic location of certain mutations conferring recombination deficiency in Escherichia coli. J. Bacteriol. 97:244-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xiao, H., V. Naktinis, and M. O'Donnell. 1995. Assembly of a chromosomal replication machine: two DNA polymerases, a clamp loader, and sliding clamps in one holoenzyme particle. IV. ATP-binding site mutants identify the clamp loader. J. Biol. Chem. 270:13378-13383. [DOI] [PubMed] [Google Scholar]
- 65.Yoshikawa, H., and N. Sueoka. 1963. Sequential replication of the Bacillus subtilis chromosome. I. Comparison of marker frequencies in exponential and stationary growth phases. Proc. Natl. Acad. Sci. USA 49:559-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yuzhakov, A., J. Turner, and M. O'Donnell. 1996. Replisome assembly reveals the basis for asymmetric function in leading and lagging strand replication. Cell 86:877-886. [DOI] [PubMed] [Google Scholar]