Abstract
Flagellum-mediated motility of Vibrio fischeri is an essential factor in the bacterium's ability to colonize its host, the Hawaiian squid Euprymna scolopes. To begin characterizing the nature of the flagellar regulon, we have cloned a gene, designated flrA, from V. fischeri that encodes a putative σ54-dependent transcriptional activator. Genetic arrangement of the flrA locus in V. fischeri is similar to motility master-regulator operons of Vibrio cholerae and Vibrio parahaemolyticus. In addition, examination of regulatory regions of a number of flagellar operons in V. fischeri revealed apparent σ54 recognition motifs, suggesting that the flagellar regulatory hierarchy is controlled by a similar mechanism to that described in V. cholerae. However, in contrast to its closest known relatives, flrA mutant strains of V. fischeri ES114 were completely abolished in swimming capability. Although flrA provided in trans restored motility to the flrA mutant, the complemented strain was unable to reach wild-type levels of symbiotic colonization in juvenile squid, suggesting a possible role for the proper expression of FlrA in regulating symbiotic colonization factors in addition to those required for motility. Comparative RNA arbitrarily primed PCR analysis of the flrA mutant and its wild-type parent revealed several differentially expressed transcripts. These results define a regulon that includes both flagellar structural genes and other genes apparently not involved in flagellum elaboration or function. Thus, the transcriptional activator FlrA plays an essential role in regulating motility, and apparently in modulating other symbiotic functions, in V. fischeri.
Animals and plants are hosts to both beneficial and pathogenic microorganisms that are typically acquired directly from their environment. Bacterial motility often plays a role in host associations, not only by enabling these microbes to gain access to desired tissues but also through a coordinated regulation of motility and other specific colonization factors. In certain pathogens, cues in the host environment elicit a bacterial response that both stimulates the production of virulence genes and represses the synthesis of those genes that encode functions no longer required for host colonization (e.g., motility) (11, 18). Additionally, the transcription of certain motility and virulence genes is known to be reciprocally coordinated directly through the flagellar regulon (3, 16, 17, 46), and the type III flagellar secretion apparatus has been shown to export virulence proteins (15, 61). Whereas in many pathogenic bacteria virulence and motility are intimately linked (reviewed in references 28 and 41), a similar relationship has not been reported in a benign association between a bacterium and its animal host.
Vibrio fischeri is the specific bacterial symbiont of the Hawaiian squid Euprymna scolopes. Symbiotic strains of V. fischeri are acquired from the environment early during the juvenile stage of E. scolopes development and are maintained within a specialized light-emitting organ whose development is triggered by the presence of the bacteria (34). Bioluminescent V. fischeri cells are housed extracellularly within three pairs of crypt spaces inside this organ, which is present throughout the life of the animal. These bacteria are thought to benefit their host by providing a source of light, which is used as a camouflage for the animal's night-foraging behavior. A program of bacterium-induced changes in development of the host has been described (37), and at least some of the bacterial signals that trigger these changes have been identified (14, 39). Similarly, several bacterial factors required for normal colonization and symbiotic competence have been elucidated (1, 12, 19-21, 55, 57, 59).
Concurrent with host developmental changes, symbiotic V. fischeri cells appear to adapt to their host environment through changes both in gene expression (K. L. Visick and E. G. Ruby, Abstr. Gen. Meet. Am. Soc. Microbiol. 1998, p. 277, 1998) and in morphology (45). Cells of V. fischeri isolated from the adult light organ are morphologically dissimilar to cells grown in culture: the symbiotic cells are typically smaller and aflagellate (45). The latter effect was hypothesized to result from a general repression of flagellum elaboration within the first 24 h of colonization and was rapidly reversed once the bacteria were removed from the light-organ environment. Subsequently, work using transposon-derived mutants defined the role of motility in host colonization; nonmotile and nonflagellated mutants cannot initiate an infection of the squid light organ (19). To further investigate the role of motility in colonization, spontaneous mutant strains of V. fischeri with altered migration patterns were isolated and found to be limited in their ability to colonize E. scolopes (36). Furthermore, a subset of these strains contained additional phenotypic defects seemingly unrelated to motility, thereby suggesting that the flagellar regulon in V. fischeri comprises nonflagellar genes as well as those required for motility. Thus, because flagellum synthesis appears to be down-regulated (45), although as shown here not entirely repressed, when V. fischeri cells are within the light-organ environment, motility and other symbiotic colonization genes may be coordinately regulated in an inverse manner, similar to that described for pathogenic associations.
In many bacterial species, flagellar gene transcription occurs in a regulatory hierarchy in which the expression of late genes (i.e., class III or IV) is dependent on the expression of early ones (i.e., class I or II). The entire flagellar regulon is ultimately dependent on a master regulator. This mechanism of regulation ensures that the secretion apparatus is functioning prior to transcription of the late flagellin and chemotaxis genes. In Vibrio cholerae, the σ54-dependent transcriptional activator FlrA is the sole class I gene (42). FlrA is required for expression of the class II genes including (i) FlrC, a second σ54-dependent activator; (ii) FliA, the alternative flagellar sigma factor σ28; and (iii) components of the flagellar basal body's MS ring-switch complex. FlrC and σ28 must be present for the expression of class III and class IV genes, respectively, which together encode the remainder of the basal body, hook structure, and flagellin subunits. Thus, flagellar transcription in V. cholerae (and likely the closely related Vibrio parahaemolyticus [33, 52]) occurs by a novel mechanism that includes features of both the σ54- and σ28-dependent flagellar transcription hierarchies of Caulobacter crescentus (60) and Salmonella enterica serovar Typhimurium (32), respectively.
