Abstract
Gelatinase (GelE), a secreted Zn-metalloprotease of Enterococcus faecalis, has been implicated as a virulence factor by both epidemiological data and animal model studies. Expression of gelE is induced at a high cell density by the fsr quorum-sensing system. In the present study, GelE was shown to be responsible for the instability of a number of Asc10 (aggregation substance) mutant proteins, implying that GelE functions to clear the bacterial cell surface of misfolded proteins. Disruption of GelE production led to increased cell chain length of E. faecalis, from a typical diplococcus morphology to chains of 5 to 10 cells. This function of GelE was also exhibited when the protein was expressed in Streptococcus pyogenes. GelE-expressing E. faecalis strains were more autolytic, suggesting that GelE affects chain length through activation of an autolysin. GelE was also essential for degradation of polymerized fibrin. GelE expression reduced the titer of cCF10, the peptide pheromone that induces conjugation of pCF10, and pCF10 had increased conjugation into non-GelE-expressing strains. These new functions attributed to GelE suggest that it acts to increase the dissemination of E. faecalis in high-density environments.
Enterococcus faecalis has become a leading cause of nosocomial infections, primarily due to its high levels of antibiotic resistance and its ability to survive in hospital environments (19). Because of its medical significance, understanding the virulence factors of this organism has become increasingly important. Gelatinase (GelE) is a secreted Zn-metalloprotease of E. faecalis subsp. liquefaciens that has been implicated as one such factor.
GelE (EC 3.4.24.30) was initially characterized 40 years ago and shown to be a Zn-metalloprotease (3), although the enzyme was not designated GelE until its gene was sequenced (39). Zn-metalloproteases of bacteria serve a wide array of functions in normal cell biology and in microbial infections (for reviews, see references 16 and 26). A biochemical analysis of purified GelE showed that it cleaved a number of substrates, including insulin-β chain, Azocoll (Calbiochem 50-100 mesh), insoluble collagen fragments, and the vasoconstrictor endothelin-1, at primarily hydrophobic amino acids (22, 23). It was also found to degrade the pheromone and inhibitor peptides involved in conjugative plasmid transfer of E. faecalis (22). The amino acid sequence of GelE (39) has a high degree of similarity to the sequences of thermolysin of Bacillus subtilis and of elastase, a known virulence factor of Pseudomonas aeruginosa (14). GelE was also shown to be produced as a zymogen with a 192-amino-acid N′ region that is cleaved to produce the active enzyme (22, 39). The gelE gene has been shown to be present more frequently in clinical isolates than in noninfectious strains (7). A gelE mutant E. faecalis strain showed reduced virulence in a mouse peritonitis model (38) and in a Caenorhabditis elegans model of bacterial virulence (37).
Recently, it has been determined that gelE is induced by a quorum-sensing system of E. faecalis encoded by the fsr locus (30, 31). This system contains three genes, fsrA, fsrB, and fsrC, which share homology with agrA, agrB, and agrC, respectively, of Staphylococcus aureus. The fsr genes have been shown to be autoregulated and to regulate the expression of gelE and a downstream serine protease gene, sprE (30), which are expressed from the same transcript (31). It was also shown that expression of the fsrB, fsrC, gelE, and sprE genes is increased in the postexponential stage of growth and is density dependent (31).
This report describes new functions of GelE. Expression of GelE is required for degradation of a number of Asc10 insertion mutant proteins. Genetic disruption of GelE expression also increased the chain length of E. faecalis, and heterologous expression of GelE in Streptococcus pyogenes decreased cellular chain length. It is hypothesized that GelE production decreases chain length through activation of an autolysin. GelE was also found to degrade polymerized fibrin. GelE expression correlated with decreased cCF10 pheromone titer, leading to less pCF10 conjugation into a GelE-expressing strain.
MATERIALS AND METHODS
Bacterial growth and nisin induction.
E. faecalis was grown at 30 or 37°C in Todd-Hewitt broth (THB) (Difco) as indicated. For aggregation analysis, cultures were grown with shaking. Otherwise, no shaking was used. Lactococcus lactis was grown in GM17 medium (M17 salts plus 0.5% glucose) (Difco) at 30°C with no shaking. Streptococcus gordonii (Challis) was grown at 37°C in THB in a candle jar. Streptococcus pyogenes 90-226 was grown in THY medium (THB plus 10% yeast extract) in a candle jar at 37°C. Escherichia coli was grown in Luria-Bertani (Difco) broth at 37°C with shaking for DNA isolation and manipulation. Gelatinase (GelE) production was measured in a plate assay by adding 3% gelatin to liquid THB agar medium. Functional GelE produced a halo that was clearly visible after overnight incubation. Erythromycin (Sigma) was used at a concentration of 200 μg/ml for E. coli and 10 μg/ml for the gram-positive species. TX5128 and TX5243 were also grown with kanamycin (Sigma) at 1 mg/ml. The nisin-inducible vectors were induced with 25 ng of nisin per ml for both broth and plate cultures.
Electrotransformation.
Competent cells of E. faecalis and L. lactis were prepared as previously described (1). Competence in S. gordonii was induced by using the competence peptide (kindly provided by L. Havarstein, Agricultural University of Norway). Two microliters of an overnight culture of S. gordonii was mixed with 100 to 200 ng of DNA and 100 ng of peptide in 200 μl of THB medium. This mixture was incubated for 8 h in a candle jar at 37°C before being plated on selective media. S. pyogenes cells were prepared for electroporation in the following manner. Fifteen milliliters of an overnight culture grown in THY plus 25 mM glycine was inoculated into 500 ml of prewarmed THY plus 25 mM glycine and allowed to grow for 3 h. The bacteria were harvested by centrifugation, resuspended in 50 ml of the culture supernatant, and incubated for 10 min at 43°C. The bacteria were then washed twice with 60 ml of 15% glycerol, resuspended in a final volume of 3 ml of 15% glycerol, and placed on ice or frozen at −80°C. Two hundred microliters of cells was mixed with 10 μg of DNA harvested from E. faecalis and electroporated (2.5 kV, 25 μF, 600 Ω) with a Bio-Rad Gene Pulser. After 1 h of incubation at 37°C, transformants were selected on THY selective solid media.
DNA manipulation and construction of pMSP3614.
Plasmids were isolated from E. coli with the Qiagen midi- or minikit as recommended by the manufacturer. Plasmids were isolated from E. faecalis, L. lactis, S. pyogenes, and S. gordonii by a slight modification of the Qiagen protocol as previously described (1). Restriction enzymes were purchased from Promega and New England Biolabs. PCR was performed with a Perkin-Elmer Gene Amp PCR system or an Eppendorf Mastercycler with BioXact DNA polymerase (Bioline). All sequencing and primer synthesis were done by the Microchemical Facility of the University of Minnesota.
