Abstract
Bacterial alginates are produced as 1-4-linked β-d-mannuronan, followed by epimerization of some of the mannuronic acid residues to α-l-guluronic acid. Here we report the isolation of four different epimerization-defective point mutants of the periplasmic Pseudomonas fluorescens mannuronan C-5-epimerase AlgG. All mutations affected amino acids conserved among AlgG-epimerases and were clustered in a part of the enzyme also sharing some sequence similarity to a group of secreted epimerases previously reported in Azotobacter vinelandii. An algG-deletion mutant was constructed and found to produce predominantly a dimer containing a 4-deoxy-l-erythro-hex-4-enepyranosyluronate residue at the nonreducing end and a mannuronic acid residue at the reducing end. The production of this dimer is the result of the activity of an alginate lyase, AlgL, whose in vivo activity is much more limited in the presence of AlgG. A strain expressing both an epimerase-defective (point mutation) and a wild-type epimerase was constructed and shown to produce two types of alginate molecules: one class being pure mannuronan and the other having the wild-type content of guluronic acid residues. This formation of two distinct classes of polymers in a genetically pure cell line can be explained by assuming that AlgG is part of a periplasmic protein complex.
Although originally described in algae (45) the polymer alginate is also produced by bacterial species belonging to the genera Pseudomonas and Azotobacter. It is a linear copolymer of 1-4-linked α-l-guluronic acid (G) and β-d-mannuronic acid (M); the latter may be O-2 and/or O-3 acetylated in bacterial alginates (43). The relative amounts and distribution of the two uronic acid residues vary among species and is also dependent on growth conditions (43). The bacteria use alginate as a part of their vegetative capsule, and it is also implicated in the formation of Pseudomonas biofilms (32). Most strains of Azotobacter produce alginates constitutively, whereas many species of Pseudomonas have downregulated their production to nondetectable amounts. However, mutants overproducing alginate can be isolated from such strains (17). Alginates from pseudomonads have never been found to contain stretches of continuous G residues (G blocks), whereas this is quite common for alginates produced by Azotobacter species and algae (43). These G blocks account for the ion-binding and gel-forming capacity of the alginates, a property that is crucial for the many industrial applications of the polymer (44). It is also biologically interesting in that it enables Azotobacter to produce a protective calcium-alginate gel coat surrounding a particular cellular resting stage designated cyst (38). The biological function of the G residues in Pseudomonas alginates and in vegetatively growing Azotobacter cells is not well understood.
When Pseudomonas aeruginosa infects the lungs of patients suffering from cystic fibrosis, spontaneous alginate-producing mutants emerge. The alginate protects the bacteria against the host's immune system and antibiotics and increases the viscosity of the fluid in the lung (27, 28). It was found that all but one of the proteins necessary for the biosynthesis, modification, and export of alginates are encoded by one operon (4). Later, a similar operon has been described for Pseudomonas syringae (34). Azotobacter vinelandii also contains a homologous gene cluster (37), but its genes are organized in several transcription units (48). Much knowledge on the regulation of the biosynthesis of alginate has also emerged during the last decade (16).
Both in bacteria and algae alginate is first synthesized as mannuronan, and the G residues are then introduced by mannuronan C-5-epimerases (47). Such enzymes were first described by Haug and Larsen (21), who found a secreted epimerase in the culture medium of A. vinelandii. During the work aimed at cloning the gene encoding this activity, it was found that A. vinelandii encodes a family of seven homologous secreted epimerases (AlgE1 to AlgE7) (8, 10, 46). The genome of P. aeruginosa is now sequenced, and this bacterium does not encode any protein homologous to the secreted epimerases of A. vinelandii. Chitnis and Ohman (3) isolated P. aeruginosa mutants, which produced pure mannuronan, and a mutation was mapped to algG in the alginate operon. Franklin et al. (12) showed algG to encode a periplasmic mannuronan C-5-epimerase. Later, it was found that A. vinelandii also encodes an active AlgG (37). Downstream of algG is algX, which is necessary for alginate production (30), algL encoding an alginate lyase (40), and algIJF, which are involved in acetylation (13, 14). The most-downstream gene of the operon, algA, encodes the bifunctional enzyme phosphomannoisomerase-d-mannose-1-phosphate guanylyl transferase, and its activity is required for alginate production (41).
In our studies of the AlgE epimerases it became important to have access to reasonable quantities of mannuronan as a substrate. We therefore decided to develop the production of this polymer in a nonpathogenic Pseudomonas species in order to avoid the problems related to handling large culture volumes of a pathogenic organism (P. aeruginosa). The Pseudomonas fluorescens strain NCIMB10525 was selected as a host for this purpose, and from chemically mutagenized cells we first isolated an alginate overproducer (designated Pf201) from this strain, by using formation of mucoid colonies on agar medium as a criterion. The colony-based G lyase assay of Chitnis and Ohman (3) was then used to isolate mannuronan producers from chemically mutagenized Pf201 cells.
The order of the genes in the alg operons characterized so far is all the same, and we therefore assumed that this was the case also for P. fluorescens. This was confirmed by cloning of alg′EGXLIJFA from strain NCIMB10525 and by inspecting the sequences of the genome of the P. fluorescens strain PfO-1, made available during the studies reported here (http://www.jgi.doe.gov/tempweb/JGI_microbial/html/index.html). In our studies of the mannuronan-producing mutants of Pf201 we discovered that AlgG plays a more complex role in alginate biosynthesis than previously anticipated. One way to interpret our results is to assume that AlgG is part of a protein complex in which it protects the newly synthesized polymer from degradation by AlgL.
MATERIALS AND METHODS
Growth of bacteria.