We are interested in how signals within the light-organ environment are processed and integrated by V. fischeri to stimulate or repress the production of flagella and nonflagellar symbiotic factors. In this study, we have begun to identify components of the flagellar regulon whose expression is dependent on a master regulator (FlrA homolog) in V. fischeri. This first demonstration of transcriptional control of V. fischeri flagellar gene expression will serve as a basis for future investigations that should lead to elucidation of the regulatory hierarchies that govern flagellum synthesis in V. fischeri.
MATERIALS AND METHODS
Bacterial strains, media, and reagents.
Wild-type V. fischeri strain ES114, isolated from E. scolopes (7), was used as the parent strain (Table 1). Escherichia coli strains DH5α (22) and CC118λpir (26) were used as hosts for plasmids with ColE1 and R6K replication origins, respectively. Mobilizable plasmid pKV111 containing a red-shifted green fluorescent protein (GFP) derivative (49) was used to label V. fischeri cells for confocal laser-scanning microscopy. V. fischeri cells were grown at 28°C in either SWT medium (7), which contains 0.5% tryptone, 0.3% yeast extract, and 0.3% glycerol in 70% seawater, or LBS medium, which contains 1% tryptone, 0.5% yeast extract, 2% NaCl, and 20 mM Tris-hydrochloride (pH 7.4). Chemotaxis and motility studies were performed with either SWT or tryptone medium, which contained 1% tryptone, 2% NaCl, and 20 mM Tris-hydrochloride (pH 7.4). Agar was added to 1.5% for solid media or to between 0.3 and 0.7% for motility media. To maintain plasmids in V. fischeri, the chloramphenicol (Cm) concentration was used at concentrations of 5 μg ml−1 of culture medium or 2 μg ml−1 of seawater for squid colonization experiments. For selection of single recombinants, kanamycin (Kn) was added at a concentration of 100 μg ml−1 of culture medium. When added to LB medium (35) for the selection of E. coli, Kn and Cm were used at concentrations of 100 and 20 μg ml−1, respectively. All chemicals were obtained from Sigma Chemical Co. (St. Louis, Mo.). Restriction enzymes and DNA ligase were obtained from New England Biolabs (Beverly, Mass.). AmpliTaq DNA polymerase was obtained from Perkin-Elmer (Branchburg, N.J.). Oligonucleotides were synthesized by Operon Technologies, Inc. (Alameda, Calif.).
TABLE 1.
Strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Relevant informationa | Source or reference |
|---|---|---|
| V. fischeri strains | ||
| ES114 | Wild-type E. scolopes isolate | 7 |
| DM126 | ES114 flrA::Knr | This study |
| DM127 | ES114 flrA::Knr | This study |
| DM128 | ES114 flrA::Knr | This study |
| DM132 | FlrA reverse-complemented derivative of DM126 | This study |
| DM159 | ES114 ΔflrA | This study |
| Plasmids | ||
| pCR2.1 | E. coli cloning vector; Knr | Invitrogen, Inc. |
| pEVS79 | Suicide vector; Cmr | 51 |
| pEVS104 | Conjugative helper plasmid | 51 |
| pLS6 | V. fischeri cloning vector; Cmr | 56 |
| pKV111 | gfp-containing derivative of pVO8 | 49 |
| pVO8 | V. fischeri cloning vector; Cmr Err | 56 |
| pDM56 | flrA PCR product in pCR2.1 | This study |
| pDM57 | 4.9-kb NheI flrA locus in pEVS79 | This study |
| pDM57-5 | flrA::Knr at nucleotide position 819 | This study |
| pDM57-6 | flrA::Knr at nucleotide position 222 | This study |
| pDM57-13 | flrA::Knr at nucleotide position 417 | This study |
| pDM58 | 2.8-kb PstI/SacI fragment in pVO8 | This study |
| pDM84 | hvnC PCR product in pCR2.1 | This study |
| pDM85 | topB PCR product in pCR2.1 | This study |
| pDM86 | flaE PCR product in pCR2.1 | This study |
| pDM108 | pDM57 HpaI deletion | This study |
| Oligonucleotides | ||
| DM13 (flrA) | GCGCGTAACATTCACTACC | |
| DM15 (flrA) | TCG(C)CGCACG(C)TTACCC GGCC | |
| DM42 (flaE) | CGATGAGACCAAACCACC | |
| DM43 (flaE) | CTAGACCACGAGTTTGCG | |
| DM44 (topB) | CTGAGGACATGCGCTATC | |
| DM45 (topB) | TTGGCCATACTTAGCGCG | |
| DM38 (hvnC) | AACGACACTACTGGCAAG | |
| DM40 (hvnC) | CACTTGGTGAGCCAAAGA |
Antibiotics used: Kn, kanamycin; Cm, chloramphenicol; Er, erythromycin.
Molecular genetic techniques and sequence analysis.
Chromosomal and plasmid DNA were isolated and purified using Qiagen spin columns following the manufacturer's suggestions (Qiagen, Valencia, Calif.). For Southern analyses, 5 μg of NheI- or HindIII-digested chromosomal DNA was separated by electrophoresis and transferred to Hybond nylon membrane (Boehringer Mannheim, Indianapolis, Ind.). Blots were hybridized with the probe overnight at 65°C, and the membranes were washed under high-stringency conditions (two 20-min washes in a solution of 0.03 M sodium citrate, 0.03 M sodium chloride, and 0.1% sodium dodecyl sulfate; pH 7.0; at 65°C), and developed using the chemiluminescent substrate CDP-Star (Boehringer Mannheim). DNA sequencing was conducted on an ABI automated DNA sequencer at the University of Hawaii Biotechnology/Molecular Biology Instrumentation Training Facility. Overlapping contiguous sequences were mapped using Sequencher (Gene Codes Corp., Ann Arbor, Mich.) and DNA Strider programs. Sequence analysis was performed using the BLAST program for database searches and the National Center for Biotechnology Information conserved-domain search program (2). Multiple sequence alignments were performed using the ClustalW program (54). Consensus binding sequences for NtrC, σ70, σ54, and σ28 were obtained from previous reports (5, 23, 25). To detect promoter binding sequences in upstream regions of flrA-regulated genes, the BioProspector program was used (31).