The gelE gene was PCR amplified from OG1RF by using a colony PCR protocol. Using a PCR optimization kit (Boehringer Mannheim), a series of reaction buffers were tested for gelE amplification from colonies of OG1RF. The 25-μl PCR mix consisted of 0.5 μM deoxynucleotide triphosphates, 0.15 μl of BioXact DNA polymerase, and a 10 pM concentration of the primers gel-f (SpeI) (5′-GAAACTAGTTAAGGAAGGAGTTAATTGTTTGATGAAG-3′) and gel-r (PstI) (5′-CTTCTGCAGTTTCATTCATTGACCAGAACAGATTC-3′). Template DNA was obtained by touching a colony of OG1RF with a pipette tip and mixing it into the PCR tube. The reaction was carried out with the following cycle: 10 min at 94°C to lyse the bacterial cells; the series 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min repeated 25 times; and then 72°C for 30 s. The products from five of the reaction buffers that had a good yield were combined and purified by using the QIAquick PCR purification system (Qiagen). This material was then ligated into pGEM-T Easy vector (Promega). The gelE gene was removed with a PstI-SpeI restriction digest and ligated (T4 DNA ligase; Gibco-BRL) into PstI-SpeI-cut pMSP3535. The ligation mix was transformed directly into competent OG1RF, and all erythromycin-resistant transformants showed nisin-inducible gelatinase production. One transformant was chosen, and the plasmid was isolated, sequenced, and designated pMSP3614.
Aggregation analysis.
Aggregation analysis was performed as previously described (41). One-milliliter aliquots of overnight nisin-induced cultures were poured into a plastic cuvette and allowed to settle for 1 h. The optical density was then read at 600 nm on a Beckman DU-70 spectrophotometer.
Western blot analysis.
A lysozyme surface extract of each induced mutant culture was prepared as previously described (13). The lysozyme extraction buffer was slightly modified to include 25 mM EDTA and 5 mg of lysozyme per ml. An equivalent volume (12 μl out of 200 μl of total lysis extract) was loaded onto a sodium dodecyl sulfate-7.5% polyacrylamide gel and transferred to a BA 85 nitrocellulose membrane (Schleicher and Schuell). Western blot analysis was performed with an antibody constructed against an N-terminal domain of Asc10 (25) at a dilution of 1/2,500. Detection was performed with the enhanced chemiluminescence protocol (Pierce).
Determination and visualization of E. faecalis chain length.
To visualize chain length, the cultures were grown overnight without shaking at 37°C. To control for the effects of nisin, all cultures received 25 ng of nisin per ml. Ninety-five microliters of the culture was mixed with 5 μl of crystal violet. The bacteria were then washed twice with 100 μl of 0.9% saline to remove excess crystal violet (4.0 g/liter). Five microliters was placed on a glass slide and covered with a coverslip. Bacteria were visualized on a Nikon Eclipse E800 microscope system at a magnification of ×80. Pictures were taken with the Nikon ACT-1 software.
The relative size of the bacteria was determined by measuring the forward scatter profiles (FSC) by flow cytometry. Overnight cultures were prepared as described above. Fifty-microliter aliquots of these cultures was mixed with 450 μl of Hank's balanced salt solution (Cellgro). A total of 50,000 events were collected on a Becton Dickinson FACScan, and the data were analyzed by using CellQuest version 3.3 software (Becton Dickinson).
Autolysis assay.
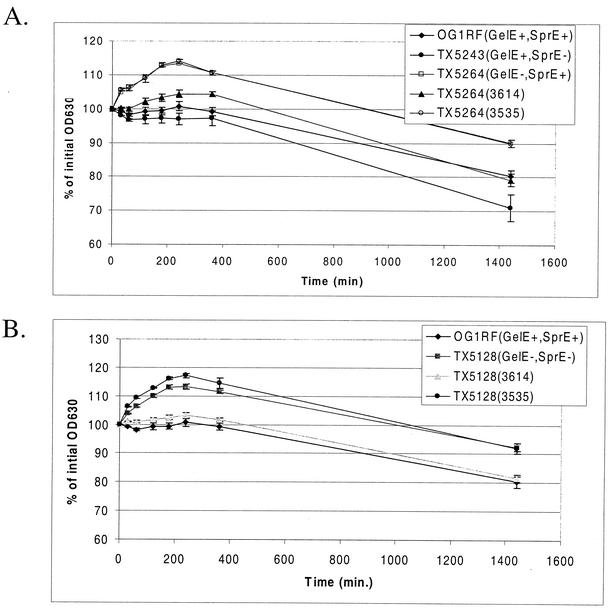
Cell autolysis was determined as described by Qin et al. (32) with minor modifications. Bacteria were grown overnight (16 h) in 1 ml of THB with nisin and the appropriate antibiotics. The bacteria were chilled on ice for 1 h, washed three times with ice-cold distilled water by pelleting in a microcentrifuge, and resuspended in a final volume of 1 ml of 10 mM sodium phosphate buffer (pH 6.8). Three wells of 200 μl of bacteria from each culture were placed in the wells of a Costar 3595 96-well plate (Corning Inc.), and the absorbance was measured on an enzyme-linked immunosorbent assay plate reader at 630 nm. The optical density at 630 nm (OD630) was measured at the times indicated in Fig. 5, and results from the three wells were averaged to give a mean OD630 for each strain at each time point. The data are expressed as a percentage of the initial OD630 value. Each data point represents the mean from four to six independent overnight cultures of each strain.
FIG. 5.
GelE-expressing strains are more autolytic. (A) Autolysis of the single protease mutants compared to that of OG1RF. TX5264 was complemented with GelE expressed from plasmid 3614 (plasmid 3535 is the vector control). (B) Autolysis of the double protease mutant TX5128 compared with that of OG1RF. TX5128 was also complemented with plasmid 3614. The data are plotted at each point as the percentage of the initial OD630. Each point represents the mean for four to six independent cultures, and the error bars depict the standard errors of the means.
Fibrin degradation.
Polymerized fibrin was generated in a Costar 96-well flat-bottom polystyrene high-binding plate (catalog no. 3590; Corning Inc.). In each well, 80 μl of fibrinogen (5 mg/ml) dissolved in 50 mM triethanolamine-100 mM NaCl (pH 7.5) was mixed with 20 μl of 100 mM CaCl2. To this mix, 2 μl of thrombin (0.5 U/ml in distilled water) was added. The fibrin was allowed to polymerize overnight at 4°C. Bacterial strains were grown overnight with nisin induction. The bacteria were pelleted, and the supernatant was filtered with a 0.45-μm-pore-size HT Tuffryn low-protein binding syringe tip filter (Gelman Laboratory). One hundred microliters of the filtered supernatant was added to the top of the fibrin and incubated at 37°C. Pictures were taken with the AlphaImager version 5.5 software and imaging system (Alpha Innotech Corp.). To quantitate the time course of degradation, the OD630 of the polymerized fibrin in the 96-well plates was analyzed on an enzyme-linked immunosorbent assay plate reader after addition of filtered culture supernatant. The relative degradation of the fibrin was determined by calculating the percent decrease in the OD630 by using the equation [(ODtime t − ODback)/(ODtime 0 − ODback)] × 100, where ODtime t is the OD630 at time t as indicated on the graph, ODback is the OD630 of a well with filtered supernatant and unpolymerized fibrinogen mix, and ODtime 0 is the initial OD630 reading. L. lactis strains NZ9800(3614) and NZ9800(3535) were induced with nisin for 48 h before filtering of the supernatant and measurement of fibrin degradation.
Pheromone supernatant titer.
