Abstract
The serine receptor (Tsr) from Escherichia coli is representative of a large family of transmembrane receptor proteins that mediate bacterial chemotaxis by influencing cell motility through signal transduction pathways. Tsr and other chemotaxis receptors form patches in the inner membrane that are often localized at the poles of the bacteria. In an effort to understand the structural constraints that dictate the packing of receptors in the plane of the membrane, we have used electron microscopy to examine ordered assemblies of Tsr in membrane extracts isolated from cells engineered to overproduce the receptor. Three types of assemblies were observed: ring-like “micelles” with a radial arrangement of receptor subunits, two-dimensional crystalline arrays with approximate hexagonal symmetry, and “zippers,” which are receptor bilayers that result from the antiparallel interdigitation of cytoplasmic domains. The registration among Tsr molecules in the micelle and zipper assemblies was sufficient for identification of the receptor domains and for determination of their contributions to the total receptor length. The overall result of this analysis is compatible with an atomic model of the receptor dimer that was constructed primarily from the X-ray crystal structures of the periplasmic and cytoplasmic domains. Significantly, the micelle and zipper structures were also observed in fixed, cryosectioned cells expressing the Tsr receptor at high abundance, suggesting that the modes of Tsr assembly found in vitro are relevant to the situation in the cell.
The serine receptor (Tsr), one of four methyl-accepting chemotaxis proteins (MCPs) that span the inner membrane of Escherichia coli, initiates responses and governs adaptation to changes in the serine concentration. MCPs belong to a large class of transducers (21, 46), which sense a variety of environmental cues and are the inputs to sensory pathways that bias cell movement toward favorable environments (12). The chemotaxis pathways belong to the two-component superfamily of signal transduction pathways (17, 42), which are chiefly found in prokaryotes. A two-component pathway consists of a sensor, which is frequently an integral membrane protein possessing kinase activity, and one or more cytoplasmic phosphate-accepting response regulator proteins. The transmembrane sensor-kinases of the chemotaxis pathways are often noncovalent complexes between MCPs (which have no enzyme activity) and two soluble cytoplasmic proteins, namely, an adaptor protein (CheW) and a kinase (CheA) (15, 39).
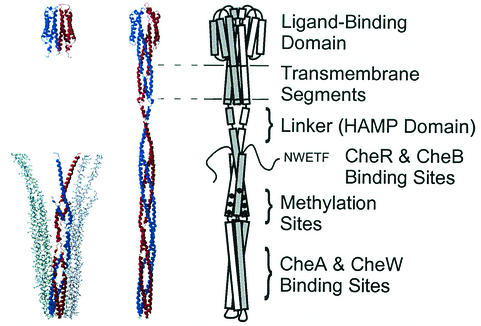
Elucidation of the structure and distribution of receptors in the membrane of the cell is integral to understanding the molecular basis of signaling by the transmembrane sensor (MCP-CheW-CheA) complexes. X-ray structure determination of the soluble domains has clearly defined the dimeric organization of the 60-kDa receptor subunits (19, 31, 45), and functional studies have helped to elucidate the role of dimer organization in the mechanism of transmembrane signaling (references 32 and 12 and references therein). Figure 1 summarizes the relationship between X-ray structure data obtained with soluble receptor fragments and an atomic model of the intact receptor and its functional domains. The dimeric organization of the periplasmic ligand-binding domain, which is evident in the X-ray structure, also places the ligand-binding pocket at the dimer interface (45). The structure of the cytoplasmic domain, which interacts with the signaling proteins and contains the sites methylated during adaptation, has proven to be an extended coiled-coil hairpin that also forms dimers and packs in the crystal as a trimer of dimers (19). Using these structures, Kim et al. have constructed a plausible model of the intact receptor dimer (19, 20). The model is largely consistent with numerous site-directed sulfhydryl cross-linking studies (reviewed in reference 13) that provide structural information in regions where high-resolution data are missing, notably in the transmembrane region, and in the flexible linker that joins the second transmembrane helix to the cytoplasmic domain (6).
FIG. 1.
An atomic model of the bacterial chemoreceptor dimer (Kim et al. [19]; middle illustration), based on high-resolution X-ray structures (left illustration) of the aspartate receptor (Tar) ligand-binding domain and the Tsr cytoplasmic fragment, which are represented here with structures of the Salmonella Tar ligand-binding domain (45) and the E. coli Tsr cytoplasmic domain fragment (19) (PDB accession numbers 1vlt and 1qu7, respectively.) The two subunits in the dimers are shown in blue and red ribbon representations. The cytoplasmic domain fragment structure also displays two more dimers (blue-green and gray stick representations), which appear in the trimer-of-dimers arrangement in the crystal. The illustration of the receptor dimer (right), in which the two subunits are different shades of gray, depicts the functional domains of the receptor.