The bacterial strains and plasmids used are described in Table 1. Escherichia coli and P. fluorescens strains were routinely grown in L broth (10 g of tryptone/liter, 5 g of yeast extract/liter, and 5 g of NaCl/liter) or on L agar (L broth containing 15 g of agar/liter) at 37 and 30°C, respectively. Matings between E. coli S17.1 and P. fluorescens strains were performed at 30°C on L agar, and selections of transconjugants were done with Pseudomonas isolation agar (PIA; Difco) with appropriate antibiotics. For transposon insertions, strain S17.1(λpir) was used as the donor. Production of P. fluorescens alginate was performed in liquid PIA medium (shake flasks) containing bacteriological peptone (20 g/liter), NaCl (5 g/liter) MgCl2 (1.4 g/liter), K2SO4 (10 g/liter), and 20 ml of 87% glycerol/liter or in PM5 medium (fermentors) containing fructose (40 g/liter), yeast extract (12 g/liter), (NH4)2SO4 (0.6 g/liter), Na2HPO4 · 2H2O (2.0 g/liter), NaCl (11.7 g/liter), and MgSO4 · 7H2O (0.3 g/liter), and clerol FBA622 (antifoam, 0.5 g/liter). The media were supplemented with proteases—Alkalase 2.4L and Neutrase 0.5L from Novo Nordisk (0.15 ml/liter each in PIA and 0.25 ml/liter each in PM5)—in order to reduce extracellular alginate-lyase activity. Antibiotics, when used in routine growth experiments, were present at the following concentrations: ampicillin, 100 to 200 μg/ml; kanamycin, 40 μg/ml; and tetracycline, 12.5 μg/ml (E. coli) or 30 μg/ml (P. fluorescens). m-Toluate was added to a final concentration of 1 mM unless otherwise stated and 60 μl of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) solution (20 mg/ml in N,N-dimethyl formamide) were added to each agar plate for detection of LacZ activity. For alginate production in shake flasks 1 to 2 vol-% inoculum from an overnight culture was transferred to a shake flask (500 ml, baffled) with 100 ml of liquid PIA medium and incubated at 25°C for 48 h in an orbital shaker (200 rpm; amplitude, 2.5 cm). For alginate production in a fermentor, 2 to 3 vol-% inoculum from an overnight culture in shake flask was transferred to a 3-liter fermentor (Applicon) containing 1.4 liters of PM5 medium. The fermentations were performed at 25°C. pH was adjusted to 7.0 to 7.2 from the start and controlled at 7.0 with NaOH (2 M). The airflow through the culture medium was 0.25 liter/liter of medium for the first 8 to 10 h; thereafter, it was increased in steps up to 0.9 to 1.0 liter/liter of medium. The dissolved oxygen was controlled at 20% of saturation by automatic control of the stirrer speed.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain, phage, or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| S17.1 | RP4 2-Tc::Mu-Km::Tn7 pro res mod+ | 42 |
| S17.1 (λpir) | λpir recA thi pro hsdR-M+ RP4 2-Tc::Mu-Km::Tn7TpRSMR | 6 |
| SURE | sbcC recB recJΔ(mcrCB-hsdSMR-mrr) endA1 gyrA96 Tcr Kmr | 19 |
| P. fluorescens | ||
| NCIMB 10525 | Nonmucoid P. fluorescens wild type | NCIMB |
| Pf201 | algG+, mucoid P. fluorescens mutant derived from NCIMB 10525 | P. Karunakaran, unpublished data |
| Pf2012 | Mannuronan-producing mutant, algG D361N | This work |
| Pf2013 | Mannuronan-producing mutant, algG G430D | This work |
| Pf20117 | Mannuronan-producing mutant, algG R408C | This work |
| Pf20118 | Mannuronan-producing mutant, algG R408C | This work |
| Pf20137 | Mannuronan-producing mutant, algG S337F | This work |
| Pf20138 | Mannuronan-producing mutant, algG S337F | This work |
| Pf201ΔalgG | algG in-frame deletion mutant | This work |
| Pf20118::TnKB10 | Derivative of Pf20118 with transposon from pKB10 | This work |
| Pf201ΔalgG::TnKB10 | Derivative of Pf201ΔalgG with transposon from pKB10 | This work |
| Pf201ΔalgG::TnCNB111 | Derivative of Pf201ΔalgG with transposon from pCNB111 | This work |
| Phages and plasmids | ||
| λ DashII | λ cloning vector | Stratagene |
| Pfλ1 | λ DashII with an ∼15 kb insert of Sau3AI partially digested genomic DNA from P. fluorescens NCIMB10525 encoding alg′EGXLIJFA | M. Gimmestad, unpublished data |
| pGEM5 | ColE1; Apr | Promega |
| pGEM11 | ColE1; Apr | Promega |
| pCVD442 | oriR6K; Apr | 7 |
| pJB3Tc20 | RK2-based vector; Apr Tcr | 2 |
| pCNB111 | oriR6K mobRP4, pUT/mini-Tn5 xylS/Pm; Apr Kmr | 49 |
| pCNB111luc | oriR6K mobRP4, pUT/mini-Tn5 xylS/Pm, luc; Apr Kmr | 49 |
| pJB3Tc20trfA | Derivative of pJB3Tc20 from which a 1.0-kb BsaAI-NdeI DNA fragment encoding TrfA was deleted | This work |
| Litmus28 | ColE1; Apr | New England Biolabs |
| pHE55 | Derivative of pJB3Tc20trfA in which a 2.6-kb PstI-XbaI DNA fragment from pCVD442 encoding SacB from B. subtilis was inserted | This work |
| pJB1002 | RK2-based vector encoding a TrfA-LacZ-fusion protein | 25 |
| pMG23 | Litmus28 in which a 1.8-kb PCR-amplified BglII-PstI DNA fragment containing algG and 135 bp of algX was inserted; the primers PfalgG3r and PfalgG4f were used for amplification | This work |
| pMG26 | pGEM11 containing a 4.6-kb SalI DNA fragment from Pfλ1 | M. Gimmestad, unpublished data |
| pMG31 | Derivative of pHE55 in which an 1.8-kb BglII-XbaI DNA fragment encoding algG from pMG23 was inserted | This work |
| pMG47 | XbaI/PstI-restricted derivative of pHE55 in which a 4.1-kb NheI-PstI DNA fragment from pJB1002 encoding the TrfA-LacZ fusion protein was inserted | This work |
| pMG48 | pMG47 with a 0.36-kb SphI-SapI DNA fragment containing the polylinker of pGEM5 | This work |
| pMG51 | Derivative of pMG26 in which a SmaI site was introduced at nucleotide position 368 in algG by using the primers algG-SmaI-1 and algG-SmaI-2 | This work |
| pMG52 | Derivative of pMG51 from which a 0.6-kb SmaI DNA fragment was deleted, creating an in-frame deletion in algG | This work |
| pMG53 | Derivative of NsiI-NcoI-restricted pMG48 in which a 2.1-kb PstI-BspHI DNA fragment from pMG52 was inserted | This work |
| pKB4 | Derivative of pMG26 from which a 3.0-kb BlpI-XhoI DNA fragment was deleted; | This work |
| pKB10 | Derivative of pCNB111luc in which luc was replaced with a 1.7-kb NdeI-NotI-restricted PCR fragment containing algG pKB4 was used as PCR template and PfalgG-NdeI-2 and M13/pUC reverse were used as primers | This work |
Tcr, Kmr, and Apr, tetracycline, kanamycin, and ampicillin resistance, respectively.
Standard techniques.
Plasmid isolation, enzymatic manipulations of DNA, and agarose gel electrophoresis was performed by the methods of Sambrook and Russell (39). The QIAquick gel extraction kit and QIAquick PCR purification kit (Qiagen) was used for DNA purifications from agarose gels and enzymatic reactions, respectively. Transformations of E. coli were performed as described by Chung et al. (5) or by use of heat shock-competent rubidium chloride-treated cells. The E. coli strains S17.1 and SURE (Stratagene) were used for standard cloning procedures. S17.1(λpir) was used as host for pCNB111 and its derivatives. PCR for cloning and allele identification was performed by using the Expand High-Fidelity PCR-system (Boehringer Mannheim) or the Pfx polymerase-Pfx PCR system (Gibco-BRL). Site-specific mutagenesis was performed by using QuickChange site-directed mutagenesis kit (Stratagene). DNA sequencing was performed by using the BigDye kit (Applied Biosystems). algG from the strains producing mannuronan was sequenced by ACGT, Inc., Northbrook, Ill.
Primers used for PCR amplifications.
The primers used were as follows: PfalgG3r (PstI), 5′-CAGGCTGCAGCACGGTTCGGC-3′; PfalgG4f (BglII), 5′-AAAAAGATCTAGTCGACTCGTACATGCACCGCG-3′; PfalgG5f (BspHI), 5′-GAGCCTGCGTCATGAACCCTCAAGC-3′; algG-SmaI-1, 5′-CACGGCATTCCCCGGGCGATCTTC-3′; algG-SmaI-2, 5′-GAAGATCGCCCGGGGAATGCCGTG-3′; PfalgG-NdeI-2, 5′-AAAAAACATATGGGAGCCTGCGCAATGAACC-3′; and M13/pUC reverse primer, 5′-AGCGGATAACAATTTCACACAGGA-3′. Nucleotides indicated in boldface are not part of the P. fluorescens wild-type sequences. Restriction endonuclease sites are underlined, and the corresponding enzymes are indicated in parentheses or as part of the primer designation.