Cloning, sequence analysis, and disruption of flrA.
Degenerate oligonucleotide primers for PCR were designed using alignments of the predicted amino acid sequences of V. cholerae flrA, V. parahaemolyticus flaK, and Pseudomonas aeruginosa fleQ (GenBank accession numbers AF014113, AF069392, and AE004540, respectively). PCR was performed as follows: 30 cycles of 95°C for 1 min, 52°C for 1 min, and 72°C for 1 min, followed by a 10-min extension at 72°C. The resulting 541-bp fragment was cloned (resulting in pDM56), sequenced to confirm its similarity to flrA and flaK, and used subsequently as a probe. A library of NheI-digested chromosomal fragments of between 5 and 6 kb was created by purifying the fragments from an agarose gel, using the Qiagen gel extraction kit and ligating them into the suicide vector pEVS79 (51), which had been similarly digested. A clone, pDM57, containing the desired flrA-containing NheI fragment, was identified by PCR and Southern analysis as described above, and the sequence of both DNA strands was determined. Flanking sequence corresponding to the flrC gene was obtained from the V. fischeri genome sequencing project (http://ergo.integratedgenomics.com/Genomes/VFI). The V. fischeri flrA gene located on plasmid pDM57 was inactivated using in vitro transposon mutagenesis (Epicentre Technologies, Madison, Wis.). The resulting plasmid pool was transformed into competent E. coli DH5α cells. Plasmids were isolated from Knr colonies, screened by restriction-digest analysis, and sequenced to confirm the location and orientation of the transposon insertion. Several clones that contained an insertion within the flrA gene were moved into wild-type V. fischeri cells by conjugal mating as previously described (51). The flrA::Knr alleles from three of these clones, pDM57-6, pDM57-5, and pDM57-13, which were found to contain an insertion within the flrA gene at codon positions 74, 139, and 273, respectively, were crossed into the chromosome of ES114 by marker exchange, generating strains DM126, DM127, and DM128, respectively (Table 1). An in-frame flrA deletion strain was constructed using the suicide plasmid pDM108, which contains the NheI fragment from pDM57, in which the DNA located between the two HpaI sites (547 nucleotides) has been removed (Fig. 1). For complementation studies, a PstI/SacI fragment containing the entire flrA gene and a portion of the downstream open reading frame (ORF) were cloned into plasmid pVO8 (56), which had been similarly digested, resulting in pDM58. The reverse-complemented strain, DM132, was created by introducing plasmid pDM57 (containing a wild-type copy of flrA) into strain DM126, selecting for Cm- and Kn-resistant single recombinants, and subsequently screening for sensitivity to both antibiotics. PCR and Southern blotting as described above confirmed the presence and location of chromosomal insertions or deletions.
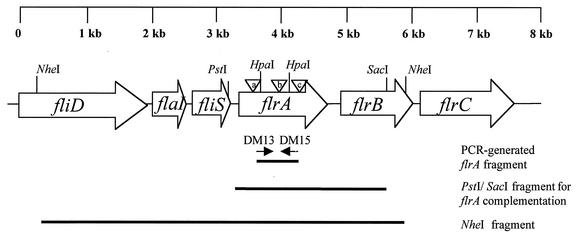
FIG. 1.
Schematic representation of the flrA locus in V. fischeri. Genes are designated by open boxes with arrows that indicate the direction of transcription. The internal fragment used to identify the flrA locus (using PCR primers DM13 and DM15), a PstI/SacI fragment inserted in pDM58 for flrA complementation, and the cloned NheI fragment are indicated by bold lines. Sequences upstream and downstream of the NheI sites were obtained from the V. fischeri genome sequencing project. The three triangles indicate the location of the TnflrA::Knr insertions (Table 1) in DM126 (a), DM127 (b), and DM128 (c). The sequence between the HpaI sites was removed to create the in-frame deletion strain DM159.
Motility assays.
Motility was measured by the movement of bacterial cells through SWT medium containing between 0.3 and 0.7% agar. The optical density at 600 nm was determined for mid-exponential-phase cultures of each strain used. Equal numbers of cells in 2 μl of medium were spotted at the centers of the plates, and the rate of movement was determined over several hours by measuring the diameter of the halo of cells that developed.
Electron microscopy.
V. fischeri cells were prepared for examination by transmission electron microscopy as described previously (36). Briefly, Formvar-coated copper grids (Ted Pella Co., Tustin, Calif.) were floated on suspensions of cells grown to mid-exponential phase (optical density = 0.4) in SWT medium and then transferred to a drop of fixative solution (2.5% glutaraldehyde and 2.5% paraformaldehyde in 0.1 M sodium cacodylate buffer, pH 7.4) for 10 min. The grids were washed twice with Nanopure (Millipore Corp., New Bedford, Mass.) water for 30 s and negatively stained for 1 min with freshly prepared and filtered 1% uranyl acetate. Grids were examined using a LEO 912 EF electron microscope at 100 kV of accelerating voltage.
Squid colonization.
Juvenile E. scolopes organisms were exposed to V. fischeri within 3 h of hatching as described previously (7), with several modifications. Animals were exposed to 4 ml of seawater containing between 4 × 103 and 2 × 104 cells of either the wild-type strain ES114 or the flrA mutant strain DM126, carrying either pVO8 (vector control) or pDM58 (flrA). Levels of colonization were determined indirectly by measuring the development of animal luminescence using an automated photometer (36). To visualize cells during the early stages of colonization, newly hatched squid were exposed to 4 × 105 cells of either the wild-type or the flrA mutant strain carrying the plasmid pKV111 (gfp). Beginning at 3 h postinoculation, animals were anaesthetized and viewed by confocal microscopy (36). Experiments to visualize motility behavior were performed as described above, except that animals were viewed by confocal microscopy to determine the location of bacteria, while their movement was determined by epifluorescence microscopy of living, whole-animal preparations.