Although numerous biochemical studies implicate the involvement of extended interactions among receptor subunits in the membrane during signaling (i.e., interactions among receptor dimers), direct structural evidence of these interactions is sparse. Biochemical properties of the signaling system that support a role for such extended interactions include (i) clustering of soluble cytoplasmic domain receptor fragments (either alone or in complexes with the kinase) (14, 28, 29), (ii) transmethylation of receptor dimers (22, 26, 44), (iii) the stoichiometry of receptor-CheW-CheA complexes and the cooperativity of ligand-mediated inhibition of kinase activity (5, 24, 25), and (iv) the interdependency of signaling and response sensitivity among receptors of different ligand specificities (1, 16). These data provide the impetus to obtain direct structural information of intact receptors in membranes. Electron and light microscopy studies conducted so far localize the receptors and cytoplasmic signaling proteins at the poles of the E. coli cell (7, 30, 41), and although these represent an important step in defining the properties of receptor complexes in the cell, the resolution is insufficient for determination of the disposition of individual subunits. Consequently, little information is available that defines, for example, the orientation of the cytoplasmic domains with respect to the plane of the membrane or the lateral distribution of subunits.
To obtain a better understanding of the arrangement of the receptors in the cell membrane, we have carried out an electron microscopic study on isolated receptor-containing membrane preparations and on cryosections of an antibody-labeled E. coli Tsr overexpressor. Images recorded from negatively stained specimens of the membrane extracts are sufficiently well defined to generate plausible interpretations of receptor subunit orientation and intersubunit arrangements. Although some of these interactions are consistent with the known packing of the soluble domains, others are novel. Moreover, arrangements of receptor subunits similar to those observed in the membrane extracts also appear to be present in whole cells, suggesting that such interactions have physiological relevance.
MATERIALS AND METHODS
Bacteria strains, plasmids, and biochemical reagents.
E. coli strain RP437 is wild type for chemotaxis (35). The plasmids pHSe5.tsrQEQE and pHSe5.tsrQQQQ (where QEQE and QQQQ denote the amino acid composition at the sites of methylation [Tsr residues 297, 304, 311, and 493]) direct IPTG (isopropyl-β-d-thiogalactopyranoside)-regulated expression of E. coli Tsr in wild-type and fully modified forms, respectively (37). The E. coli strain HCB721 [relevant genotype: Δtsr(7021) trg::Tn10 Δ(cheA-cheY)::XhoI(Tn5) (9)] does not express the receptors Tar, Tsr, Trg, and Tap or the cytoplasmic signaling proteins CheA and CheW (found in receptor complexes) or CheR and CheB (the enzymes involved in altering the level of receptor covalent modification). HCB721 transformed with pHSe5.tsr maintains overproduced Tsr in the genetically coded state of covalent modification. The detergents, Tween 80, octyl glucoside (OG), and nonylglucoside (NG), were obtained from Sigma-Aldrich (St. Louis, Mo.) and Anatrace (Maumee, Ohio).
Preparation and electron microscope analysis of inner membrane samples.
Inner membranes containing TsrQEQE were prepared from HCB721/pHSe5.tsrQEQE by osmotic shock, isolated on sucrose gradients as described previously (27, 34), and stored at −80°C in pH 8.0 buffer (10 mM Tris HCl, 5% [wt/vol] glycerol, 1 mM phenylmethylsulfonyl fluoride, 1 mM EDTA). The inner membranes were treated with detergent in the manner described by Corless et al. (10) prior to electron microscopic analysis. Although a range of conditions (noted in parentheses as follows) were tested, detergent treatment typically consisted of a 4-h incubation (2 to 20 h) of 5 μM Tsr (3 to 30 μM) at 20°C with a Tween 80 concentration of 0.3% (wt/vol) (0.03 to 2.0%) and sometimes also OG or NG (0.001 to 0.1%) in a pH 7.5, 50 mM Tris buffer with 100 mM NaCl, 10% (wt/vol) glycerol, 1 mM EDTA, and 1 mM AEBSF [4-(2-aminoethyl)-benzene sulfonyl fluoride]. No degradation of Tsr was observed on sodium dodecyl sulfate (SDS)-polyacrylamide electrophoresis gels following these treatments. To determine the extent to which the detergent treatment extracted lipid and protein from the membrane, some samples were subjected to centrifugation (30 min at 180,000 × g) and the protein and phospholipid contents in the supernatant and pellet fractions were measured by the Lowry (Bio-Rad DC protein assay kit; catalog no. 500-0111) and phosphate (8) assays, respectively. The fraction of Tsr in the pellet and the supernatant following detergent treatment was also estimated by densitometric analysis of Coomassie-stained SDS gels. For electron microscopy, the preparations were deposited on glow-discharged, carbon-coated grids. At 30 s after the application of a membrane sample (3 μl), grids were prepared for microscopy by two (45 s) washes in detergent-free, low-ionic-strength Tris buffer, stained in 1% uranyl acetate (10 s), and blotted dry. Images were recorded using a Tecnai 12 electron microscope operating at 120 kV and equipped either with a tungsten filament or an LaB6 crystal and a Gatan 2k × 2k charge-coupled device camera.
Cell growth, fixation, and immunogold electron microscopy of frozen sections.