Construction of a suicide vector for gene replacement studies.
Originally, we intended to select for double crossovers by using the sacB marker from Bacillus subtilis, but this system was not found to be reliable in our strain of P. fluorescens. We found, however, that the E. coli lacZ gene could be used for a similar purpose, based on previous reports demonstrating that this marker can be used in P. fluorescens (20). A new suicide vector designated pMG47 was therefore constructed by replacing the sacB of pHE55 (lacking the essential plasmid replication initiation gene trfA) by a particular trfA-lacZ fusion construct previously shown to express β-galactosidase but not a functional replication-initiation protein (24). To simplify further cloning steps, new cloning sites were finally introduced into pMG47, generating pMG48 (Table 1). P. fluorescens strains with this vector with inserted genomic DNA integrated into their chromosome were tetracycline resistant and formed blue colonies in the presence of X-Gal. Cells lacking the vector formed white colonies in the presence of X-Gal. These characteristics could therefore be used to easily follow the outcomes of both steps (vector integration and loss of the integrated copy) in double-crossover experiments.
Alginate quantification.
Culture samples were diluted about 10-fold in 0.2 M NaCl in order to reduce viscosity and centrifuged to remove bacterial cells. The alginates in the cell-free supernatants were deacetylated by mild alkaline treatment as described previously (11). Alginates were quantified by using the M-specific lyase from Abalone and G-specific lyase from Klebsiella aerogenes as described earlier (33). Isolation of deacetylated alginate from culture supernatants was performed by adding an equal volume of isopropanol. The precipitate was collected by centrifugation and washed with both 70 and 96% ethanol.
Measurements of lyase activity and G-specific degradation of alginate with lyase.
For measurements of intracellular alginate lyase activity, bacterial cells were collected by centrifugation, resuspended in buffer (Tris-HCl [50 mM], NaCl [0.25 M]; pH 7.5) to an optical density at 660 nm of 3 to 10, and sonicated. The lyase activities in these extracts were determined by measuring the degradation rate of mannuronan by using a Ubbelodhe capillary viscometer (Scott-Geräte instrument no. 53620/II). The mannuronan substrate was dissolved (1 mg/ml) in 12.5 mM Tris-HCl-62.5 mM NaCl (pH 7.5), and 4 ml of this solution was mixed with 0.4 ml or extract (diluted if necessary) and added to the Ubbelodhe. M-specific lyase from Abalone was used as a standard (33). One unit of lyase activity was defined as described by Ertesvåg et al. (9). The time for the solution to pass the capillary of the Ubbelodhe was measured every 2 min over a period of 1 h. The analyses were performed at 25°C. The G-specific degradation of alginate was measured in the Ubbelodhe as described above by mixing 0.1 ml of G lyase (0.06 U/ml) with 4 ml of alginate substrate (12.5 mM Tris-HCl, 62.5 mM NaCl [pH 7.5], 1 mg of alginate/ml).
1H-NMR spectroscopy.
Alginate samples were collected as described for alginate quantification, except that the deacetylated alginates were precipitated by acid instead of isopropanol. HCl was added until the pH of the sample was 2. The alginates were then collected by centrifugation, washed in 70 and 96% ethanol, redissolved in distilled water, and neutralized by NaOH. To reduce the viscosity of the polymers for NMR analyses the samples were degraded by mild acid hydrolysis to a final average degree of polymerization of about 35, neutralized, and freeze-dried (11). The samples were dissolved in D2O (10 mg/ml), and the nuclear magnetic resonance (NMR) spectra were obtained by using a Bruker 300-MHz spectrometer. Integration of the spectra and further calculations and assignment of peaks were performed as described earlier (9, 18).
Capillary electrophoresis.
An Applied Biosystems HPCE model 270A-HT with Turbochrom Navigator (4.0) software was used. The fused silica column (72 cm, 50 cm to detector, 50-μm inner diameter) was from Supelco (St. Louis, Mo.). All runs were performed at 30°C. Samples were loaded under vacuum at a pressure of 16.9 kPa (1.5 s). Before sample injection, the capillary was conditioned for 4 min with 50 mM tetraborate (pH 8.0), followed by a 2-min washing with 0.1 M NaOH (vacuum pressure of 67.6 kPa). The detection wavelength was 232 nm, and the voltage was 15 kV.
Electrospray ionization mass spectrometry.
Samples were diluted in 5 mM ammonium acetate (pH 9) and analyzed by direct infusion (0.6 ml/h) into an Agilent MSDTrap SL mass spectrometer equipped with an electrospray ion source and operated in negative-ion mode. The drying gas flow was 5 liters/min, the drying gas temperature was 325°C, and the nebulizer pressure was 15 lb/in2. The capillary voltage was 3,500 V with an endplate offset of −500 V.
Nucleotide sequence accession numbers.
The DNA sequence reported here has been submitted to GenBank under accession number AF527790. P. aeruginosa and A. vinelandii algG and algE4 and Sphingomonas sp. aly are listed under GenBank accession numbers U27829, X87973, L39096, and AB011415, respectively. The sequence data of the alginate biosynthetic cluster from P. fluorescens strain PfO-1 were obtained from The DOE Joint Genome Institute (JGI) at http://www.jgi.doe.gov/tempweb/JGI_microbial/html/index.html. Sequence data of algG from Pseudomonas putida KT2440 and P. syringae were obtained from The Institute for Genomic Research (TIGR) website (http://www.tigr.org) as TIGR 1604881/13538 and TIGR 317/5336, respectively.
RESULTS
The mannuronan production is caused by point mutations in algG.
Six mutants forming mucoid colonies in the presence of G lyase were isolated from strain Pf201, and NMR spectroscopy analyses of the corresponding alginates showed that they all produced mannuronan (not shown). To verify that this phenotype was caused by mutations in algG, the algG alleles in the mutants were replaced by the wild-type allele. For this purpose a gene replacement vector, pMG31, encoding wild-type algG and the first 135 bp of the downstream algX was constructed (Table 1). The plasmid was conjugated into each of the mannuronan-producing mutants, and the transconjugants were selected on PIA medium containing tetracycline. As expected, nonmucoid colonies appeared due to the disruption of the alginate biosynthetic operon as pMG31 recombined into algG. These transconjugants were grown in two to six sequential liquid overnight cultures in the absence of tetracycline to allow loss of the integrated plasmid. Diluted cultures were plated on PIA and screened for mucoid revertants, which were then restreaked on L-agar containing G lyase. All six mutants could be reverted by this procedure.
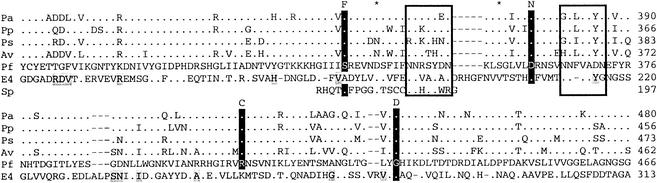
Based on the nucleotide sequences of the cloned alg genes, the predicted mutant algG genes could be PCR amplified from the six mannuronan-producing strains and then subjected to DNA sequencing. By comparison to the wild-type sequence, four different mutations were identified: (i) S337F, TCC to TTC (strains Pf20137 and Pf20138); (ii) D361N, GAC to AAC (strain Pf2012); (iii) R408C, CGT to TGT (strains Pf20117 and Pf20118); and (iv) G430D, GGC to GAC (strain Pf2013). The mannuronan production in the mutants was therefore caused by single amino acid substitutions in algG in each mutant. By comparing these results with the known sequences of AlgG homologues from different species it was found that the four substituted amino acids were conserved in all of the deduced AlgG proteins reported thus far (Fig. 1).