RNA preparations.
Cells from exponential-phase cultures of different V. fischeri strains were harvested, and RNA was extracted by using RNAzol B (Tel-Test, Friendswood, Tex.), a guanidinium thiocyanate-phenol-based reagent, according to the manufacturer's instructions. RNA concentration was determined using a UV spectrophotometer, and its quality was assessed on an agarose gel prior to experiments.
RAP-PCR.
RNA arbitrarily primed PCR (RAP-PCR) was performed essentially as described previously (6) except that a mixture of 10-mer oligonucleotides with G:C contents of 50% were used as arbitrary primers (Genosys Biotechnologies, The Woodlands, Tex.). Following identification of putative regulated fragments, 32P-labeled DNA bands of interest were excised from the dried gel by using a scalpel and placed into a microcentrifuge tube containing 50 μl of Tris buffer (10 mM Tris-HCl, 1 mM EDTA; pH 8.0) for overnight elution at room temperature. A portion of the eluted fragment was used in a subsequent PCR containing the original primers used in RAP-PCR amplification. Following this secondary PCR, fragments were cloned using the TOPO-TA cloning kit (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. Clones containing inserts of the predicted size were then sequenced to determine their orientation and homology.
Confirmation of FlrA-regulated genes by quantitative RT-PCR.
Quantitative PCR was performed by amplifying a portion of each gene corresponding to the putative differentially expressed RAP products. One nanogram of total RNA isolated from either ES114 or DM126 was the substrate in reverse transcription-PCR (RT-PCR) using either Access RT-PCR (Promega, Madison, Wis.) or TaqMan (Perkin-Elmer). Three-step reactions were 40 cycles of 95°C for 15 s, 58°C for 5 s, and 72°C for 10 s, and the formation of products was monitored continuously during the 72°C incubation step. Standard curves were generated using serial dilutions of plasmid DNA-containing sequence of the individual RAP products (pDM84 for hvnC, pDM85 for topB, and pDM86 for flaE). Quantitative reactions were repeated a minimum of three times with independent RNA extracts. A semiquantitative analysis was performed in which serial dilutions of templates were used in RT-PCRs.
Nucleotide sequence accession number.
The DNA sequence of the flrA locus reported here has been submitted to GenBank, and its accession number is AY142201. The V. fischeri DNA sequence of FlrC and flagellar promoters can be obtained from the V. fischeri genomic sequencing project website at http://ergo.integratedgenomics.com/Genomes/VFI.
RESULTS
Motility behavior of V. fischeri cells during colonization of E. scolopes.
Because V. fischeri must use flagellum-mediated motility to initiate colonization of E. scolopes, we were interested in determining whether symbiotic V. fischeri cells retain flagellar structure and function in the light organ. Confocal microscopy of GFP-expressing cells of V. fischeri revealed that 45% of animals that had been colonized for over 12 h contained motile bacteria within the deep regions of one or more of the three pairs of light-organ crypts. Motile cells were observed most often (15 of 37 animals) in crypt 3, possibly due to the easier visualization of bacteria within this crypt, which is more superficially located (38). The presence of motile cells was always observed in at least a subset of animals that were colonized for 12, 24, and 35 h. In fact, at 36 h postinoculation, motile cells could be seen in almost half of the colonized animals, suggesting that a subset of the populating cells continues to express motility genes and their products even after the colonization has become well established.
Identification and characterization of a motility master regulator from V. fischeri.
Expression of motility genes is often regulated through a transcriptional hierarchy that is ultimately controlled by a master regulator (e.g., FlrA in V. cholerae). In this study we determined whether a FlrA-like protein similarly controls flagellar synthesis in V. fischeri. A flrA homolog from V. fischeri was identified using PCR with degenerate oligonucleotide primers designed to recognize sequences coding for conserved residues of σ54 transcriptional regulators (PCR primers DM13 and DM15) (Fig. 1 and 2). A 541-bp fragment was amplified from chromosomal DNA of V. fischeri, and its sequence was found to be similar to those encoding NtrC-like transcriptional activators. The amplified fragment was labeled and used as a probe in Southern analysis to identify a 4.9-kb NheI fragment of V. fischeri DNA. Sequence analysis of the NheI fragment in pDM57 revealed the presence of several ORFs, including the NtrC-like one (Fig. 1). This ORF potentially encodes a polypeptide of 493 amino acids with a calculated molecular mass of approximately 55,363 Da. Analysis of the predicted amino acid sequence revealed a putative σ54 interaction domain located between amino acids 137 and 356 that also contains an ATP-binding motif typical of σ54-dependent activators (Fig. 2). A possible helix-turn-helix DNA-binding domain characteristic of transcriptional regulators also was identified in the C-terminal region of the predicted protein (Fig. 2). Analysis of the surrounding regions revealed the presence of additional flagellar gene homologs (Fig. 1); thus, this ORF was designated flrA (for flagellar regulator protein A), following the nomenclature used to describe the homolog in V. cholerae. The predicted amino acid sequence of FlrA could be aligned with the flagellar regulators FlrA of V. cholerae (68% identity), FlaK of V. parahaemolyticus (69% identity), and FleQ of P. aeruginosa (67% identity) (Fig. 2). Analysis of the hydropathic characteristics of the FlrA protein by the method of Kyte and Doolittle (30), using the PSORT program, suggested that FlrA is hydrophilic, lacking any long stretches of hydrophobic residues characteristic of transmembrane segments. Therefore, we predict that FlrA, like its counterparts, is a soluble, cytoplasmic protein.
FIG. 2.