Cultures were grown overnight in M9 plus appropriate antibiotics. They were then subcultured (1:10) in fresh medium (5 ml) without antibiotics in the presence of nisin and grown for 6 h. Culture supernatant was collected, and 100 μl was used in the clumping assay (5). A 16-h culture of OG1RF(pCF10) was used as the indicator strain in the clumping assay. The titer of cCF10 detected is reported as the reciprocal of the dilution in which clumping was last detected.
Conjugation.
Overnight cultures of OG1SSp(pCF10), OG1RF, and TX5128 were grown overnight in THB with tetracycline (15 μg/ml), no antibiotics, and kanamycin (1,000 μg/ml), respectively. The overnight cultures were inoculated 1:50 into fresh THB with no antibiotics and grown without shaking for 2 h. From these cultures, 0.5 ml of OG1SSp(pCF10) was mixed with 0.5 ml of OG1RF or TX5128 and incubated for 30 min at 37°C. Transconjugants and donor cell CFU were determined by serial dilution with selection on TH agar plates containing rifampin (200 μg/ml) and tetracycline (15 μg/ml) and TH plates containing spectinomycin (1,000 μg/ml) and tetracycline (15 μg/ml), respectively.
RESULTS
GelE degrades misfolded Asc10.
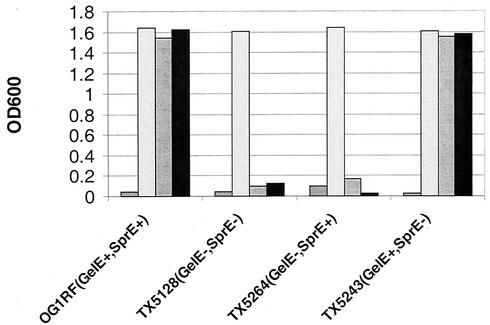
In a previous study, 23 in-frame 31-amino-acid insertion mutations were constructed in the prgB gene, which encodes the surface protein Asc10, the aggregation substance encoded by pCF10. Of these, two mutations, Ω2049 and Ω2421, were able to mediate aggregation of the E. faecalis strain OG1RF at 30 but not 37°C (41). Western blot analysis determined that cells expressing these proteins expressed detectable Asc10 on the cell surface at 30 but not 37°C, suggesting the defect at 37°C was due to protein instability.
To further elucidate the mechanism of this instability, we expressed these two mutant Asc10 proteins in a collection of gelatinase (GelE) and serine protease (SprE) mutants of E. faecalis (Table 1). As previously observed, expression of either insertion mutation in OG1RF (GelE+ SprE+) grown overnight at 37°C resulted in no aggregation, but expression of wild-type Asc10 under the same conditions resulted in high levels of aggregation (Fig. 1). Expression of Ω2049 and Ω2421 in the GelE− SprE− double protease mutant strain TX5128 resulted in wild-type levels of aggregation at 37°C (Fig. 1). To identify which protease is responsible for the mutant protein instability, the insertion mutations were expressed in two E. faecalis strains that were deficient in one of the proteases. Expression of Ω2049 and Ω2421 in the in-frame GelE deletion mutant TX5264 resulted in aggregation at 37°C; however, these mutant proteins failed to elicit aggregation in the SprE− mutant TX5243. Thus, GelE and not SprE is responsible for the instability of Ω2049 and Ω2421 at 37°C.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Bacteria | ||
| E. coli CC118 | C. Manoil | |
| E. faecalis | ||
| OG1RF | GelE+ SprE+ | 10 |
| OG1SSp | 10 | |
| TX5128 | GelE−, SprE−; mγδ insertion in gelE | 38 |
| TX5264 | GelE− SprE+; internal in-frame deletion in gelE | 29 |
| TX5243 | GelE+ SprE−; sprE insertion mutation | 30 |
| S. gordonii DLT (Challis) | Naturally competent | 27 |
| S. pyogenes 90-226 | 6 | |
| L. lactis NZ9800 | 21 | |
| Plasmids | ||
| pCF10 | Pheromone-inducible conjugative plasmid | 11 |
| pMSP3535 | Nisin-inducible cloning shuttle vector | 4 |
| pMSP7517 | Nisin-inducible Asc10 in pMSP3535 | 17 |
| pMSP3614 | Nisin-inducible gelE in pMSP3535 | This study |
| pCWΩ1074 | 31-amino-acid insertion at base 1074 in prgB gene of pMSP7517 | 41 |
| pCWΩ1077 | 31-amino-acid insertion at base 1077 in prgB gene of pMSP7517 | 41 |
| pCWΩ1317 | 31-amino-acid insertion at base 1317 in prgB gene of pMSP7517 | 41 |
| pCWΩ1419 | 31-amino-acid insertion at base 1419 in prgB gene of pMSP7517 | 41 |
| pCWΩ1551 | 31-amino-acid insertion at base 1551 in prgB gene of pMSP7517 | 41 |
| pCWΩ1638 | 31-amino-acid insertion at base 1638 in prgB gene of pMSP7517 | 41 |
| pCWΩ2049 | 31-amino-acid insertion at base 2049 in prgB gene of pMSP7517 | 41 |
| pCWΩ2421 | 31-amino-acid insertion at base 2421 in prgB gene of pMSP7517 | 41 |
| pCWΩr2760 | 31-amino-acid insertion at base 2760 in prgB gene of pMSP7517 | 41 |
| pCWΩ3102 | 31-amino-acid insertion at base 3102 in prgB gene of pMSP7517 | 41 |
| pCWΩr3183 | 31-amino-acid insertion at base 3183 in prgB gene of pMSP7517 | 41 |
FIG. 1.
GelE is required for loss of aggregation of Ω2049 and Ω2421 at 37°C. Aggregation of wild-type Asc10 (7517), the vector control (3535), and two temperature-sensitive aggregation insertion mutants (2049 and 2421) grown at 37°C in four E. faecalis strains expressing all combinations of GelE and SprE was determined by OD. 2049 and 2421 expressed in the GelE− strains TX5128 (GelE− SprE−) and TX5264 (GelE− SprE+) resulted in functional aggregation. However, expression of 2049 and 2421 in the GelE+ strains OG1RF (GelE+ SprE+) and TX5243 (GelE+ SprE−) strain did not aggregate. Protease expression had no effect on the aggregation mediated by wild-type Asc10. Bars, from left to right, 7517, 3535, 2049, and 2421.