FIG. 1.
Alignment of the central part of mannuronan C-5-epimerases. The AlgG epimerases are from the following sources: Pa, P. aeruginosa; Pp, P. putida; Ps, P. syringae; Av, A. vinelandii; and Pf, P. fluorescens. E4 is the secreted epimerase AlgE4 from A. vinelandii. “Sp” is the alginate lyase ALYIII from a Sphingomonas sp. The numbers relate to the start of the deduced proteins. Amino acids identical to those in the P. fluorescens AlgG-sequence are shown as dots, and gaps are indicated as horizontal lines. AlgE4 amino acids identical to the corresponding residues in one or more of the AlgG proteins apart from P. fluorescens AlgG are shaded gray. The conserved motifs (see the text) are boxed. The four amino acids shown to be essential for epimerase activity in P. fluorescens are highlighted in black, whereas the corresponding amino acids in the epimerization-defective mutants are indicated above the alignment. Asterisks mark the amino acids S356 and D342. These two residues are found in the same positions relative to the motifs as the corresponding pair S337 and D361, which both have been found to be critical for epimerization.
Construction of an algG deletion mutant.
NMR spectroscopy has limitations in detecting very low G contents and, since the mannuronan production was caused by point mutations in algG, we decided to construct a deletion in this gene to make sure that we could produce a totally pure homopolymer. For this purpose, ca. 40% of algG in pMG26 (Table 1) were deleted in frame, and the flanking sequences were transferred to pMG48, generating pMG53. This internal deletion in algG corresponds to a deletion of amino acid residues 125 to 337 in the protein. pMG53 was transferred to P. fluorescens Pf201 by conjugation, and transconjugants were selected on PIA medium containing X-Gal and tetracycline. Most of the resistant colonies were blue (lacZ+) and nonmucoid (no alginate polymer production), indicating that pMG53 had been incorporated into the alginate biosynthetic genes, as expected. One of these transconjugants was grown in a series of overnight liquid cultures in the absence of tetracycline to allow the vector to recombine out of the chromosome. Such recombinants were expected to form white mucoid colonies (ΔalgG or wild-type algG, tetracycline-sensitive ΔlacZ mutants) and should therefore be easily identified. Of 4,200 colonies inspected, only 0.2% were white and mucoid but, interestingly, 13% were white and nonmucoid. The remaining colonies were still blue and nonmucoid. The majority of the nonmucoid white colonies were found to still be tetracycline resistant, and PCR analyses with primers PfalgG3r and PfalgG5f showed that they still contained both alleles and had lost only parts of the integrated plasmid. However, a similar analysis of one white and tetracycline-sensitive strain, designated Pf201ΔalgG, showed that it carried the desired algG deletion.
Surprisingly, strain Pf201ΔalgG formed nonmucoid colonies on agar medium, even though the algG deletion was designed to be in frame. A similar experiment was carried out in parallel on another P. fluorescens strain, and this mutant was recently also reported to form nonmucoid colonies on agar medium (31). Here we are aiming at understanding the reasons for this phenotype, which correlated with the observation that no alginate could be recovered by standard isopropanol precipitation from the liquid culture medium of Pf201ΔalgG-grown cells.
To make sure that the nonmucoid phenotype was caused by lack of AlgG only, we first measured the intracellular activity of algL, located downstream of algG, and found it to be similar in Pf201ΔalgG and in Pf201 (results not shown). Thus, polar effects did not appear to be relevant to explain the phenotype. Further confirmation was obtained by complementing Pf201ΔalgG with wild-type algG. In our strain of P. fluorescens this could not be done with a plasmid encoding AlgG because of problems with a high frequency of plasmid integration by homologous recombination into the corresponding site in the alg operon. However, we were instead able to use a previously constructed transposon system for complementation. This system is based on the inducible broad-host range Pm promoter previously used to express the luc reporter gene in E. coli (49). Preliminary studies of this system in P. fluorescens showed that expression was very efficient, since it was observed that even in the absence of inducer the luc expression level was similar to that of the corresponding induced E. coli cells (data not shown). The wild-type algG gene was therefore inserted into this transposon present in the suicide plasmid pCNB111, generating plasmid pKB10, which was conjugated into Pf201ΔalgG. Transconjugants with chromosomal insertions of the transposon (TnKB10) displayed a strongly mucoid phenotype on agar medium containing the Pm inducer m-toluate, and the mucoid phenotype was much less prominent on the same medium lacking the inducer (not shown). Analyses of the alginates produced in liquid media by one such transconjugant, Pf201ΔalgG::TnKB10, also showed that a significant amount (2.5 g/liter) of high-molecular-weight polymer was produced even in the absence of inducer (Table 2). This is probably due to the relatively high background level of expression from the Pm promoter in P. fluorescens, which also was observed for the luciferase control experiments. Still, in the presence of inducer more alginate (4.7 g/liter) was produced, and the product contained ca. 30% G. As a negative control in these experiments we used a Pf201ΔalgG strain in which the corresponding transposon lacking algG was inserted into the chromosome. As expected, this strain, designated Pf201ΔalgG::TnCNB111, did not produce alginate polymer neither in the absence nor in the presence of m-toluate (Table 2). It could therefore be concluded that AlgG wild-type alone is sufficient to complement the phenotype of strain Pf201ΔalgG.
TABLE 2.
Alginate production and composition by different mutant strains grown in liquid PIA
| Strain | m-Toluate concn (mM) | Mean alginate concn (g/liter)a ± SD | FGb |
|---|---|---|---|
| Pf201 | 0 | 4.7 ± 0.4 | 0.33 |
| Pf20118 | 0 | 4.5 ± 0.2 | 0.0 |
| Pf201ΔalgG | 0 | 0.0c | |
| Pf201ΔalgG::TnCNB111 | 0 | 0.0c | |
| Pf201ΔalgG::TnCNB111 | 0.1 | 0.0c | |
| Pf201ΔalgG::TnKB10 | 0 | 2.5 ± 0.2 | NDd |
| Pf201ΔalgG::TnKB10 | 0.025 | 4.7 ± 0.4 | 0.30 |
| Pf20118::TnKB10 | 0 | 4.6 ± 0.3 | 0.06 |
| Pf20118::TnKB10 | 0.01 | 4.9 ± 0.1 | 0.12 |
| Pf20118::TnKB10 | 0.025 | 4.9 ± 0.5 | 0.18 |
| Pf20118::TnKB10 | 0.1 | 4.9 ± 0.2 | 0.23 |
| Pf20118::TnKB10 | 0.5 | 4.7 ± 0.3 | 0.25 |
| Pf20118::TnKB10 | 3.0 | 4.4 ± 0.3 | 0.28 |
Alginate production was measured from three independent cultures for each strain. The alginate concentrations are given as mean values of eight analyses of one typical sample; the other two samples were within the limits of variation shown. Standard deviations were calculated using the following formula: {[nΣx2 − (Σx)2]/n(n − 1)}0.5.