Amino acid sequence alignment of V. fischeri flrA (VfFlrA) and the motility regulators found in V. cholerae (VcFlrA), V. parahaemolyticus (VpFlaK), and P. aeruginosa (PaFleQ). Features and characteristic motifs are indicated. The predicted ATP-binding site and helix-turn-helix motifs are enclosed in boxes, and the conserved central domain is indicated by an overline. The oligonucleotide primers DM13 and DM15, used to identify the flrA homolog in V. fischeri, correspond to the amino acid regions below the arrows.
Downstream of flrA was a putative operon containing two ORFs (Fig. 1) whose predicted protein sequences most closely resemble FlrBC of V. cholerae (72 and 84% identity, respectively) (29). Similar to FlrB, the V. fischeri homolog contains a carboxy-terminal domain that defines it as a member of the two-component family of bacterial signal transducers, and it contains an N-terminal PAS domain often found in sensor kinases. Members of this family act to transduce signals via phosphorylation (27). FlrB in V. cholerae is the sensor kinase component that, when activated, phosphorylates its cognate response regulator, FlrC (10). Upstream of flrA were two ORFs that could encode proteins corresponding to FliS of E. coli, which is thought to function as a chaperone for the export of flagellin proteins (4), and the FlaI protein of V. parahaemolyticus, which is of unknown function (52). Sequence analysis upstream of flaI revealed a large ORF corresponding to the fliD gene that encodes a hook-associated protein. The gene organization of the V. fischeri flrA locus is identical to that found in V. cholerae and V. parahaemolyticus and can be aligned with that found in P. aeruginosa.
Sequence analysis of flagellar promoters.
When examined, the predicted promoter region of the flrA gene did not reveal the presence of specific motifs for NtrC, σ70, σ54, or σ28 binding. In contrast, examination of the predicted promoter region upstream of flrB revealed a possible σ54-binding consensus sequence that is similar to that found upstream of flrB in V. cholerae, flaK in V. parahaemolyticus, and fleQ in P. aeruginosa (Fig. 3). The proposed σ54-binding sequence of V. fischeri flrB varies from the consensus sequence at only 3 of 12 nucleotide positions (Fig. 3). In addition, we searched the available genome sequence of V. fischeri upstream of predicted flagellar operons and identified those genes that are similarly preceded by putative σ54 recognition motifs. Such predicted promoters were found upstream of homologs of genes that are known in V. cholerae to be activated by the transcriptional regulatory proteins FlrA or FlrC, both of which require σ54 for activity (Fig. 3). Thus, by analogy, transcription of these flagellar operons is likely to be controlled by a similar mechanism in V. fischeri.
FIG. 3.
Nucleotide sequence alignment of putative σ54 promoter elements located upstream of loci containing predicted V. fischeri motility-related genes. The bacterial consensus σ54 sequence is given for comparison (5). The general binding motif is in bold, and conserved residues are underlined.
Construction and analysis of flrA mutant strains.
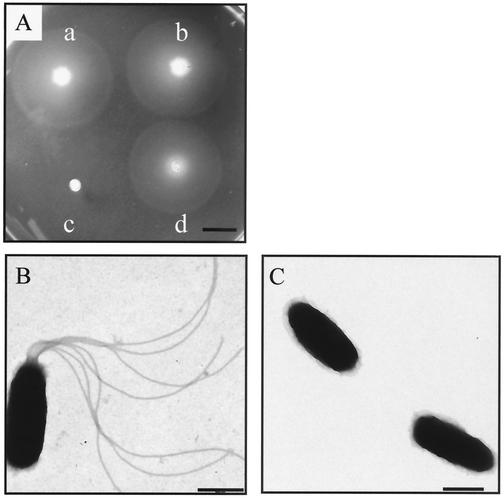
To determine whether FlrA functions as a motility regulator in V. fischeri, chromosomal flrA mutants were constructed in V. fischeri ES114 by gene replacement. Three mutants, DM126, DM127, and DM128 (Table 1), were marked with a Knr cassette at unique sites and in different orientations (Fig. 1) relative to flrA (in the same direction of transcription in strain DM126 and in the reverse orientation in DM127 and DM128). An in-frame deletion strain, DM159, was constructed by removing approximately 40% of the gene sequence, including the region corresponding to the predicted ATP-binding site and conserved central domain essential for FlrA function. The mutant flrA alleles of these strains were confirmed by Southern blot analysis (data not shown). The flrA mutant strains of V. fischeri, unlike those in V. cholerae and V. parahaemolyticus, were nonmotile in soft agar motility plates even after an extended incubation of 5 days at 24°C (Fig. 4A). Furthermore, motile cells were never observed when the flrA mutant strains were viewed by light microscopy. Transmission electron microscopy confirmed that the loss of motility was due to an inability to synthesize flagella and that flrA mutant cells are otherwise morphologically similar to wild-type cells (Fig. 4B and C). To confirm that the nonmotile phenotype of the flrA strains was indeed due to inactivation of the flrA gene, a wild-type copy of this gene was introduced into the flrA mutant strains DM126 and DM159 on pDM58. Both motility and flagellum synthesis was restored by the flrA gene provided in trans, while the vector alone (pVO8) did not complement (Fig. 4A and data not shown). Furthermore, transmission electron microscopy analysis revealed that the number and length of flagella produced by the complemented mutant strain DM126 was indistinguishable from that produced by wild-type cells (data not shown). Because altered motility behavior in 0.7% agar confers a disadvantage in squid colonization (36), we confirmed that the complemented mutant strain displayed the same rate of movement as the wild type in motility plates containing between 0.3 and 0.7% agar (Fig. 4A; data for 0.4 to 0.7% agar are not shown). Complementation of the additional flrA mutant strains (DM127, DM128, and DM159) gave similar results. Thus, we did not find evidence for any difference in motility behavior among the complemented flrA mutant and wild-type strains.
FIG. 4.