Overnight T-broth (1% tryptone, 0.5% NaCl) cultures (2 ml) were inoculated with colonies from freshly grown LB plates, which had been streaked with cells from frozen permanents of HCB721/pHSe5.tsrQQQQ. T-broth cultures (50 ml) were inoculated with 50 μl of the overnight culture and were grown at 35°C until they entered exponential phase (optical density at 650 nm of ∼0.1), at which time tsr expression was induced with 1 mM IPTG. Cells were grown for 2 h, harvested, and then fixed at room temperature for 2.5 h in a mixture of 2% paraformaldehyde and 0.2% glutaraldehyde in PHEM buffer {120 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], 50 mM HEPES [pH 6.9], 4 mM MgCl2, 20 mM EGTA}. The fixed cells were collected by centrifugation, resuspended in a minimum of PHEM buffer, and gently mixed with an equal amount of molten gelatin (2%) at 37°C. After solidifying at room temperature, the cell-containing gelatin pellets were cut into 1-mm cubes and infiltrated with a 2.3 M sucrose solution in 0.1 M sodium phosphate buffer (pH 7.4). Cubes of gelatin were frozen to the surface of special aluminum pins by being plunged into liquid nitrogen. By using an α-Tsr antibody directed against the highly conserved signaling domain of Tsr (1), sections of the frozen cubes were cut in a freezing ultramicrotome at −100°C and immunogold labeled as described previously (36). Sections were examined as described above for the membrane preparations.
RESULTS
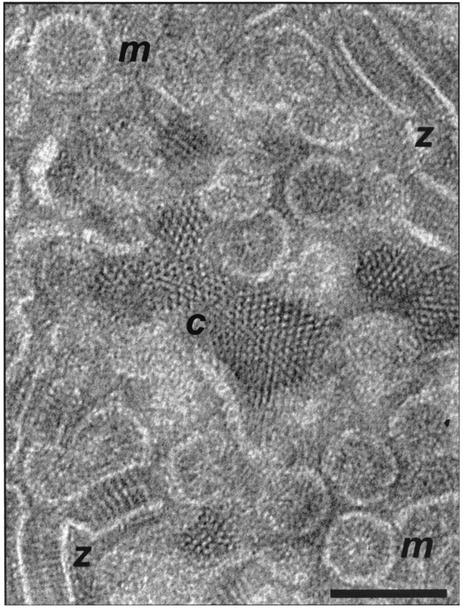
The detergent-treated inner membranes isolated from E. coli cells induced to overexpress the wild-type form of Tsr (TsrQEQE) were observed by conventional transmission electron microscopy of negatively stained specimens. Regular assemblies of receptor subunits were found; Fig. 2 presents a collection of the assemblies seen in a large membrane aggregate. Three distinct modes of assembly can be identified, which are referred to as crystalline (c), micellar (m), and zippered (z) by virtue of the visual similarity each bears to its namesake. Hybrid structures (e.g., micelle-zipper and micelle-crystallite assemblies) were also observed, which serve to indicate how the different forms join and/or interconvert. We attribute these structures to the Tsr protein, since they are not observed in membranes isolated from control cells from which the major chemoreceptor genes (tar, tsr, trg, and tap) were deleted and since SDS-polyacrylamide gel electrophoresis analysis of the inner membrane protein composition isolated from the overexpressor demonstrates that Tsr proteins are present in an overwhelming majority. Treatment with Tween 80 solubilized ca. 65% of the membrane phospholipid and less than 10% of Tsr, but as a consequence, the large membrane aggregates were often separated so that the crystalline, micellar, and zippered structures were observable as individual entities. The orientation and gross conformation of the Tsr subunits are readily apparent in the separated assemblies. As described below, observations of the three types of subunit assembly allow us to conclude that (i) there is significant agreement between the overall structure of the Tsr subunit in the membrane samples and the model of the chemoreceptor dimer (judged on the basis of X-ray structure data of the periplasmic and cytoplasmic domains) (Fig. 1) (19) and (ii) the packing interactions among Tsr dimers in the membrane assemblies, while compatible with the trimer-of-dimer arrangement, also display a novel and specific interdigitating interaction in the zipper-like assembly.
FIG. 2.
Electron micrograph from a negatively stained inner-membrane sample (isolated from an E. coli strain that overexpresses TsrQEQE) which was treated with 0.3% Tween 80. Zippers (z), micelles (m), and crystalline patches (c), which are three types of structures that are characteristic of these preparations, can be seen in this micrograph. Scale bar, 100 nm.
Ordered two-dimensional assemblies.
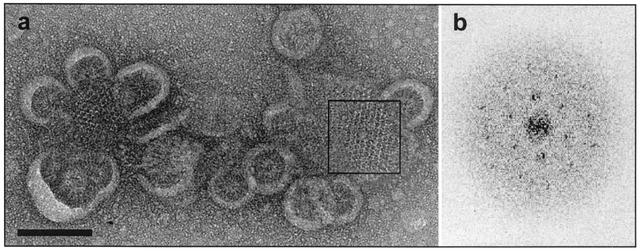
Figure 3a, an electron microscope image of a negatively stained specimen of detergent-treated Tsr inner membrane, displays two examples of ordered receptor arrays connected to micellar structures. The arrays in Fig. 3a can be plausibly interpreted as projections of membrane patches viewed from a perspective perpendicular to the plane of the membranes. Although the arrays were limited in extent (∼103 Å), it was possible to conduct a preliminary analysis of the subunit arrangement in the membrane by generating a Fourier transform (Fig. 3b) of the boxed region in Fig. 3a, which was found to be an approximately hexagonal lattice with a unit cell dimension of 75 Å and a corresponding unit cell area of ∼5,000 Å2. An estimate of the number of Tsr receptor subunits that is compatible with this unit cell area is based on the following reasoning. The average cross-sectional area observed in a well-ordered two-dimensional protein crystal for an α-helix perpendicular to the membrane plane is ∼180 Å2 (18). Since the ligand-binding domain of the Tsr receptor subunit is a bundle of four helices (Fig. 1) that are likely to be nearly perpendicular to the membrane, each receptor subunit has an approximate cross-sectional area of 800 Å2 in loosely packed crystals like those observed in our samples. Thus, each unit cell is large enough to accommodate six (or fewer) Tsr molecules, i.e., three dimers, which is consistent with the trimer-of-dimer packing suggested by Kim et al. (20).