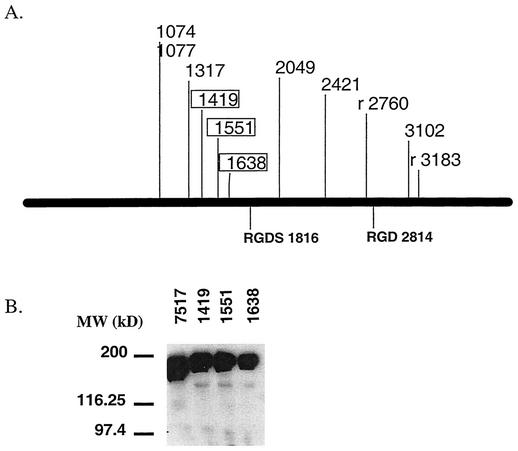
To test whether the loss of aggregation of the rest of the insertions was due to increased protease susceptibility, all 23 Asc10 insertion mutations were expressed in TX5128. As previously described, the insertions shown in Fig. 2A result in reduced or abolished aggregation when expressed in the protease-positive strain OG1RF (41). However, the only insertions that maintain abolished aggregation in TX5128 are Ω1419, Ω1551, and Ω1638 (Fig. 2A). To confirm that these insertions are properly localized to the cell surface, a lysozyme surface extraction of Ω1419, Ω1551, and Ω1638 expressed in TX5128 was performed, and the extract was subjected to Western blotting with an Asc10-specific polyclonal antibody (25). As seen in Fig. 2B, all of these insertions express full-length protein at amounts equivalent to those of wild-type Asc10 (lane 7517). The slight mobility retardation of the mutant proteins is due to the 31-amino-acid insertion. It should be noted that Ω1317 expressed in TX5128 may have a slight defect in aggregation, as it exhibits large, visible aggregates that appear to be less dense than those seen with wild-type Asc10. Also, Ω96 and Ωs3599 are not included in Fig. 2, even though they also fail to aggregate in both OG1RF and TX5128, as Ω96 is located in the Asc10 signal sequence and likely interferes with protein secretion, while Ωs3599 does not have a functional cell wall anchor, as previously reported (41).
FIG. 2.
Expression of Asc10 insertion mutants in TX5128. (A) The insertions that resulted in aggregation defects when expressed in OG1RF are shown on a linear map of the gene encoding Asc10. The boxed insertions are those that maintained an aggregation defect when expressed in TX5128. (B) A Western blot of lysozyme surface extracts of insertions in the putative Asc10 aggregation domain expressed in TX5128 confirms full-length protein expression. (7517, wild-type Asc10). Molecular mass markers are indicated to the left of the blot.
GelE shortens E. faecalis chains.
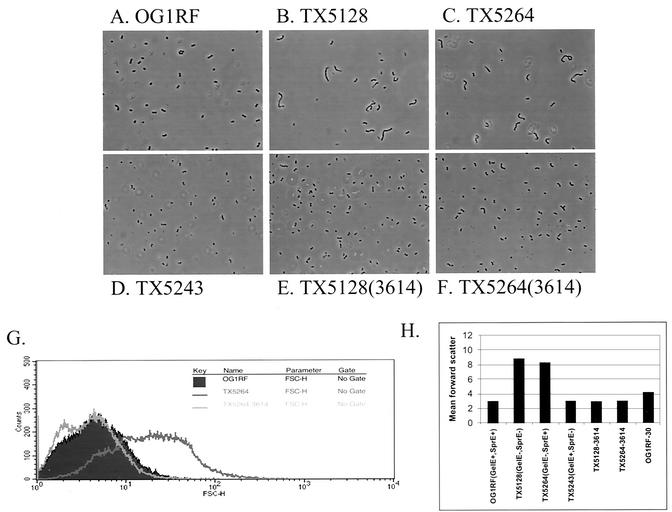
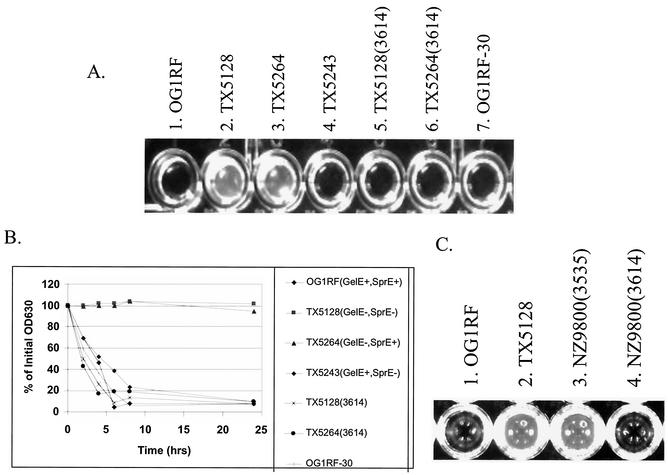
While working with these strains, it was observed that some of the mutants had cell chain lengths longer than that of OG1RF. OG1RF forms a diplococcus morphology that is typical for E. faecalis (Fig. 3A). However, examination of the two GelE-negative strains, TX5128 (Fig. 3B) and TX5264 (Fig. 3C), revealed many bacterial cells present in chains of 5 to 10 cells. The sprE mutant TX5243 (Fig. 3D) appeared similar to OG1RF. To complement the gelE mutations, the gelE gene was PCR amplified from the chromosome of OG1RF and cloned into a transcriptional fusion with the nisA promoter in the nisin-inducible vector pMSP3535(3535) to generate the vector pMSP3614(3614). Overexpression of GelE in TX5128 and TX5264 restored these strains to the diplococcus morphology (Fig. 3E and F). Induction of 3535 alone in these strains had no effect (data not shown).
FIG. 3.
(A to D) GelE− E. faecalis strains form small chains. Strains that lack GelE production form chains of 5 to 10 cells not usually seen with OG1RF. After crystal violet staining, the cells were visualized at a magnification of ×80 with a phase contrast microscope. (A) OG1RF(GelE+ SprE+); (B) TX5128(GelE− SprE−); (C) TX5264(GelE− SprE+); (D) TX5243(GelE+ SprE−). (E and F) Induction of GelE from a complementing plasmid (3614) restores normal diplococcus morphology. (E) TX5128(3614); (F) TX5264(3614). (G) The relative cell sizes of the bacterial populations were determined by analyzing the FSC of 50,000 events on a flow cytometer. The clear population shift of TX5264 (dark line) versus OG1RF (filled) is evident. Expression of GelE in TX5264 (light line) from 3614 complements the increased cell size and restores it back to OG1RF levels. (H) The mean FSC of three independent cultures of each strain was measured (error bars indicate standard errors of the means). OG1RF-30, OG1RF grown overnight at 30°C.
To quantitate these findings, the FSC of these six strains were determined by using flow cytometry. An example of a typical FSC histogram plot is shown in Fig. 3G. The filled-in plot represents the FSC of 50,000 events from an overnight OG1RF broth culture. The clear population shift in TX5264 is shown by the dark line. Expression of GelE in TX5264 restored the population to a size range similar to that of OG1RF, as can be seen by the light line. The mean FSC of each of the strains confirmed that the size difference between the GelE-expressing strains and the gelE mutant strains is significant and can be seen on a population-wide scale (Fig. 3H).
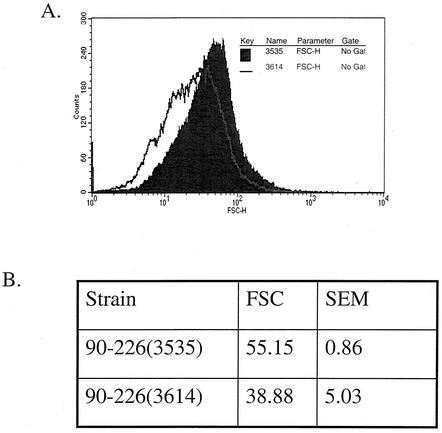
The GelE expression vector pMSP3614 was electroporated into S. pyogenes strain 90-226 to determine if the ability of GelE to reduce chain length is specific to E. faecalis. Induction of GelE expression in 90-226 resulted in a decrease of the chain length but not a complete reduction to single cells of diplococci (Fig. 4). This suggests that the mechanism of chain length reduction mediated by GelE in E. faecalis may be conserved in S. pyogenes strain 90-226.