Motility phenotypes and transmission electron micrographs of the flrA mutant and complementing strains. (A) Motility patterns of wild-type strain ES114 (a), ES114 carrying pDM58 (flrA) (b), DM126 (c), and DM126 carrying pDM58 (flrA) (d) after 18 h in soft agar. Growth of the mutant strains in soft agar was similar to that of the wild-type parent. Bar, 10 mm. (B and C) Transmission electron micrographs of individual cells of ES114 (B) and the flrA mutant strain, DM126 (C). Cells of the complemented flrA mutant strain (DM126 carrying pDM58) were indistinguishable from those seen in panel B. Bars, 0.5 μm.
Light-organ colonization assays.
As expected, the flrA mutant DM126 was unable to initiate light-organ colonization (data not shown) even when presented to juvenile animals at inoculum concentrations 1,000-fold greater than that typically used to initiate an infection by wild-type cells. Because certain behaviors including motility (19) and aggregation on host-derived mucous (40) are thought to contribute to colonization, we investigated more closely the flrA mutant cells as they attempted to initiate infection. The flrA mutant cells formed aggregates of tens of cells when presented to the animal at an inoculum dose of 4 × 105 cells, while wild-type cells typically formed aggregates of hundreds of cells (40). In addition, a few (<10) flrA mutant cells were observed within the pores, ducts, and crypts of the juvenile light organ in about 20% (3 of 14) of the animals viewed within the first few hours of inoculation. Presumably, these events did not result in sustained colonization, perhaps because these early colonizers are subsequently eliminated (39).
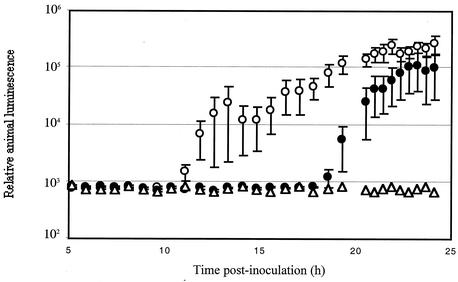
Because a wild-type copy of flrA provided in trans could restore normal motility to the flrA mutant (Fig. 4), we determined whether complementation of motility was enough to restore wild-type levels of colonization. When grown in culture, the luminescence and growth of strain DM126, or DM126 carrying either pVO8 or pDM58, were identical to those of wild-type cells (data not shown). However, when animals were exposed to the flrA mutant strain DM126 carrying pDM58, the complemented mutant was delayed in colonization and was unable to achieve either the levels of luminescence or colonization characteristic of the wild-type strain (Fig. 5 and Table 2). Similarly, the in-frame deletion strain carrying pDM58 was impaired in its ability to colonize (Table 2). In contrast, wild-type strain ES114 carrying either pDM58 or pVO8 colonized animals normally. The complemented mutants were also less effective at initiating colonization; fewer than one half of the animals exposed to the complementing mutant strains became luminous over a 60-h time period (Table 2). In contrast, the reverse-complemented mutant DM132, with its single chromosomal copy of flrA, achieved wild-type levels of colonization (data not shown). These results suggest that colonization by V. fischeri is dependent on either the proper location or copy number of the flrA gene.
FIG. 5.
Colonization of squid by V. fischeri strains as followed by the development of luminescence. Newly hatched juvenile E. scolopes were exposed to seawater containing either no V. fischeri (triangles), ES114 pDM58 (open circles), or DM126 pDM58 (closed circles). Bioluminescence emission was measured over time and represents an average of 10 animals for each treatment. Error bars indicate the standard errors of the means. Similar results were obtained in three separate experiments.
TABLE 2.
Squid colonization by flrA mutant and complementing strains
| Strain | No. of animals | % Colo- nizeda | Symbiotic luminescenceb
|
CFU/light organc (104) | |||
|---|---|---|---|---|---|---|---|
| 12 h | 24 h | 36 h | 48 h | ||||
| ES114 pVO8 | 15 | 100 | 100 | 100 | 100 | 100 | 14 ± 2 |
| DM126 pVO8 | 15 | 0 | <0.1 | <0.1 | <0.1 | <0.1 | <0.001 |
| DM159 pVO8 | 10 | 0 | <0.1 | <0.1 | <0.1 | <0.1 | NDd |
| ES114 pDM58 | 30 | 94 | 100 | 100 | 100 | 100 | 9 ± 1 |
| DM126 pDM58 | 30 | 46 | <0.1 | 34 | 59 | 27 | 0.16 ± 0.5 |
| DM159 pDM58 | 20 | 35 | ND | 27 | ND | 27 | 0.9 ± 0.2 |
Colonized animals were defined as those displaying luminescence at 24 h.
Relative to the levels of the cognate ES114 strain (set at 100).
Colonization levels determined at 48 h postinoculation.
ND, not determined.
Identification of FlrA-regulated genes by RAP-PCR fingerprinting of RNA.
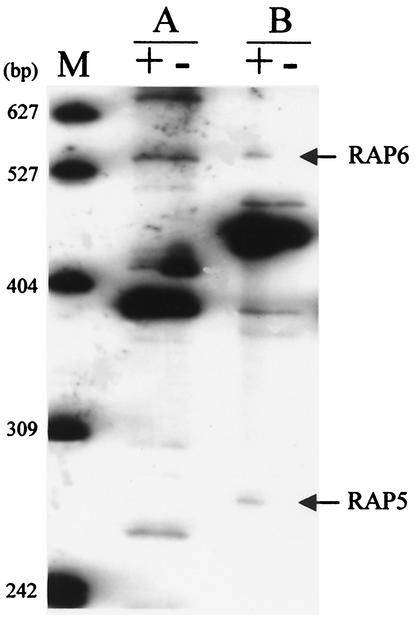
Because we were interested in the possibility that FlrA regulates additional colonization factors, we utilized a differential display approach to search for FlrA-regulated genes. Total RNA isolated from strains ES114 and DM126 grown in the laboratory to the exponential phase was subjected to RAP-PCR. Analyses using six unique primer combinations yielded a total of 69 clearly visible PCR products, 17 of which were differentially expressed. Of these, eight were isolated from the RAP-PCR polyacrylamide gel (e.g., Fig. 6) and subjected to further analysis as described in Materials and Methods. Eight of the differentially expressed transcripts (RAP1 to RAP8) were sequenced and analyzed for homologs in the database (Table 3). To confirm the results obtained with RAP-PCR, we performed quantitative and semiquantitative RT-PCR using gene-specific PCR primers on three of the transcripts. The pattern of FlrA-regulated gene expression matched that seen in the original RAP-PCR analysis (Table 3), although with RAP3 and RAP4 differential expression was found to be low and variable. Examination of the upstream regions of the predicted RAP-PCR products showed no evidence for known regulatory sequences.