FIG. 3.
(a) An electron micrograph of a negatively stained, ordered two-dimensional array of Tsr in a membrane sample that was incubated with detergent (Tween 80 = 0.08% [wt/vol], [OG] = 0.1% [wt/vol]) for 12 h. Scale bar, 100 nm. (b) The optical transform generated from the region of ordered Tsr molecules (corresponding to the boxed region in panel a) roughly corresponds to a lattice with constants a = ∼75 Å, b = ∼75 Å, and γ = ∼60°.
Micellar assemblies.
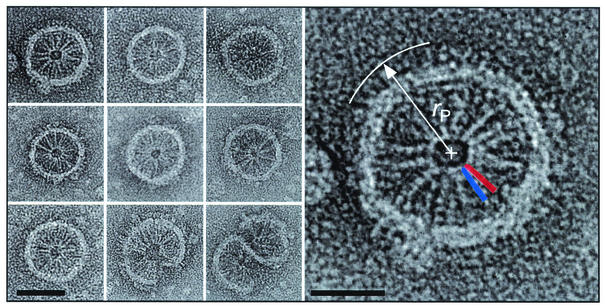
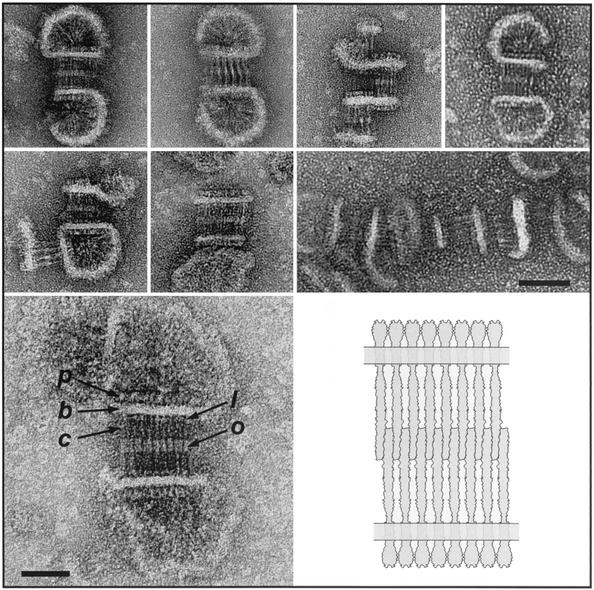
In the crystal structure of the Tsr cytoplasmic domain (Fig. 1), the trimer-of-dimers arrangement is due to subunit interactions near the coiled-coil hairpin. Similarly, in the micellar assemblies presented in Fig. 4, the cytoplasmic domains are analogous to the spokes of a wheel, which connect at the hub through subunit interactions that have a strong resemblance to the trimer-of-dimer interaction. The resemblance is depicted in the enlarged image of this micellar structure, shown at the right in Fig. 4, in which silhouettes of two dimers are placed at the hub. The cytoplasmic domains radiate outward and are bounded by an annulus that is probably composed of lipid, detergent, and receptor transmembrane segments. In some cases, the periplasmic domains can be discerned outside the lipid ring, as expected from this packing arrangement. Characteristic radial dimensions were measured for several micellar structures, including those shown in Fig. 4, and are summarized as averages in angstroms ± standard deviations as follows: radius of hairpin contact (rH), 52 ± 5 (n = 11); radius at the membrane inner edge (rI), 245 ± 13 (11); radius at the membrane outer edge (rO), 300 ± 16 (11); and outermost radius (rP), 362 ± 32 (3). Estimates of the characteristic dimensions of the receptor, e.g., the end-to-end length, were determined as differences of two radial dimensions, e.g., rP − rH, on each micellar assembly individually and then averaged. The result of this analysis is presented in Table 1 for both the micellar and (as discussed below) the zippered assemblies. Dimensions estimated from the electron micrographs of the two types of assemblies are in good agreement. The measured estimate for the end-to-end length of the receptor (310 Å) is ∼20% shorter than the length of the receptor dimer model described by Kim et al. (19).
FIG. 4.
Nine panels at left: a gallery of isolated Tsr micellar assemblies. Scale bar, 50 nm. Right panel: An enlarged micellar assembly of Tsr, illustrating how characteristic radial dimensions are determined (in this case, rP). Also superimposed on this assembly are scaled silhouettes of two Tsr cytoplasmic domain dimers, which are patterned after the trimer-of-dimer organization of subunits observed in the crystal structure of the Tsr cytoplasmic domain fragment (residues 286 to 526 [19]) and represent about 70% of the C-terminal portion of the Tsr protein that is located in the cytoplasm. The individual dimers are rendered as blue and red cigar shapes in which the pair of dimers forms a V-shaped assembly. Scale bar, 25 nm.