FIG. 4.
GelE expression in S. pyogenes shortens bacterial chains. Induction of GelE (3614) in strain 90-226 showed a significant (P = 0.032, homoscedastic Student's t test) reduction in chain length versus the vector control (3535) as measured by flow cytometry. (A) Representative histogram of the shift in FSC. (B) Mean FSC and standard error of the mean (SEM) from three independent cultures.
GelE-producing strains increase E. faecalis autolysis.
E. faecalis has been predicted to have two distinct autolysins (12), one with activity against its own cell wall (muramidase-1) (9, 20, 36) and the other with activity against Micrococcus lysodeikticus cell wall (muramidase-2) (2, 8, 9). In the initial characterization of the muramidase-1 autolysin, the Zn-metalloprotease (GelE) characterized from S. faecalis (E. faecalis) subsp. liquefaciens strain 31 (20) was found to speed the lysis of E. faecalis cell walls by activating muramidase-1 activity (36). Also, protease inhibitors inhibit conversion of the 130-kDa latent muramidase-1 to the 87-kDa active form (36). As autolysins have been shown to be critical in the determination of chain length (15, 32, 34, 40, 42), it was possible that the loss of GelE production in TX5128 and TX5264 resulted in less maturation of the muramidase-1 autolysin, leading to increased chain length. To test this hypothesis, the autolysis of stationary-phase cultures of OG1RF and the protease mutants was measured (Fig. 5).
During the first 6 h of the experiment, the OD630 values of TX5264 (GelE− SprE+) actually increased. This result is likely due to incomplete removal of all nutrients from the medium and dilution of waste products during washing, resulting in low levels of bacterial division. However, by 24 h, the OD630 of TX5264 had decreased to 90% of the initial value (Fig. 5A). Cultures of the GelE-producing strains OG1RF and TX5243 showed no significant increase at the early time points of the experiments and decreased to 80 and 71% of the initial values by 24 h. Induction of the gelE gene from plasmid 3614 in TX5264 resulted in a curve similar to that for OG1RF, while results for the vector control, TX5264 (3535), were almost identical to those for TX5264 alone (Fig. 5A). TX5128 (GelE− SprE−) showed results equivalent to those for TX5264 (Fig. 5B).
GelE degrades supernatant cCF10.
Biochemical characterization of purified GelE showed that it was able to degrade purified E. faecalis pheromone and inhibitor peptides (22). To determine if the degradation of pheromone by GelE had any biological relevance in bacterial cultures, the pheromone titers in the supernatants of stationary-phase cultures of E. faecalis OG1RF (GelE+ SprE+) and the protease mutant strains were determined. For these assays, unconcentrated supernatant was serially diluted twofold and analyzed for the ability to aggregate an E. faecalis pCF10 indicator strain. The pheromone titer is presented as the reciprocal of the highest dilution that was able to induce aggregation in the indicator strain. OG1RF showed no cCF10 activity at any dilution (Table 2). TX5128, the double protease mutant, showed a cCF10 titer of 4, while TX5264, the GelE specific mutant, had a titer of only 2. TX5243, the SprE mutant, had no detectable cCF10. Complementation of GelE in TX5128 and TX5264 completely abolished detectable cCF10 in the supernatant (Table 2). This experiment was repeated twice and gave the same result.
TABLE 2.
Protease expression reduces cCF10 in the culture supernatant
| Strain | cCF10 activitya
|
|
|---|---|---|
| Expt 1 | Expt 2 | |
| OG1RF (GelE+ SprE+) | 0 | 0 |
| TX5128 (GelE− SprE−) | 4 | 4 |
| TX5264 (GelE− SprE+) | 2 | 2 |
| TX5243 (GelE+ SprE−) | 0 | 0 |
| TX5128(3614) | 0 | 0 |
| TX5264(3614) | 0 | 0 |
Unconcentrated supernatants (grown for 6 h from a 10% overnight inoculum) from the strains listed were diluted twofold, and cCF10 activity is reported as the inverse of the largest dilution that was able to aggregate an OG1RF(pCF10) indicator strain. The experiment was repeated twice and gave the same results. A zero indicates that no active cCF10 was observed at any dilution examined.
Conjugation of pCF10 into GelE-expressing strains is less frequent.
To determine if the degradation of supernatant cCF10 had any detectable biological consequences, the conjugation of pCF10 from an OG1SSp donor strain into OG1RF and TX5128 was determined. For these experiments, OG1SSp(pCF10) was not induced with exogenous cCF10. Donor cells were incubated with recipient cells at a 1:1 ratio for 30 min. In three independent experiments, conjugation of pCF10 into TX5128 was found to occur at a 2.3- ± 0.4-fold higher level than that into OG1RF (Table 3).
TABLE 3.
pCF10 conjugates more efficiently into TX5128 than OG1RFa
| Expt | Transconjugants/donor cell
|
Fold increase (TX5128/OG1RF) | |
|---|---|---|---|
| TX5128 (GelE− SprE−) | OG1RF (GelE+ SprE+) | ||
| 1 | 6.88 × 10−5 | 3.13 × 10−5 | 2.2 |
| 2 | 7.94 × 10−5 | 2.88 × 10−5 | 2.8 |
| 3 | 5.00 × 10−4 | 2.45 × 10−4 | 2.0 |
TX5128 and OG1RF served as recipients, while OG1SSp(pCF10) was the donor cell.
GelE degrades polymerized fibrin.
Previous work analyzing adherence of E. faecalis to polymerized fibrin led to the observation that a secreted factor produced by OG1RF could degrade fibrin. To determine if GelE or SprE is responsible for this phenotype, the filtered supernatants of stationary-phase cultures of the protease mutants were examined for the ability to degrade fibrin. After a 24-h incubation at 37°C, both OG1RF and TX5243 were able to completely degrade the fibrin (Fig. 6A, cultures 1 and 4), while the non-GelE-expressing strains, TX5128 and TX5264, were unable to degrade the fibrin even up to 24 h of incubation (Fig. 6A, cultures 2 and 3). Nisin-induced expression of GelE from plasmid 3614 restored the fibrin degradation ability of TX5128 and TX5264 (Fig. 6A, cultures 5 and 6) to levels similar to those for OG1RF. Induction of plasmid 3535, the vector control, in these strains had no effect (data not shown).
FIG. 6.
GelE degrades polymerized fibrin. (A) Complete fibrin degradation by the supernatants of strains that express GelE at 24 h. Cultures were grown overnight in the presence of 25 ng of nisin per ml at 37°C, with the exception of well 7, which was grown at 30°C. The supernatants of these cultures were filtered with a 0.45-μm-pore-size syringe tip filter, and a equal volume was added to the polymerized fibrin (time zero). Wells: 1, OG1RF (GelE+ SprE+); 2, TX5128 (GelE− SprE−); 3, TX5264 (GelE− SprE+); 4, TX5243 (GelE+ SprE−); 5, TX5128(3614); 6, TX5264(3614); 7, OG1RF grown at 30°C. (B) The rate of fibrin degradation was also measured by determining the decrease in OD630 of the wells after addition of culture supernatants over the course of 24 h. A representative time course of fibrin degradation is shown. (C) GelE expressed heterologously in L. lactis NZ9800 degrades fibrin at 24 h.