FIG. 6.
RAP-PCR fingerprinting comparison of the wild type and the flrA mutant. Total cellular RNA isolated from ES114 (+) or DM126 (−) cells was subjected to RAP-PCR. Shown are results for two different primer pairs (A and B), as well as molecular size markers (M). Arrows indicate differentially expressed bands that subsequently were removed for cloning and sequencing.
TABLE 3.
Products of the V. fischeri FlrA-regulated genes identified by RAP-PCR
| Clone | Product length (bp) | Closest relativea | FlrA+ condition | Fold regulation relative to FlrA+ |
|---|---|---|---|---|
| RAP1, 2 | 731, 1,000 | V. anguillarum flagellin subunit protein | + | >100 |
| RAP3, 4 | 468, 1,000 | V. cholerae topoisomerase | + | 2-4b |
| RAP5, 6 | 280, 557 | V. fischeri halovibrin A | − | 10 |
| RAP7 | 298 | V. cholerae phosphoglycerate kinase | − | NDc |
| RAP8 | 547 | V. cholerae potassium efflux protein | − | ND |
Sequence similarity indexes with P value of <10−10.
In one of four experiments there was no induction.
ND, not determined.
Four products were induced in the presence of FlrA. RAP1 and RAP2 encode polypeptides that are homologous to flagellin subunit proteins (FlaE). RAP3 and RAP4 contained overlapping sequences that are homologous to the topoisomerase III protein, TopB, of V. cholerae (24). In contrast, four other RAP products were induced in the absence of FlrA. RAP5 and RAP6 contained overlapping sequences predicted to encode a homolog of two previously identified halovibrins of V. fischeri (50). RAP7 contained sequence homologous to the amino terminus of the glycolytic enzyme phosphoglycerate kinase, pgk. The RAP7 transcript contained sequence that aligned with approximately 189 nucleotides upstream of the predicted start site for pgk, including a predicted intergenic region and 15 nucleotides of the upstream coding region predicted to be d-erythrose-4-phosphate dehydrogenase. Thus, these ORFs are probably cotranscribed as in other organisms (9). Finally, RAP8 was predicted to encode a polypeptide similar to the glutathione-regulated potassium efflux system protein (KefB) of V. cholerae (24).
DISCUSSION
Studies using nonmotile mutants of V. fischeri have shown that motility is essential to initiate colonization of the squid host E. scolopes; however, the genetic bases for their defects have not been determined (19). In addition, although it is likely that V. fischeri responds to cues within the light-organ environment to modulate flagellum synthesis (45), nothing is known either about these signals or about the mechanisms of flagellar gene transcription in this species. Thus, an investigation of flagellar gene regulation in V. fischeri would enhance our understanding of the biology of their symbiosis.
Flagellar regulation in Vibrio species.
We describe here the cloning, sequencing, and characterization of flrA, a flagellar transcriptional regulator gene in V. fischeri. FlrA belongs to the same subclass of transcriptional regulators as its homolog in V. cholerae, which has been shown to control the expression of flagellar genes transcribed by the alternative sigma factor, σ54 (RpoN). In V. cholerae, transcription of flagellar genes is arranged in a hierarchical fashion comprised of four classes (42). The σ54-dependent transcriptional activator FlrA is the sole class I gene and is required for expression of class II, III, and IV genes, which together encode the flagellar secretion apparatus, flagellin subunits, and regulatory proteins (42). Analysis of the flrA locus in V. fischeri revealed a similar genetic arrangement to that of V. cholerae and V. parahaemolyticus (Fig. 1). In addition, putative σ54 consensus binding sequences within the regulatory regions of flagellar operons were identified (Fig. 3), and transcription of a flagellin gene (flaA) by FlrA is dependent on the presence of σ54 (K. L. Visick and D. S. Millikan, unpublished data). Furthermore, the flrA mutants generated in this study, as well as an rpoN mutant (A. J. Wolfe, E. J. Simel, and K. L. Visick, Abstr. Gen. Meet. Am. Soc. Microbiol. 1999, p. 388, 1999), are nonmotile. Thus, it is likely that V. fischeri motility is governed by a regulatory hierarchy similar to that described for V. cholerae and V. parahaemolyticus.
Interestingly, in V. fischeri, unlike V. cholerae and V. parahaemolyticus, loss of flrA function results in a completely nonmotile phenotype (Fig. 4A). An in-frame deletion mutant of flrA in V. cholerae has been reported to display an unusual behavior; after prolonged incubation in soft agar, a small number of ΔflrA cells moved from the point of inoculation and produced a star-like pattern (29). Similarly, insertional disruptions of the flrA homolog in V. parahaemolyticus at the position of the sequence encoding either amino acid 37 or 216 resulted in a slow-motility phenotype (52). In contrast, while they contained deletions, or insertions at positions similar to those of the V. parahaemolyticus mutants, the flrA mutants described in this study (Table 1) were completely nonmotile. One might hypothesize that there is read-through transcription of the downstream class II genes flrBC or flaLM in the V. cholerae and V. parahaemolyticus mutants (33). However, this notion is inconsistent with the most recently published model of regulation in V. cholerae, in which FlrA and σ54 together coordinate transcription of additional required structural components independently of FlrBC (42). Further studies are necessary to more clearly define the flagellar regulon in V. fischeri and to compare it to that of other Vibrio species.