TABLE 1.
Receptor dimensions determined from micelle and zipper assemblies
| Dimensiona | Avg dimension ± SD in Å (n)
|
|
|---|---|---|
| Micelles | Zippers | |
| Length of ligand binding domain (p) | 62 ± 8 (3) | 60 ± 6 (13) |
| Lipid membrane thickness (b) | 55 ± 5 (11) | 50 ± 4 (13) |
| Length of linker region (l) | 33 ± 3 (13) | |
| Overlap length (o) | 65 ± 6 (10) | |
| Length of cytoplasmic domain (c) | 193 ± 11 (11) | 189 ± 6 (17) |
| Overall length of receptor | 310 ± 12 (3) | 300 ± 11 (18) |
See Fig. 5 legend for explanation of dimension notation.
Zipper assemblies.
Two prominent features of the zippered assemblies shown in Fig. 5 are the bilateral symmetry and the parallel striations perpendicular to the zipper axis; these features allow us to deduce that the zippered structure represents a bilayer of interdigitating receptors. The transmembrane region of the receptor serves to maintain the alignment among the individual subunits, which lie alongside one another in ribbon-like fashion. The alignment among the subunits facilitates the identification of the receptor domains and makes clear the correspondence to the domains in the receptor dimer model (Fig. 1). The regularity of the overlap (65 ± 6 Å [Table 1]) implies the existence of specific contacts between the cytoplasmic domains in opposing leaflets and gives rise to the double-layer arrangement. The cross-sectional areas of the periplasmic and cytoplasmic domains in the receptor dimer are determined largely by the number of α-helices in each domain (eight and four helices, respectively). Interdigitation effectively doubles the close-packed area on the cytoplasmic side of the membrane (from four helices to eight), and as a result, the cross-sectional areas on the two sides of the membrane are expected to be essentially equivalent, facilitating parallel alignment of the opposing leaflets in the receptor bilayer (as depicted in the illustration in Fig. 5).
FIG. 5.
Top seven panels: a gallery of isolated zippered assemblies. Scale bar, 50 nm. Lower left panel: An electron micrograph of an isolated Tsr bilayer specimen that is labeled in one leaflet of the twin bilayer membrane to illustrate the topology of the receptor subunits. p, periplasmic ligand-binding domain; b, membrane bilayer; c, cytoplasmic domain. Domain assignments in the other leaflet are apparent by symmetry. Two other features are labeled: a zone of interdigitation, or overlap (o), and a feature highlighted by stain that may represent the linker domain (l). Scale bar, 25 nm. Lower right panel: An illustration of a zipper assembly, serving to show only the “bilayer of bilayers” topology and the antiparallel nature of cytoplasmic domain interdigitation.
The interdigitating interaction may prove to be a feature common to all MCP assemblies of this type, owing to the high degree of sequence conservation near the hairpin (21). Electron microscopic observations of two other MCPs are consistent with the presence of an interdigitating interaction. First, the periodic striations across the Tsr zippered assemblies resemble the striations seen in reconstituted lamellar microcrystals of the E. coli ribose/galactose receptor (Trg) (4). Second, the arrangement of the Salmonella aspartate receptor (Tar) cytoplasmic domain within soluble supramolecular signaling complexes (that also contain CheW and CheA [14]) is compatible with a similar type of interdigitating interaction.
Measurements of the other dimensions in representative zippered assemblies of Tsr, including an estimate of the probable end-to-end length of the receptor, are provided in Table 1. As in the case of micellar assemblies, the measured receptor length is ∼20% shorter than that described for the model of Kim et al. (19) (Fig. 1). Given the known limitations for the interpretation of biological macromolecular structure from negatively stained electron microscope specimens, it is difficult to assert with certainty the reason for the difference in the two estimates, but based on several pieces of information, a more compact arrangement of the polypeptide in the linker region (Fig. 1) provides a plausible explanation for the discrepancy between the measured end-to-end length and the end-to-end length predicted in the model. First, detailed structural information about the linker is unavailable, although it is known by a conserved sequence motif (2) and an amphipathic α-helical character (6). Second, while the model plausibly depicts the linker as an extension of the second transmembrane α-helix (given the available structural information), it is a relatively uncompact conformation that reduces the cross-section in this portion of the receptor dimer to that of two α-helices. Third, the zippered assemblies depicted in Fig. 5 often show a region highlighted by the stain at a location in the cytoplasmic domain that is consistent with the position of the linker domain in the primary sequence of MCPs (Fig. 1) (2). Finally, a comparison of the measured (Table 1) versus model-generated lengths (19) of the periplasmic, transmembrane, and cytoplasmic domains shows that the agreement is poorest between the two lengths estimated for the cytoplasmic domain (190 Å [Table 1] versus 260 Å [19]).
To further assess whether or not this difference is meaningful, we tested for specimen shrinkage under our experimental conditions by measuring the accurately known repeat dimension in the tail striations of T4 phage (33). These experiments showed that the shrinkage of the tail striation repeat was no more than ∼5% (data not shown), although we cannot exclude the possibility that there is a different extent of shrinkage in the Tsr membrane assembly specimens. Taking all of these observations together, we suggest that the highlighted feature in the zippered assemblies (Fig. 5) reflects a more compact structure in the polypeptide chain than the extended α-helix depicted in the model. Apart from this one (but potentially important) difference, the dimensions determined from the electron micrographs are in reasonable agreement with the dimensions of the model, lending support both to the model and to our interpretation of the subunit arrangements in the electron microscope images.