To get a sense of the kinetics of the fibrin degradation, the decrease in OD630 that corresponds to the clearing of the fibrin was measured, and the percent degradation from 0 to 24 h was calculated (see Materials and Methods). A representative example of this experiment is shown in Fig. 6B. Degradation began almost immediately in strains that produce GelE, and complete degradation was seen at 8 h. The non-GelE-expressing strains, TX5128 and TX5264, showed no degradation even at 24 h. After multiple repetitions (n = 6), no statistical difference (P > 0.05) was seen between any of the fibrin-degrading strains.
One possibility is that GelE is required for fibrin degradation through maturation of another E. faecalis protease. In this case, GelE expressed heterologously would not be predicted to degrade fibrin. To determine if GelE expressed heterologously could mediate fibrin degradation, plasmid 3614 and the vector control plasmid 3535 were transformed into L. lactis strain NZ9800. When the cells were plated on gelatin plates with nisin, a light halo formed around NZ9800(3614), indicating GelE production, while no halo was seen with NZ9800(3535). Filtered supernatants from nisin-induced overnight cultures of NZ9800(3614) and NZ9800(3535) were added to wells of polymerized fibrin. As shown in Fig. 6C, at 24 h, the supernatant of GelE-expressing L. lactis completely degraded the polymerized fibrin, suggesting that GelE directly degrades fibrin.
GelE is expressed at 30°C.
As previously described, expression of the Asc10 insertion mutants Ω2049 and Ω2421 in OG1RF at 37°C resulted in no aggregation; however, expression of these same proteins in OG1RF at 30°C did result in aggregation and increased protein stability (41). This result suggested that GelE might not be expressed at 30°C. To address this issue, OG1RF was grown on a gelatinase plate at 30°C overnight. A halo typical of functional GelE production was clearly visible (data not shown). Also, OG1RF grown overnight in a broth culture at 30°C showed the same diplococcus morphology seen at 37°C (data not shown). The relative size of the E. faecalis cells in this culture, as determine by flow cytometry, was slightly greater than that of OG1RF grown at 37°C (Fig. 3H) but much less than that of the GelE mutants. Filtered supernatant of this broth culture was also able to degrade polymerized fibrin comparably to that from an OG1RF culture grown at 37°C (Fig. 6A, culture 7, and B). Clearly, the stability of Ω2049 and Ω2421 when grown at 30°C is not due to a lack of GelE expression.
GelE is functionally processed in three gram-positive heterologous hosts.
Purification of the active GelE protease (22) and sequencing of the gelE gene (39) led to the observation that GelE is first produced as a zymogen with a 192-amino-acid N-terminal fragment that must be cleaved to generate active protease. Two homologous Zn-metalloproteases, elastase of P. aeruginosa and the Vibrio vulnificus metalloprotease (designated VVP), are both matured by autocatalytic cleavage of the N-terminal propeptide (26). Induction of the gelE gene from plasmid 3614 in L. lactis strain NZ9800 resulted in functional GelE production as evidenced by halo production on a gelatinase plate (data not shown) and fibrin degradation (Fig. 5C). This result suggests that the GelE zymogen is also processed in L. lactis. Likewise, induction of plasmid 3614 in S. gordonii Challis and S. pyogenes 90-226 produced functional GelE (data not shown). However, attempts to produce GelE heterologously in E. coli were unsuccessful, as previously shown (39), but this defect in GelE expression is likely due to inefficient recognition of the gram-positive signal sequence of GelE by E. coli. These results show that the processing mechanism of GelE is not specific to E. faecalis and suggest the GelE zymogen may be autoproteolytic.
DISCUSSION
This paper describes the observation and analysis of a number of new functions for GelE, a secreted Zn-metalloprotease of E. faecalis. GelE appears to play a housekeeping role on the bacterial cell surface by removing misfolded surface proteins and reducing cCF10 activity in the culture supernatant. Disruptions in GelE expression resulted in increased cell chain length in E. faecalis, and expression of GelE in S. pyogenes shortened cellular chain length. Finally, GelE expression is necessary for the degradation of polymerized fibrin.
The initial observation that two in-frame 31-amino-acid insertion mutations in Asc10, Ω2049 and Ω2421, were selectively degraded at 37 but not 30°C (41) led us to examine the involvement of the secreted proteases GelE and SprE. The data obtained by using defined, isogenic mutant strains clearly show that GelE is necessary, and that SprE is dispensable, for this degradation. As functional GelE was shown to be expressed at 30°C, the stability of these insertions at 30°C is likely due to alternate folding. Degradation of misfolded surface proteins by GelE could serve an important biological role by removing nonfunctional proteins that are occupying valuable cell surface area.
Although only Ω2049 and Ω2421 showed temperature-specific degradation when expressed in OG1RF, a number of other 31-amino-acid insertion mutations in Asc10 showed partial or deficient aggregation at all temperatures (Fig. 2A) (41). To address whether the loss of aggregation of these insertions was due to increased susceptibility to secreted proteases, all of the Asc10 insertion mutant proteins were expressed in TX5128. Only three insertions, Ω1419, Ω1551, and Ω1638, maintained deficient aggregation when expressed in TX5128, even though good levels of full-length protein of these mutants were observed on the cell surface. Thus, it can be concluded that these three insertions lead to a specific loss of aggregation rather than decreased protein expression due to increased protease susceptibility. These data highlight that future studies examining derivatives of E. faecalis surface proteins should be performed with non-protease-expressing strains (for example, TX5128, OG1X, or FA2-2) to lessen the experimental confound of decreased protein stability on the cell surface.
The data presented in Fig. 3 show that GelE production was required for the diplococcal state of E. faecalis. As purified GelE was shown to activate a muramiadase-1 autolysin from E. faecalis in vitro (36) and GelE-producing strains were shown to have increased autolysis (Fig. 5), it is likely that GelE mediates its effects on bacterial chain length through maturation of the muramidase-1 autolysin of E. faecalis. In support of this hypothesis, Qin et al. (32) found that inactivation of the muramidase-2 autolysin resulted in an increased chain length of 2 to 10 cells, similar to the findings of this report. Heterologous expression of GelE from plasmid 3614 in S. pyogenes also led to reduced chain length, suggesting that these species have a conserved autolysin that can be activated by GelE.
Most studies using animal models attempt to introduce the same number of bacteria by equalizing the input CFU. However, for any bacteria that form chains, the CFU count will underestimate the actual number of bacterial cells. For example, we have found that TX5128 produces fewer CFU per milliliter than OG1RF at equivalent OD values (unpublished observation). Thus, future studies using E. faecalis strains with different protease expression patterns should be cautious with regard to the pleiotropic effect of cellular chain length. A more accurate method to introduce equivalent numbers of bacteria with different chain lengths is to equalize the OD600 values of the input cultures.