FG, fraction of G-residues.
No alginate isolated by isopropanol precipitation.
ND, not determined.
The absence of the AlgG protein leads predominantly to production of an unsaturated disaccharide originating from alginate lyase activity.
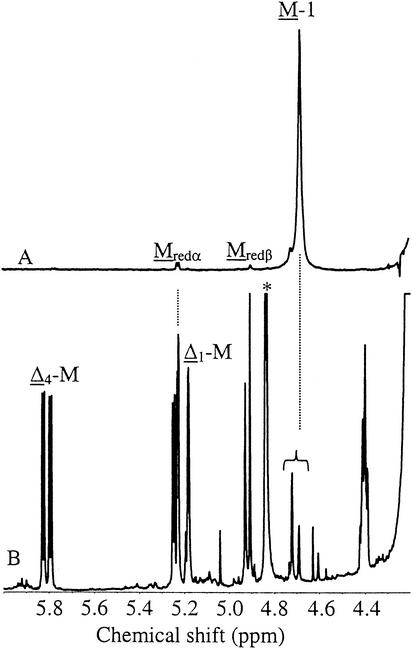
In our studies aiming at understanding the reasons for the Pf201ΔalgG phenotype it was discovered that the growth medium of cultures of this strain absorbed light at 230 nm to an extent that significantly exceeded that of the corresponding algG wild-type and strains Pf201 and Pf20118 (Table 3). The absorption was also much stronger than the medium background from the nonmucoid parent wild-type strain. Absorption at 230 nm is known to be the result of AlgL-mediated degradation of alginate due to the double bond formed in this reaction (9). Interestingly, complete degradation by externally added lyase of the alginate produced by strain Pf201 and Pf20118 gave rise to A230 signals of the same magnitude as that from strain Pf201ΔalgG without added lyase (16.1, 16.6, and 14.9, respectively). It therefore seemed possible that the nonmucoid phenotype of Pf201ΔalgG was caused by extensive degradation of the alginate produced, thereby also explaining the lack of precipitation in the presence of isopropanol. To investigate this, the medium supernatant from Pf201ΔalgG grown for 2 days in PM5 medium was freeze-dried and analyzed by 1H-NMR (Fig. 2). The spectrum clearly showed that extensive AlgL-mediated degradation had taken place, as seen by the strong signals from the double bonds (Δ4-M and Δ1-M) and from the reducing ends (Mredα plus Mredβ). The M-1 signals from internal mannuronic acid residues were much weaker than the signals from end-residues, indicating that the degradation product might be primarily a dimer. Similar results were found in a parallel study on P. aeruginosa (22). This was surprising since it has been shown previously that when AlgL from A. vinelandii was used to completely degrade mannuronan in vitro (9), the average degree of polymerization of the endproducts was about three. Similarly, Rehm (35) found that AlgL from P. aeruginosa produced far more trimers than dimers from a polymeric alginate containing only 5% G residues. It appears that the P. fluorescens lyase either acts in another way on its substrate compared to that of the P. aeruginosa and A. vinelandii lyases or that its mode of action is different in vivo from what it is in vitro.
TABLE 3.
Unsaturated ends in culture medium (liquid PIA) measured at A230 before and after treatment with M lyasea
| Strain | Mean A230 ± SD on:
|
|
|---|---|---|
| Untreated growth medium | Medium plus M lyase and G lyase | |
| NCIMB10525 | 10.4 ± 0.4 | 10.1 ± 0.4 |
| Pf201 | 10.3 ± 0.1 | 16.1 ± 0.2 |
| Pf20118 | 10.6 ± 0.2 | 16.6 ± 0.3 |
| Pf201ΔalgG | 14.9 ± 0.3 | 14.4 ± 0.4 |
Each value is the mean of six analyses of the same sample, and standard deviations were calculated as described in Table 2. The high values observed for the wild type are the result of a high background absorption in the growth medium. This value was not subtracted from the measured values.
FIG. 2.
1H-NMR of the oligouronides produced by Pf201ΔalgG (B) compared to the spectrum of mannuronan produced by Pf20118 (A). Cells were grown in PM5 medium. The monomers from which the signals originate are underlined. Note that the M-1 peak is split in panel B. The origin of the signal at ca. 4.85 ppm (∗) is unknown, but the signal is too intense to originate from the oligouronides.
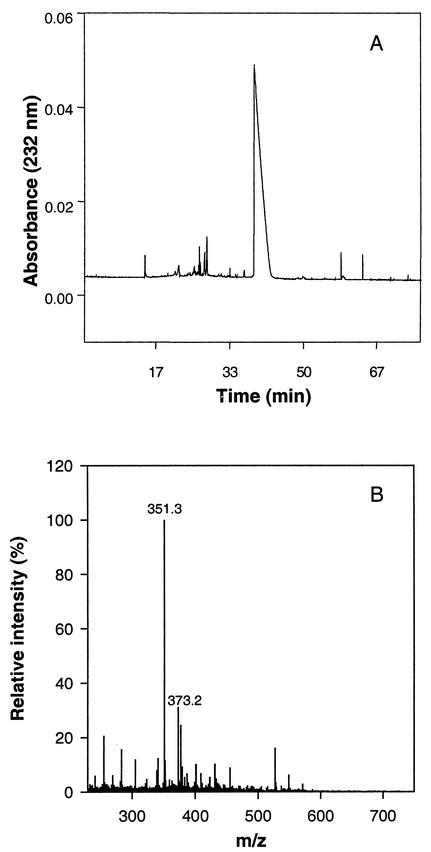
Since the NMR spectrum described above contained some unidentified signals from other components in the growth medium, we considered it necessary to examine the alginate degradation products produced by Pf201ΔalgG more closely. This was done by first size fractionating them by capillary electrophoresis. Interestingly, a single peak dominated in this analysis, one which separated the oligomers of alginate very well (Fig. 3A). The culture supernatant was further analyzed by mass spectroscopy (Fig. 3B), and the dominant peaks have masses corresponding to ΔM-1H+ (i.e., 351.2) and ΔM+Na+-2H+ (i.e., 373.2), confirming that Pf201ΔalgG predominantly produces dimeric oligomannuronic acids that are unsaturated at their nonreducing ends. These experiments therefore clearly demonstrated that AlgG, in addition to its epimerization activity, plays a role in protecting the alginate polymer from AlgL-mediated degradation and that the in vivo mode of action of the lyase in the absence of AlgG is such that it predominantly forms unsaturated dimers.
FIG. 3.
Electrophoretic mobility and mass spectroscopy analysis of oligouronides produced by Pf201ΔalgG. Cells were grown in PM5 medium. (A) Separation by capillary electrophoresis; (B) analyses by mass spectroscopy. 351.2 is the expected mass for ΔM-1H+, and 373.2 is the expected mass for ΔM+Na+-2H+.
Culture supernatants from strain Pf201, Pf20118, and uninduced Pf201ΔalgG::TnKB10 (from the experiments shown in Table 2) were then analyzed by mass spectroscopy. The analyses showed that strain Pf201ΔalgG::TnKB10 produced nearly as much dimers as polymer. Pf201 and Pf20118 also produced dimers, but in much smaller quantities.
AlgG is probably part of a periplasmic protein complex necessary for alginate production.