In situ imaging in cell sections.
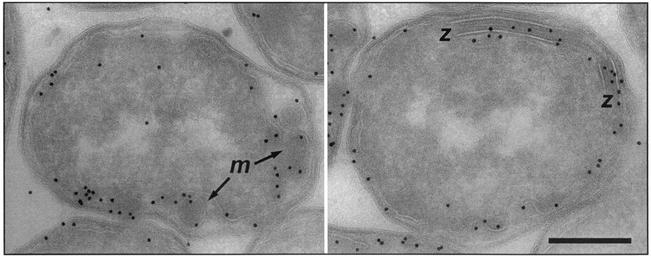
To determine whether the assemblies observed in the extracted membranes also occur in the intact cell, we recorded images from E. coli wild-type and Tsr overexpressor strains which had been fixed with glutaraldehyde, sectioned under cryogenic conditions, and labeled with an α-Tsr antibody that binds to the highly conserved cytoplasmic signaling domain. Projection images of wild-type E. coli sections (data not shown) exhibited specific labeling of receptors localized mainly at the cell poles, in agreement with previous observations (30). In the Tsr overexpressor, receptor labeling is distributed around the periphery of the cell and labeled regions also project into the interior of the cell (Fig. 6), suggesting that the cell becomes filled with membranous structures. Moreover, the images from the Tsr overexpressor clearly possess features that bear a striking resemblance to the zippered and micellar assemblies seen in the isolated membrane preparation (Fig. 6), which demonstrates that the assemblies observed in vitro have relevance for receptor subunit interactions in vivo. A detailed investigation (using electron microscope tomography) of the three-dimensional arrangement of zippered and micellar assemblies in the cell is to be presented elsewhere (J. Lefman et al., submitted for publication).
FIG. 6.
Images recorded from stained 70-nm sections of the E. coli Tsr overexpressor (HCB721/pHSe5.tsrQQQQ induced with 1 mM IPTG) after immunogold labeling of Tsr (10-nm gold particles). Regions identified with zippered (z) (right panel) and micellar (m) (left panel) assemblies are indicated. Scale bar, 200 nm.
DISCUSSION
The work described here demonstrates that membranes containing Tsr can arrange themselves into a variety of structures with strong interactions between neighboring subunits and, in the case of the zippered assemblies, nonlocal interactions between membranes. In other examples of E. coli membrane protein overexpression (3, 11, 43), extended membrane structures form that are enriched in the overexpressed protein. Often, these protein-lipid assemblies are long cylinders, an organization that is compatible with proteins in which the major portion of the polypeptide is embedded within the lipid bilayer. In contrast, a prominent feature of the Tsr-containing membrane preparations is the extensive interaction among the extramembranous portions of the receptor subunits, in particular the cytoplasmic domains. Many of these interactions are compatible with the subunit interactions in the X-ray structures of the soluble domains (19, 45). Specifically, the hexagonal arrangement of subunits in ordered arrays and the hairpin contacts observed in micellar assemblies are consistent with the trimer-of-dimer interaction found in the crystal structure of the Tsr cytoplasmic domain.
The interdigitating cytoplasmic domain in the zippered assemblies represents a novel subunit interaction that may be common to all MCPs, since it occurs within a region of high sequence identity (21) and it is compatible with electron microscope observations on detergent-solubilized, reconstituted Trg microcrystals (4) and soluble signaling complexes containing the Tar cytoplasmic fragment (14). The occurrence of interdigitation in cryosections of the Tsr overexpressor strain demonstrates that this type of interaction can also occur in the cell when Tsr is expressed at a high level. The extent to which the interactions observed with the elevated Tsr expression levels reflect the behavior of receptors at normal (wild-type) levels of expression remains to be determined. This issue has relevance to biochemical investigations of receptor function, which are often conducted either with cells that have elevated levels of receptor or with membranes isolated from these cells (5, 15, 22-27).
The notion that the receptor protein in the overexpressed state is functional in vivo has support from the observation that at moderate levels of Tar overexpression (but normal levels of the cytoplasmic signaling proteins), the increase in the adaptation time of a cell in response to an aspartate stimulus is approximately commensurate with the degree of overexpression (38). This property is consistent with the interpretation that in the overexpressed state, the majority of the receptors in these cells can be methylated. Still, the relationship between overexpression and biochemical function is complex, particularly with respect to receptor-mediated kinase activation and regulation, in which the subunit stoichiometry in the receptor-kinase complex may have a significant influence on the signaling properties of the complex. With the growing appreciation of the importance of receptor arrays in signaling (1, 16, 20, 23, 40), these electron microscopic observations contribute to and illustrate an approach for the further development of our understanding of the architecture of signaling complexes in the cell.
Acknowledgments
We thank John S. Parkinson (University of Utah) for the gift of Tsr antisera, Erik Bos (Netherlands Cancer Institute, Amsterdam) for assistance in preparing the E. coli cryosections, Sung-Ho Kim (University of California at Berkeley) for the Tsr dimer model coordinates, and David DeRosier (Brandeis University) for helpful discussions.