Secretion of GelE in the supernatant has a significant impact on active pheromone levels. Typically, the culture supernatant of OG1RF must be concentrated to measure active cCF10 by the clumping assay (5). However, TX5128 had detectable cCF10 in unconcentrated supernatant up to a fourfold dilution. Interestingly, active cCF10 was observed only in a twofold dilution of the supernatant of TX5264, suggesting that SprE, in the absence of GelE, may have cCF10 degradation activity. However, both TX5243 and TX5128 expressing only GelE showed no detectable cCF10 in the absence of SprE. Thus, GelE production in the absence of SprE is sufficient to reduce the titer of active cCF10 to undetectable levels in unconcentrated supernatant. The decrease of cCF10 could also be measured by a slight, but reproducible, 2.3-fold increase of uninduced pCF10 transfer into the protease mutant strain TX5128 versus OG1RF. As CFU counts underestimate the number of TX5128 cells, this fold difference is likely larger than 2.3. Future studies on the biology of the pheromone-inducible plasmid systems should take into account the effect of GelE and SprE on pheromone levels.
GelE-expressing strains were also able to degrade polymerized fibrin. Expression of GelE in TX5128, the double protease mutant, restored the fibrin degradation activity of the supernatant to wild-type levels, clearly indicating that SprE is not essential for this process. Heterologous GelE expression in L. lactis was also able to mediate fibrin degradation. Thus, degradation is likely direct and not through modification of another E. faecalis factor. Degradation of fibrin by members of the elastase protease family is not unprecedented, as fibrinolysis was also observed in the Zn-metalloprotease, VVP, of V. vulnificus (26).
Fibrin cleavage by GelE has important implications for the pathogenesis of E. faecalis. Secreted bacterial proteases that damage host tissue allow bacterial migration and spread. Blood infections of E. faecalis and enterococcal vegetations formed during endocarditis are likely coated with polymerized fibrin (24). One could envision activation of the fsr system in the vegetation, leading to the induction of GelE expression. The degradation of the fibrin layer surrounding the bacteria would allow further dissemination of the organism.
Our laboratory, which has done extensive rabbit endocarditis experiments with E. faecalis (18, 35), uses strain OG1SSp (a weakly protease-producing strain) for these studies, as OG1RF is more virulent and often causes lethality before the conclusion of the experiment (unpublished observation). Expression of Asc10, which has been shown to increase vegetation size in the endocarditis model (18, 35), is rapidly induced in blood and likely facilitates the early steps of vegetation formation. As the bacteria grow to a high cell density inside the vegetation, expression of GelE in OG1RF could lead to seeding of E. faecalis back into the bloodstream through degradation of fibrin and host attachment proteins including Asc10.
Rasmussen and Bjorck have presented a two-step model to explain the proteolytic state of S. pyogenes during an infection (33). The first phase is dominated by surface-attached proteins and low bacterial proteolytic activity that lead to host attachment. This phase presumably reflects the in vitro logarithmic growth state. In the second phase, represented by in vitro stationary-phase growth states with high bacterial density and low-nutrient conditions, the bacterial cell surface has a high level of proteolytic activity that aids bacterial dissemination by cleaving bacterial attachment proteins and host tissue proteins. The induction of GelE during postexponential to stationary phase by the fsr system and the ability of the GelE-expressing strains to degrade misfolded surface proteins, fibrin, and collagen strongly fit with this infection model. Activation of the muramidase-1 autolysin leading to dechaining may also be a mechanism to increase dissemination in high bacterial densities. The reduction in supernatant pheromone would also increase bacterial dissemination by reducing bacterial aggregation of sex pheromone plasmid-containing E. faecalis. This model not only is applicable to in vivo situations but also could be applied to high-density concentrations of E. faecalis in the environment.
The degradation of fibrin by GelE supports its potential role during a host infection. However, the observations that GelE plays a housekeeping role in degrading misfolded surface proteins and supernatant pheromone and is necessary for the diplococcal state of E. faecalis suggest that it also has important non-virulence-related roles in the biology of E. faecalis.
Acknowledgments
This work was supported by NIH grant HL-51987 to G.M.D. C.M.W. was supported by NIH training grant 5 T32 AI07421-5.
REFERENCES
- 1.Bae, T., B. Kozlowicz, and G. M. Dunny. 2002. Two targets in pCF10 DNA for PrgX binding: their role in production of Qa and prgX mRNA and in regulation of pheromone-inducible conjugation. J. Mol. Biol. 315:995-1007. [DOI] [PubMed] [Google Scholar]
- 2.Beliveau, C., C. Potvin, J. Trudel, A. Asselin, and G. Bellemare. 1991. Cloning, sequencing, and expression in Escherichia coli of a Streptococcus faecalis autolysin. J. Bacteriol. 173:5619-5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bleiweis, A. S., and L. N. Zimmerman. 1964. Properties of proteinase from Streptococcus faecalis var. liquefaciens. J. Bacteriol. 98:653-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryan, E. M., T. Bae, H. Kleerebezem, and G. M. Dunny. 2000. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44:183-190. [DOI] [PubMed] [Google Scholar]
- 5.Buttaro, B. A., M. H. Antiporta, and G. M. Dunny. 2000. Cell-associated pheromone peptide (cCF10) production and pheromone inhibition in Enterococcus faecalis. J. Bacteriol. 182:4926-4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cleary, P. P., E. L. Kaplan, J. P. Handley, A. Wlazlo, M. H. Kim, A. R. Hauser, and P. M. Schlievert. 1992. Clonal basis for resurgence of serious Streptococcus pyogenes disease in the 1980s. Lancet 339:518-521. [DOI] [PubMed] [Google Scholar]
- 7.Coque, T. M., J. E. Patterson, J. M. Steckelberg, and B. E. Murray. 1995. Incidence of hemolysin, gelatinase, and aggregation substance among enterococci isolated from patients with endocarditis and other infections and from feces of hospitalized and community-based persons. J. Infect. Dis. 171:1223-1229. [DOI] [PubMed] [Google Scholar]
- 8.Dolinger, D. L., L. Daneo-Moore, and G. D. Shockman. 1989. The second peptidoglycan hydrolase of Streptococcus faecium ATCC 9790 covalently binds penicillin. J. Bacteriol. 171:4355-4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dolinger, D. L., V. L. Schramm, and G. D. Shockman. 1988. Covalent modification of the β-1,4-N-actylmuramoylhydrolase of Streptococcus faecium with 5-mercaptouridine monophosphate. Proc. Natl. Acad. Sci. USA 85:6667-6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunny, G. M., B. L. Brown, and D. B. Clewell. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc. Natl. Acad. Sci. USA 75:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunny, G. M., M. Yuhasz, and E. Ehrenfeld. 1982. Genetic and physiological analysis of conjugation in Streptococcus faecalis. J. Bacteriol. 151:855-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fontana, R. A., M. Boaretti, A. Grossato, E. A. Tonin, M. M. Lleo, and G. Satta. 1990. Paradoxical response of Enterococcus faecalis to the bactericidal activity of penicillin is associated with reduced activity of one autolysin. Antimicrob. Agents Chemother. 34:314-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galli, D., F. Lottspeich, and R. Wirth. 1990. Sequence analysis of Enterococcus faecalis aggregation substance encoded by the sex pheromone plasmid pAD1. Mol. Microbiol. 4:895-904. [DOI] [PubMed] [Google Scholar]