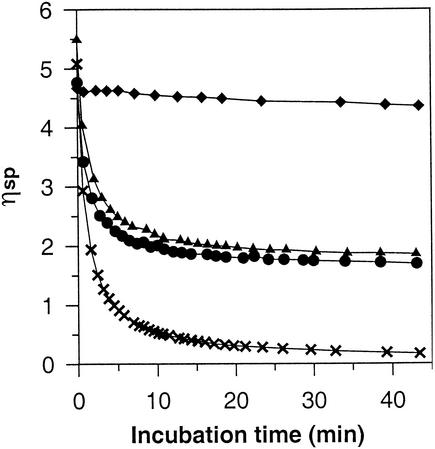
The data described above suggested that AlgG and AlgL may be parts of a protein scaffold, and this hypothesis led to another interesting prediction that could be tested experimentally. In the epimerization-defective algG mutants the complexes are presumably formed, and if wild-type AlgG is expressed under such conditions one would predict that some scaffolds (containing mutant AlgG) would produce mannuronan, whereas those containing wild-type AlgG would produce epimerized alginates. The relative amounts of each alginate type would depend on the expression levels of each of the two versions of AlgG. The Pm promoter used in the complementation experiment has the additional advantage that it can be activated at different levels dependent of the concentration of inducer added (49), and we could therefore test our hypothesis by inserting transposon TnKB10 into the chromosome of the mannuronan-producing strain Pf20118. When the resulting transposon insertion strain, Pf20118::TnKB10, was grown in the presence of increasing amounts of inducer, the relative fraction of G residues in the alginate also increased (Table 2). Since AlgG does not epimerize M residues next to G residues, the substrate rapidly accumulates M residues inaccessible to epimerization as the reaction proceeds. In strain Pf20118::TnKB10 wild-type AlgG must, in addition, compete with the corresponding mutant version, and this also leads to a nonlinear relationship between the amount of wild-type AlgG and the G content. High-molecular-weight alginates form viscous solutions, and this viscosity is extremely sensitive to cuts in the polymer chain. Therefore, even at very low fractions of randomly distributed G residues, the intrinsic viscosity should drop dramatically if the solution is treated with a G-specific lyase. In contrast, if the G residues were confined to only a subfraction of the molecules, the intrinsic viscosity would not drop below the limit determined by the amount and molecular weight of the mannuronan fraction. As can be seen from Fig. 4, alginate produced by strain Pf201 (wild-type algG) was rapidly degraded by the G lyase, whereas the mannuronan produced by the mutant strain Pf20118 was hardly affected at all. When equal amounts of these two alginates were mixed, the viscosity was rapidly reduced, but not below the value determined by the amount of mannuronan present. Lastly, alginate containing 18% G from Pf20118::TnKB10 induced by 0.025 mM m-toluate was analyzed. As predicted, this alginate behaved just like the mixture of mannuronan and wild-type alginate. In contrast, addition of M lyase eliminated the viscosity in all of the samples (not shown). This clearly demonstrated that the two versions of AlgG are acting on distinctly different polymer strands, and this observation is consistent with the scaffold hypothesis.
FIG. 4.
Degradation of alginates by G-specific lyase measured as decrease in viscosity. The bacteria were grown in liquid PIA medium and collected by isopropanol precipitation. The relative viscosity ηsp is defined as ηsp = (ts − to)/to, where ts is the time (s) for a given volume of sample to pass the Ubbelodhe capillary and to is the time (s) for the same amount of the solvent. The alginates used were from Pf201 (33% G) (×), Pf20118 (100% M) (⧫), Pf20118::TnKB10 (18% G) (▴), and Pf201 and Pf20118 (17% G total) (•).
DISCUSSION
Unlike most other heteropolymers, alginates are synthesized by first producing a homopolymer, mannuronan, and some of the mannuronic acid residues are then epimerized to guluronic acid (21). The introduction of single G residues affects the flexibility of the polymer (44). It probably also affects the degree of acetylation, since only M residues are acetylated (43). It has been shown previously in P. aeruginosa that point mutations in algG blocking epimerization lead to the production of mannuronan but did not seem to otherwise affect polymer production (3). That finding is in agreement with the results presented here and, by comparing the four algG mutations with the sequences of AlgG homologues from different species, it was found that all of the four substituted amino acids were conserved in all of the deduced AlgG proteins reported so far (Fig. 1).
It has earlier been proposed that the reaction mechanism of mannuronan C-5-epimerases and alginate lyases share the first step (extraction of the proton at C-5) (15). In a recent study describing the three-dimensional structure of one of the Sphingomonas lyases complexed to alginate a particular motif (NNHSY) was reported to be implicated in the binding of the M residue at the catalytic site (50, 51). This motif has been shown to be shared by the periplasmic and the secreted epimerases and by some of the M-specific alginate lyases (9). Interestingly, AlgG of P. fluorescens has two expanded copies (NNRSYDN and NNFVADN) sharing some similarity to this motif (Fig. 1). Two of the point mutations of the epimerase-negative mutants reported here are found immediately N terminal to each of these motifs (S337F and D361N). Both the serine and the aspartic acid seem to be conserved at the same relative position to both copies of the conserved motif. These observations support the conclusion that the motifs and residues discussed above might have an important function in binding or catalysis in AlgG.
It is well known that many of the genes in the alg operon are required for alginate polymer formation, but their exact biochemical functions are in many cases only partly understood. Alg8 and Alg44 have been found to be membrane proteins necessary for alginate production (26, 29) and, based on sequence alignment studies, Alg8 is proposed to be the polymerase (29). AlgX has also been shown to be necessary for alginate biosynthesis (30), although both its function and location are unknown. AlgK has been proposed to facilitate periplasmic transport of alginate (1), and deletion mutants of algK have been shown to make small oligouronides with unsaturated nonreducing ends similar to those found for algG deletion reported here (22, 23). Finally, AlgE has been reported to be an outer membrane pore necessary for alginate export (36). Even without considering the results of the experiments reported here it appeared possible that some or all of these proteins form some kind of scaffold that leads the newly formed alginate polymer from the cytoplasmic membrane, through the periplasmic space and outer membrane, ending up in the extracellular environment. By the experiments reported here we feel that such a hypothesis has been significantly strengthened, and a possible model that is consistent with available experimental data is shown in Fig. 5. In this model AlgX is not included since its location is unknown.
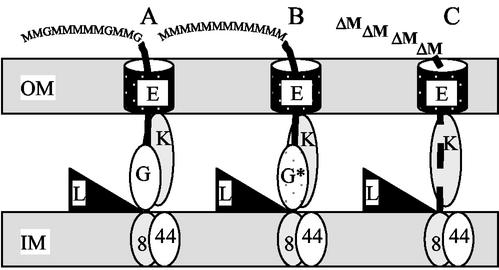
FIG. 5.
Hypothetical model of the alginate biosynthesis complex. Only the proteins known to be located in the cell envelope and to be essential for alginate production are shown. (A) Wild-type complex; (B) complex with an epimerization-defective AlgG; (C) complex without AlgG. OM, outer membrane; IM, inner membrane; 8, Alg8; 44, Alg44; L, AlgL; K, AlgK; G, AlgG; G*, epimerization-deficient copy of AlgG; E, AlgE.