This work was supported by grants to R.M.W. from the NIH (RO1 NIGMS53210) and to S.S. from the intramural program at the National Cancer Institute, NIH.
REFERENCES
- 1.Ames, P., C. A. Studdert, R. H. Reiser, and J. S. Parkinson. 2002. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:7060-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravind, L., and C. P. Ponting. 1999. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 176:111-116. [DOI] [PubMed] [Google Scholar]
- 3.Arechaga, I., B. Miroux, S. Karrasch, R Huijbregts, B. de Kruijff, M. J. Runswick, and J. E. Walker. 2000. Characterisation of new intracellular membranes in Escherichia coli accompanying large scale over-production of the b subunit of F(1)F(o) ATP synthase. FEBS Lett. 482:215-219. [DOI] [PubMed] [Google Scholar]
- 4.Barnakov, A. N., K. H. Downing, and G. L. Hazelbauer. 1994. Studies of the structural organization of a bacterial chemoreceptor by electron microscopy. J. Struct. Biol. 112:117-124. [DOI] [PubMed] [Google Scholar]
- 5.Bornhorst, J. A., and J. J. Falke. 2000. Attractant regulation of the aspartate receptor-kinase complex: limited cooperative interactions between receptors and effects of the receptor modification state. Biochemistry 39:9486-9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler, S. L., and J. J. Falke. 1998. Cysteine and disulfide scanning reveals two amphiphilic helices in the linker region of the aspartate chemoreceptor. Biochemistry 37:10746-10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantwell, B. J., R. R. Draheim, R. B. Weart, C. Nguyen, R. C. Stewart, and M. D. Manson. 2003. CheZ phosphatase localizes to chemoreceptor patches via CheA-short. J. Bacteriol. 185:2354-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, P. S., T. Y. Toribara, and H. Warner. 1956. Microdetermination of phosphorous. Anal. Chem. 28:1756-1758. [Google Scholar]
- 9.Conley, M. P., A. J. Wolfe, D. F. Blair, and H. C. Berg. 1989. Both CheA and CheW are required for reconstitution of chemotactic signaling in Escherichia coli. J. Bacteriol. 171:5190-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corless, J. M., D. R. McCaslin, and B. L. Scott. 1982. Two-dimensional rhodopsin crystals from disk membranes of frog retinal rod outer segments. Proc. Natl. Acad. Sci. USA 79:1116-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elmes, M. L., D. G. Scraba, and J. H. Weiner. 1986. Isolation and characterization of tubular organelles induced by fumarate reductase overproduction in Escherichia coli. J. Gen. Microbiol. 132:1429-1439. [DOI] [PubMed] [Google Scholar]
- 12.Falke, J. J., and G. L. Hazelbauer. 2001. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 26:257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falke, J. J., and S. H. Kim. 2000. Structure of a conserved receptor domain that regulates kinase activity: the cytoplasmic domain of bacterial taxis receptors. Curr. Opin. Struct. Biol. 10:462-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francis, N. R., M. N. Levit, T. R. Shiakh, L. A. Melanson, J. B. Stock, and D. L. DeRosier. 2002. Subunit organization in a soluble complex of Tar, CheW, and CheA by electron microscopy. J. Biol. Chem. 277:36755-36759. [DOI] [PubMed] [Google Scholar]
- 15.Gegner, J. A., D. R. Graham, A. F. Roth, and F. W. Dahlquist. 1992. Assembly of an MCP receptor, CheW, and kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell 70:975-982. [DOI] [PubMed] [Google Scholar]
- 16.Gestwicki, J. E., and L. L. Kiessling. 2002. Inter-receptor communication through arrays of bacterial chemoreceptors. Nature 415:81-84. [DOI] [PubMed] [Google Scholar]
- 17.Grebe, T. W., and J. B. Stock. 1999. The histidine protein kinase superfamily. Adv. Microb. Physiol. 41:139-227. [DOI] [PubMed] [Google Scholar]
- 18.Heymann, J. A., R. Sarker, T. Hirai, D. Shi, J. L. Milne, P. C. Maloney, and S. Subramaniam. 2001. Projection structure and molecular architecture of OxlT, a bacterial membrane transporter. EMBO J. 20:4408-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, K. K., H. Yokota, and S. H. Kim. 1999. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature 400:787-792. [DOI] [PubMed] [Google Scholar]
- 20.Kim, S. H., W. Wang, and K. K. Kim. 2002. Dynamic and clustering model of bacterial chemotaxis receptors: structural basis for signaling and high sensitivity. Proc. Natl. Acad. Sci. USA 99:11611-11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Moual, H., and D. E. Koshland, Jr. 1996. Molecular evolution of the C-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis. J. Mol. Biol. 261:568-585. [DOI] [PubMed] [Google Scholar]
- 22.Le Moual, H., T. Quang, and D. E. Koshland, Jr. 1997. Methylation of the Escherichia coli chemotaxis receptors: intra- and interdimer mechanisms. Biochemistry 36:13441-13448. [DOI] [PubMed] [Google Scholar]