- 14.Galloway, D. R. 1991. Pseudomonas aeruginosa elastase and elastolysis revisited: recent developments. Mol. Microbiol. 5:2315-2321. [DOI] [PubMed] [Google Scholar]
- 15.Garcia, P., M. P. Gonzalez, E. Garcia, R. Lopez, and J. L. Garcia. 1999. LytB, a novel pneumococcal murein hydrolase essential for cell separation. Mol. Microbiol. 31:1275-1281. [DOI] [PubMed] [Google Scholar]
- 16.Hase, C. C., and R. A. Finkelstein. 1993. Bacterial extracellular zinc-containing metalloproteases. Microbiol. Rev. 57:823-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirt, H., S. L. Erlandsen, and G. M. Dunny. 2000. Heterologous inducible expression of Enterococcus faecalis pCF10 aggregation substance Asc10 in Lactococcus lactis and Streptococcus gordonii contributes to cell hydrophobicity and adhesion to fibrin. J. Bacteriol. 182:2299-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirt, H., P. M. Schlievert, and G. M. Dunny. 2002. In vivo induction of virulence and antibiotic resistance transfer in Enterococcus faecalis mediated by the sex pheromone-sensing system of pCF10. Infect. Immun. 70:716-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jett, B. D., M. M. Huycke, and M. S. Gilmore. 1994. Virulence of enterococci. Clin. Microbiol. Rev. 7:462-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawamura, T., and G. D. Shockman. 1983. Purification and some properties of the endogenous, autolytic N-acetylmuramolylhydrolase of Streptococcus faecium, a bacterial glycoenzyme. J. Biol. Chem. 258:9514-9521. [PubMed] [Google Scholar]
- 21.Kuipers, O. P., M. M. Beerthuizen, R. J. Siezen, and W. M. de Vos. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis: requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281-291. [DOI] [PubMed] [Google Scholar]
- 22.Makinen, P., D. B. Clewell, F. An, and K. K. Makinen. 1989. Purification and substrate specificity of a strongly hydrophobic extracellular metalloendopeptidase (“gelatinase”) from Streptococcus faecalis (strain OG1-10.). J. Biol. Chem. 264:3325-3334. [PubMed] [Google Scholar]
- 23.Makinen, P., and K. K. Makinen. 1994. The Enterococcus faecalis extracellular metalloendopeptiadase (EC 3.4.24.30; coccolysin) inactivates human endothelin at bonds involving hydrophobic amino acid residues. Biochem. Biophys. Res. Commun. 200:981-985. [DOI] [PubMed] [Google Scholar]
- 24.McCormick, J., H. Hirt, G. M. Dunny, and P. M. Schlievert. 2000. Pathogenic mechanisms of enterococcal endocarditis. Curr. Infect. Dis. Rep. 2:315-321. [DOI] [PubMed] [Google Scholar]
- 25.McCormick, J. K., H. Hirt, C. M. Waters, T. J. Tripp, G. M. Dunny, and P. M. Schlievert. 2001. Antibodies to a surface-exposed, N-terminal domain of aggregation substance are not protective in the rabbit model of Enterococcus faecalis infective endocarditis. Infect. Immun. 69:3305-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyoshi, S., and S. Shinoda. 2000. Microbial metalloproteases and pathogenesis. Microb. Infect. 2:91-98. [DOI] [PubMed] [Google Scholar]
- 27.Pakula, R., and W. Walczak. 1963. On the nature of transformable streptococci. J. Gen. Microbiol. 31:125-133. [DOI] [PubMed] [Google Scholar]
- 28.Poquet, I., V. Saint, E. Seznec, N. Simoes, A. Bolotin, and A. Gruss. 2000. HtrA in the unique surface housekeeping protease in Lactococcus lactis and is required for natural protein processing. Mol. Microbiol. 35:1042-1051. [DOI] [PubMed] [Google Scholar]
- 29.Qin, X. 2000. Identification and characterization of virulence in Enterococcus faecalis. Ph.D. thesis. University of Texas Graduate School of Biomedical Sciences, Houston.
- 30.Qin, X., K. V. Singh, G. M. Weinstock, and B. M. Murray. 2000. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect. Immun. 68:2579-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin, X., K. V. Singh, G. M. Weinstock, and B. M. Murray. 2001. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J. Bacteriol. 183:3372-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qin, X., K. V. Singh, Y. Xu, G. M. Weinstock, and B. E. Murray. 1998. Effect of disruption of a gene encoding an autolysin of Enterococcus faecalis OG1RF. Antimicrob. Agents Chemother. 42:2883-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rasmussen, M., and L. Bjorck. 2002. Proteolysis and its regulation at the surface of Streptococcus pyogenes. Mol. Microbiol. 43:537-544. [DOI] [PubMed] [Google Scholar]
- 34.Sanchez-Puelles, J. M., C. Ronda, J. L. Garcia, P. Garcia, R. Lopez, and E. Garcia. 1986. Searching for autolysin functions: characterization of a pneumococcal mutant deleted in the lytA gene. Eur. J. Biochem. 158:289-293. [DOI] [PubMed] [Google Scholar]
- 35.Schlievert, P. M., P. J. Gahr, A. P. Assimacopoulos, M. M. Dinges, J. A. Stoehr, H. Hirt, and G. M. Dunny. 1998. Aggregation and binding substances enhance pathogenicity in rabbit models of Enterococcus faecalis endocarditis. Infect. Immun. 66:218-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shockman, G. D., and M. C. Cheney. 1969. Autolytic enzyme system of Streptococcus faecalis (V). J. Bacteriol. 98:1199-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sifri, C. D., E. Mylonakis, K. V. Singh, X. Qin, D. A. Garsin, B. M. Murray, F. M. Ausubel, and S. B. Calderwood. 2002. Virulence effect of Enterococcus faecalis protease genes and the quorum-sensing locus fsr in Caenorhabditis elegans and mice. Infect. Immun. 70:5647-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh, K. V., X. Qin, G. M. Weinstock, and B. M. Murray. 1998. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. J. Infect. Dis. 178:1416-1420. [DOI] [PubMed] [Google Scholar]
- 39.Su, Y. A., M. C. Sulavik, P. He, K. K. Makinen, P. Makinen, S. Fiedler, R. Wirth, and D. B. Clewell. 1991. Nucleotide sequence of the gelatinase gene (gelE) from Enterococcus faecalis subsp. liquefaciens. Infect. Immun 59:415-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomasz, A. 1968. Biological consequence of the replacement of choline by ethanolamine in the cell wall of pneumococcus: chain formation, loss of transformability, and loss of autolysis. Proc. Natl. Acad. Sci. USA 59:86-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waters, C. M., and G. M. Dunny. 2001. Analysis of functional domains of the Enterococcus faecalis pheromone-induced surface protein aggregation substance. J. Bacteriol. 183:5659-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yother, J., K. Leopold, J. White, and W. Fischer. 1998. Generation and properties of a Streptococcus pneumoniae mutant which does not require choline or analogs for growth. J. Bacteriol. 180:2093-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]