In the wild type (Fig. 5A) one might envision that the proteins described above form a scaffold extending from the cytoplasmic membrane (polymerization), through the periplasm (modification and transport) and an outer membrane pore. Under these conditions AlgG is located such that it protects the newly formed strand from most, but not all, of the potential activity of AlgL. The alginate strands consequently become very long. When wild-type AlgG is exchanged with an epimerization-deficient AlgG point mutant, the protein structure is almost unaffected, such that protection against AlgL still works. The product obviously lacks G residues (Fig. 5B). When AlgG is removed from the complex, on the other hand (Fig. 5C), the polymer is no longer protected from the activity of AlgL, which is present in excess relative to the amount of polymer produced. The reason a dimer is formed could be that the organization of the complex enables AlgL to act on the polymer end as it protrudes from the cytoplasmic membrane. This also explains why a different product pattern would be obtained if alginate and lyase simply were mixed in a test tube.
Based on the results described above it appeared probable that construction of a lyase-deficient mutant of strain Pf201ΔalgG would lead to a strain in which alginate polymer formation was restored. This hypothesis has been tested, but it turned out that inactivation of algL (in-frame deletion or point mutation) is lethal to the cells. One can get mutants with these genotypes, but they have acquired additional mutations that turn off alginate synthesis. Such mutants could therefore not be complemented by algL wild type.
The protein scaffold hypothesis is also consistent with the ability of Pf20118::TnKB10 to simultaneously make two distinct populations of alginates, by predicting that these cells contain two different types of protein complexes (Fig. 5A and B). Since the production of AlgG from the Pm promoter is added to the production of epimerization-defective AlgG from the alg operon, the induced cells presumably contain AlgG in surplus. If AlgG were not part of a protein complex it is difficult to see how about half of the polymer produced can be pure mannuronan, whereas the remaining half contains wild-type levels of G residues. The hypothesis is also consistent with the observed simultaneous production of polymer and dimer by strain Pf201ΔalgG::TnKB10.
The scaffold model presented above must for the time being be considered as a working hypothesis, but it has the advantage that it is consistent with the experimental data. We also find it difficult to envision an alternative model that appears equally likely and does not involve any form of protein complex formation.
Acknowledgments
This work was funded by a grant from the Norwegian Research Council; by FMC Biopolymers AS; by Veterans Administration Medical Research Funds (D.E.O.); and Public Health Service grant AI-19146 (D.E.O.).
We thank Cristiana Campa, University of Trieste, for analyzing the oligomer by capillary electrophoresis and mass spectroscopy. We are deeply grateful to Randi Aune for performing fermentations and analysis of alginates, and to Wenche Iren Strand for recording the 1H-NMR spectra.
REFERENCES
- 1.Aarons, S. J., I. W. Sutherland, A. M. Chakrabarty, and M. P. Gallagher. 1997. A novel gene, algK, from the alginate biosynthesis cluster of Pseudomonas aeruginosa. Microbiology 143:641-652. [DOI] [PubMed] [Google Scholar]
- 2.Blatny, J. M., T. Brautaset, H. C. Winther-Larsen, K. Haugan, and S. Valla. 1997. Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. Appl. Environ. Microbiol. 63:370-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chitnis, C. E., and D. E. Ohman. 1990. Cloning of Pseudomonas aeruginosa algG, which controls alginate structure. J. Bacteriol. 172:2894-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chitnis, C. E., and D. E. Ohman. 1993. Genetic analysis of the alginate biosynthetic gene cluster of Pseudomonas aeruginosa shows evidence of an operonic structure. Mol. Microbiol. 8:583-593. [DOI] [PubMed] [Google Scholar]
- 5.Chung, C. T., S. L. Niemela, and R. H. Miller. 1989. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. USA 86:2172-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Lorenzo, V., I. Cases, M. Herrero, and K. N. Timmis. 1993. Early and late response of TOL promoters to pathway inducers: identification of postexponential promoters in Pseudomonas putida with lacZ-tet bicistronic reporters. J. Bacteriol. 175:6902-6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ertesvåg, H., B. Doseth, B. Larsen, G. Skjåk-Bræk, and S. Valla. 1994. Cloning and expression of an Azotobacter vinelandii mannuronan C-5-epimerase gene. J. Bacteriol. 176:2846-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ertesvåg, H., F. Erlien, G. Skjåk-Bræk, B. H. Rehm, and S. Valla. 1998. Biochemical properties and substrate specificities of a recombinantly produced Azotobacter vinelandii alginate lyase. J. Bacteriol. 180:3779-3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ertesvåg, H., H. K. Høidal, I. K. Hals, A. Rian, B. Doseth, and S. Valla. 1995. A family of modular type mannuronan C-5-epimerase genes controls alginate structure in Azotobacter vinelandii. Mol. Microbiol. 16:719-731. [DOI] [PubMed] [Google Scholar]
- 11.Ertesvåg, H., and G. Skjåk-Bræk. 1999. Modification of alginate using mannuronan C-5-epimerases, p.71-78. In C. Bucke (ed.), Methods in biotechnology 10: carbohydrate bio/technology protocols. Humana Press, Inc., Totowa, N.J.
- 12.Franklin, M. J., C. E. Chitnis, P. Gacesa, A. Sonesson, D. C. White, and D. E. Ohman. 1994. Pseudomonas aeruginosa AlgG is a polymer level alginate C5-mannuronan epimerase. J. Bacteriol. 176:1821-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franklin, M. J., and D. E. Ohman. 1996. Identification of algI and algJ in the Pseudomonas aeruginosa alginate biosynthetic gene cluster which are required for alginate O acetylation. J. Bacteriol. 178:2186-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franklin, M. J., and D. E. Ohman. 2002. Mutant analysis and cellular localization of the AlgI, AlgJ, and AlgF proteins required for O acetylation of alginate in Pseudomonas aeruginosa. J. Bacteriol. 184:3000-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gacesa, P. 1987. Alginate-modifying enzymes: a proposed unified mechanism of action for the lyases and epimerases. FEBS Lett. 212:199-202. [Google Scholar]
- 16.Gacesa, P. 1998. Bacterial alginate biosynthesis-recent progress and future prospects. Microbiology 144:1133-1143. [DOI] [PubMed] [Google Scholar]
- 17.Govan, J. R., J. A. Fyfe, and T. R. Jarman. 1981. Isolation of alginate-producing mutants of Pseudomonas fluorescens, Pseudomonas putida, and Pseudomonas mendocina. J. Gen. Microbiol. 125:217-220. [DOI] [PubMed] [Google Scholar]
- 18.Grasdalen, H. 1983. High-field, 1-H-n.m.r. spectroscopy of alginate. Sequential structure and linkage-conformation. Carbohydr. Res. 118:255-260. [Google Scholar]
- 19.Greener, A. 1990. E. coli SURE[trade] strain: clone “uncloneable” DNA. Strategies 3:5-6. [Google Scholar]