- 23.Levit, M. N., T. W. Grebe, and J. B. Stock. 2002. Organization of the receptor-kinase signaling array that regulates Escherichia coli chemotaxis. J. Biol. Chem. 277:36748-36754. [DOI] [PubMed] [Google Scholar]
- 24.Levit, M. N., and J. B. Stock. 2002. Receptor methylation controls the magnitude of stimulus-response coupling in bacterial chemotaxis. J. Biol. Chem. 277:36760-36765. [DOI] [PubMed] [Google Scholar]
- 25.Li, G., and R. M. Weis. 2000. Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli. Cell 100:357-365. [DOI] [PubMed] [Google Scholar]
- 26.Li, J., G. Li, and R. M. Weis. 1997. The serine chemoreceptor from Escherichia coli is methylated through an inter-dimer process. Biochemistry 36:11851-11857. [DOI] [PubMed] [Google Scholar]
- 27.Lin, L.-N., J. Li, J. F. Brandts, and R. M. Weis. 1994. The serine receptor of bacterial chemotaxis exhibits half-site saturation for serine binding. Biochemistry 33:6564-6570. [DOI] [PubMed] [Google Scholar]
- 28.Liu, Y., M. Levit, R. Lurz, M. G. Surette, and J. B. Stock. 1997. Receptor-mediated protein kinase activation and the mechanism of transmembrane signaling in bacterial chemotaxis. EMBO J. 16:7231-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long, D. G., and R. M. Weis. 1992. Oligomerization of the cytoplasmic fragment from the aspartate receptor of Escherichia coli. Biochemistry 31:9904-9911. [DOI] [PubMed] [Google Scholar]
- 30.Maddock, J. R., and L. Shapiro. 1993. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 259:1717-1723. [DOI] [PubMed] [Google Scholar]
- 31.Milburn, M. V., G. G. Privé, D. L. Milligan, W. G. Scott, J. Yeh, J. Jancarik, D. E. Koshland, Jr., and S. H. Kim. 1991. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science 254:1342-1347. [DOI] [PubMed] [Google Scholar]
- 32.Milligan, D. L., and D. E. Koshland, Jr. 1988. Site-directed cross-linking. Establishing the dimeric structure of the aspartate receptor of bacterial chemotaxis. J. Biol. Chem. 263:6268-6275. [PubMed] [Google Scholar]
- 33.Moody, M. F., and L. Makowski. 1981. X-ray diffraction study of tail-tubes from bacteriophage T2L. J. Mol. Biol. 150:217-244. [DOI] [PubMed] [Google Scholar]
- 34.Osborn, M. J., and R. Munson. 1974. Separation of inner (cytoplasmic) and outer membranes of Gram-negative bacteria. Methods Enzymol. 31:642-653. [DOI] [PubMed] [Google Scholar]
- 35.Parkinson, J. S., and S. E. Houts. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 151:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peters, P. J., and W. Hunziker. 2001. Subcellular localization of Rab17 by cryo-immunogold electron microscopy in epithelial cells grown on polycarbonate filters. Methods Enzymol. 329:210-225. [DOI] [PubMed] [Google Scholar]
- 37.Rice, M. S., and F. W. Dahlquist. 1991. Sites of deamidation and methylation in Tsr, a bacterial chemotaxis sensory transducer. J. Biol. Chem. 266:9746-9753. [PubMed] [Google Scholar]
- 38.Russo, A. F., and D. E. Koshland, Jr. 1983. Separation of signal transduction and adaptation functions of the aspartate receptor in bacterial sensing. Science 220:1016-1020. [DOI] [PubMed] [Google Scholar]
- 39.Schuster, S. C., R. V. Swanson, L. A. Alex, R. B. Bourret, and M. I. Simon. 1993. Assembly and function of a quaternary signal transduction complex monitored by surface plasmon resonance. Nature 365:343-347. [DOI] [PubMed] [Google Scholar]
- 40.Shimizu, T. S., N. Le Novere, M. D. Levin, A. J. Beavil, B. J. Sutton, and D. Bray. 2000. Molecular model of a lattice of signalling proteins involved in bacterial chemotaxis. Nat. Cell Biol. 2:792-796. [DOI] [PubMed] [Google Scholar]
- 41.Sourjik, V., and H. C. Berg. 2000. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol. Microbiol. 37:740-751. [DOI] [PubMed] [Google Scholar]
- 42.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 43.Wilkison, W. O., J. P. Walsh, J. M. Corless, and R. M. Bell. 1986. Crystalline arrays of the Escherichia coli sn-glycerol-3-phosphate acyltransferase, an integral membrane protein. J. Biol. Chem. 261:9951-9958. [PubMed] [Google Scholar]
- 44.Wu, J., J. Li, G. Li, D. G. Long, and R. M. Weis. 1996. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry 35:4984-4993. [DOI] [PubMed] [Google Scholar]
- 45.Yeh, J. I., H. P. Biemann, G. G. Privé, J. Pandit, D. E. Koshland, Jr., and S. H. Kim. 1996. High-resolution structures of the ligand binding domain of the wild-type bacterial aspartate receptor. J. Mol. Biol. 262:186-201. [DOI] [PubMed] [Google Scholar]
- 46.Zhulin, I. B. 2001. The superfamily of chemotaxis transducers: from physiology to genomics and back. Adv. Microb. Physiol. 45:157-198. [DOI] [PubMed] [Google Scholar]