- 20.Hansen, L. H., S. J. Sorensen, and L. B. Jensen. 1997. Chromosomal insertion of the entire Escherichia coli lactose operon, into two strains of Pseudomonas, using a modified mini-Tn5 delivery system. Gene 186:167-173. [DOI] [PubMed] [Google Scholar]
- 21.Haug, A., and B. Larsen. 1969. Biosynthesis of alginate: epimerization of d-mannuronic to l-guluronic acid residues in the polymer chain. Biochim. Biophys. Acta 192:557-559. [DOI] [PubMed] [Google Scholar]
- 22.Jain, S., M. J. Franklin, H. Ertesvåg, S. Valla, and D. E. Ohman. 2003. The dual roles of AlgG in C-5-epimerization and secretion of alginate polymers in Pseudomonas aeruginosa. Mol. Microbiol. 47:1123-1133. [DOI] [PubMed] [Google Scholar]
- 23.Jain, S., and D. E. Ohman. 1998. Deletion of algK in mucoid Pseudomonas aeruginosa blocks alginate polymer formation and results in uronic acid secretion. J. Bacteriol. 180:634-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karunakaran, P., J. M. Blatny, H. Ertesvåg, and S. Valla. 1998. Species-dependent phenotypes of replication-temperature-sensitive trfA mutants of plasmid RK2: a codon-neutral base substitution stimulates temperature sensitivity by leading to reduced levels of trfA expression. J. Bacteriol. 180:3793-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karunakaran, P., D. T. Endresen, H. Ertesvåg, J. M. Blatny, and S. Valla. 1999. A small derivative of the broad-host-range plasmid RK2 which can be switched from an replicating to a non-replicating state as a response to an externally added inducer. FEMS Microbiol. Lett. 180:221-227. [DOI] [PubMed] [Google Scholar]
- 26.Maharaj, R., T. B. May, S. K. Wang, and A. M. Chakrabarty. 1993. Sequence of the alg8 and alg44 genes involved in the synthesis of alginate by Pseudomonas aeruginosa. Gene 136:267-269. [DOI] [PubMed] [Google Scholar]
- 27.May, T. B., and A. M. Chakrabarty. 1994. Pseudomonas aeruginosa: genes and enzymes of alginate synthesis. Trends Microbiol. 2:151-157. [DOI] [PubMed] [Google Scholar]
- 28.May, T. B., D. Shinabarger, R. Maharaj, J. Kato, L. Chu, J. D. DeVault, S. Roychoudhury, N. A. Zielinski, A. Berry, R. K. Rothmel, T. K. Misra, and A. M. Chakrabarty. 1991. Alginate synthesis by Pseudomonas aeruginosa: a key pathogenic factor in chronic pulmonary infections of cystic fibrosis patients. Clin. Microbiol. Rev. 4:191-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mejia-Ruiz, H., J. Guzman, S. Moreno, G. Soberon-Chavez, and G. Espin. 1997. The Azotobacter vinelandii alg8 and alg44 genes are essential for alginate synthesis and can be transcribed from an algD-independent promoter. Gene 199:271-277. [DOI] [PubMed] [Google Scholar]
- 30.Monday, S. R., and N. L. Schiller. 1996. Alginate synthesis in Pseudomonas aeruginosa: the role of AlgL (alginate lyase) and AlgX. J. Bacteriol. 178:625-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morea, A., K. Mathee, M. J. Franklin, A. Giacomini, M. O'Regan, and D. E. Ohman. 2001. Characterization of algG encoding C5-epimerase in the alginate biosynthetic gene cluster of Pseudomonas fluorescens. Gene 278:107-114. [DOI] [PubMed] [Google Scholar]
- 32.Nivens, D. E., D. E. Ohman, J. Williams, and M. J. Franklin. 2001. Role of alginate and its O acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J. Bacteriol. 183:1047-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Østgaard, K. 1992. Enzymatic microassay for the determination and characterization of alginates. Carbohydr. Polymers 19:51-59. [Google Scholar]
- 34.Penaloza-Vazquez, A., S. P. Kidambi, A. M. Chakrabarty, and C. L. Bender. 1997. Characterization of the alginate biosynthetic gene cluster in Pseudomonas syringae pv. syringae. J. Bacteriol. 179:4464-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rehm, B. H. 1998. Alginate lyase from Pseudomonas aeruginosa CF1/M1 prefers the hexameric oligomannuronate as substrate. FEMS Microbiol. Lett. 165:175-180. [DOI] [PubMed] [Google Scholar]
- 36.Rehm, B. H., G. Boheim, J. Tommassen, and U. K. Winkler. 1994. Overexpression of algE in Escherichia coli: subcellular localization, purification, and ion channel properties. J. Bacteriol. 176:5639-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rehm, B. H., H. Ertesvåg, and S. Valla. 1996. A new Azotobacter vinelandii mannuronan C-5-epimerase gene (algG) is part of an alg gene cluster physically organized in a manner similar to that in Pseudomonas aeruginosa. J. Bacteriol. 178:5884-5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sadoff, H. L. 1975. Encystment and germination in Azotobacter vinelandii. Bacteriol. Rev. 39:516-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook, J., and D. Russell. 2000. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, New York, N.Y.
- 40.Schiller, N. L., S. R. Monday, C. M. Boyd, N. T. Keen, and D. E. Ohman. 1993. Characterization of the Pseudomonas aeruginosa alginate lyase gene (algL): cloning, sequencing, and expression in Escherichia coli. J. Bacteriol. 175:4780-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shinabarger, D., A. Berry, T. B. May, R. Rothmel, A. Fialho, and A. M. Chakrabarty. 1991. Purification and characterization of phosphomannose isomerase-guanosine diphospho-d-mannose pyrophosphorylase: a bifunctional enzyme in the alginate biosynthetic pathway of Pseudomonas aeruginosa. J. Biol. Chem. 266:2080-2088. [PubMed] [Google Scholar]
- 42.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 43.Skjåk-Bræk, G., H. Grasdalen, and B. Larsen. 1986. Monomer sequence and acetylation pattern in some bacterial alginates. Carbohydr. Res. 154:239-250. [DOI] [PubMed] [Google Scholar]
- 44.Smidsrød, O., and K. I. Draget. 1996. Chemistry and physical properties of alginates. Carbohydr. Eur. 14:6-13. [Google Scholar]
- 45.Stanford, E. C. C. 1883. On algin: a new substance obtained from some of the commoner species of marine algae. Chem. News 96:254-257. [Google Scholar]
- 46.Svanem, B. I., G. Skjåk-Bræk, H. Ertesvåg, and S. Valla. 1999. Cloning and expression of three new Azotobacter vinelandii genes closely related to a previously described gene family encoding mannuronan C-5-epimerases. J. Bacteriol. 181:68-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valla, S., J. Li, H. Ertesvåg, T. Barbeyron, and U. Lindahl. 2001. Hexuronyl C5-epimerases in alginate and glycosaminoglycan biosynthesis. Biochimie 83:819-830. [DOI] [PubMed] [Google Scholar]
- 48.Vazquez, A., S. Moreno, J. Guzman, A. Alvarado, and G. Espin. 1999. Transcriptional organization of the Azotobacter vinelandii algGXLVIFA genes: characterization of algF mutants. Gene 232:217-222. [DOI] [PubMed] [Google Scholar]
- 49.Winther-Larsen, H. C., K. D. Josefsen, T. Brautaset, and S. Valla. 2000. Parameters affecting gene expression from the Pm promoter in gram-negative bacteria. Metab. Eng. 2:79-91. [DOI] [PubMed] [Google Scholar]
- 50.Yoon, H. J., W. Hashimoto, O. Miyake, K. Murata, and B. Mikami. 2001. Crystal structure of alginate lyase A1-III complexed with trisaccharide product at 2.0 Å resolution. J. Mol. Biol. 307:9-16. [DOI] [PubMed] [Google Scholar]
- 51.Yoon, H. J., B. Mikami, W. Hashimoto, and K. Murata. 1999. Crystal structure of alginate lyase A1-III from Sphingomonas species A1 at 1.78 Å resolution. J. Mol. Biol. 290:505-514. [DOI] [PubMed] [Google Scholar]