Host colonization by flrA mutants.
In this study, we present evidence that at least some of the cells populating the light organ remain motile well beyond the initiation of the association. Perhaps having subpopulations involved in motility behavior confers a symbiotic advantage on V. fischeri. In certain pathogenic bacteria, motility must be switched off in order for the organism to express virulence genes and cause disease (11, 47), while other bacteria in persistent associations continue to elaborate flagella throughout colonization (13). It would be interesting to determine if either of these mechanisms plays a role in the symbiotic light-organ association described here. From our studies, the motility regulator FlrA in V. fischeri controls the expression of genes that are seemingly unrelated to motility (Table 3). Future work will address whether these genes encode proteins required for symbiotic colonization and whether colonization is dependent on the repression of flagellar synthesis.
The observation that flrA mutants were unable to initiate colonization of juvenile squids was not surprising, because motility is essential for colonization (19). However, it was unexpected that complemented mutant strains were unable to maintain a normal persistent association. Unlike results from complementation of mutations in other genes (12, 21, 55, 59), both the flrA-complemented insertion and in-frame deletion mutants were less effective at colonizing the squid light organ. That is, when animals were exposed to either of the complemented mutant strains, fewer became luminous, and those that did became colonized to only 2% of the level of animals inoculated with the wild-type strain carrying the complementing plasmid (Table 2). In contrast, motility of the complemented mutant strains in culture was comparable to that of the wild-type strain (Fig. 4A). These results suggest several possible explanations: (i) proper expression of a particular motility behavior, undetectable in our in vitro assay, is necessary for normal colonization; (ii) the presence of multiple copies of flrA, without a functional copy in the chromosome, results in expression of the flagellar regulon under inappropriate conditions; or (iii) FlrA regulation of additional colonization factors is adversely affected by multiple copies of the flrA gene in trans. Unlike previous studies in which an increase in flagellum number contributed to a defect in V. fischeri colonization (36), the complemented flrA mutant strain exhibited a similar number of flagella to the wild type. Furthermore, whereas these hyperflagellated strains continued to overexpress flagella even when grown on a solid surface (36), the complemented flrA mutant apparently repressed flagellar elaboration to the same degree as the wild-type strain. In this report, we demonstrate that FlrA regulates the expression of genes that are seemingly unrelated to motility (Table 3). It would be interesting to determine whether these gene products are required for symbiotic colonization and whether their expression is sensitive to flrA gene dosage or location. In any case, it is apparent that symbiotic colonization by V. fischeri is dependent on the proper expression of the flagellar regulon.
In V. cholerae, both flrA and flrC mutants were defective for colonization to a greater extent than a nonmotile flaA (encoding an essential flagellin subunit) strain, suggesting that FlrA and/or FlrC are responsible for the expression of additional, unknown colonization genes in this organism (10). However, it is not possible to make such a direct comparison between upstream (flrA) and downstream motility mutants in V. fischeri, because motility is an absolute requirement for its colonization behavior. Nevertheless, by using inducible constructs (58) to complement these mutants, we hope to evaluate whether expression of these genes is important after colonization has been initiated. Such studies may indicate that the FlrA-regulated genes identified in this study play a central role in squid colonization.
FlrA-regulated genes in V. fischeri.
We used RAP-PCR to identify a subset of differentially expressed genes in V. fischeri. As predicted, this approach identified a transcript containing sequence similarity to genes encoding flagellin subunits. Additional FlrA-regulated genes known to be required for motility are likely to be identified with a greater number of primer combinations. Our results not only provide evidence of differentially expressed genes in the flagellar regulon but also suggest useful candidates for positive controls in future microarray expression studies. Because FlrA is located at the top of the flagellar hierarchy of gene transcription in Vibrio species, it is possible that some of the genes identified in this study are directly controlled by FlrC or other downstream regulators whose expression is modulated by the presence of FlrA.
Two of the differentially expressed fragments identified in this study are similar at the amino acid level to the DNA-relaxing enzyme TopB (Table 3). In E. coli, the NtrC-activator protein-binding site can be substituted with an element containing an intrinsic supercoil structure (8). Thus, the expression of flagellar genes by NtrC-like regulators (e.g., FlrA) might depend on the supercoil state of some promoters. For example, transcription of the flaB flagellin of Helicobacter pylori is sensitive to DNA supercoiling and may be coordinately regulated with the topA gene (48, 53). Furthermore, FlgR, the FlrA homolog in H. pylori, is cotranscribed with gyrA, the gene encoding subunit A of DNA gyrase, suggesting that genes whose products control DNA topology are coordinately regulated with flagellar genes in this organism as well.
Another differentially expressed transcript in the V. fischeri flrA mutant encodes a polypeptide (here designated HvnC) whose closest homolog in GenBank is the halovibrin protein HvnA of V. fischeri. It has been speculated that this protein and its ortholog HvnB act as signaling molecules (43, 44). A subsequent study determined that HvnA and HvnB are secreted NAD+ glycohydrolases (NADases) that cleave NAD+, producing free, reactive ADP-ribose (50). Cell-free culture supernatants of an hvnA hvnB double mutant in V. fischeri lack any detectable NADase activity (50); thus, the discovery of this third halovibrin-like gene was unexpected, and its protein product apparently either displays no detectable NADase activity or is expressed at very low levels under laboratory culture conditions. Furthermore, whereas the double mutant was capable of infecting squids normally, it is interesting to speculate that perhaps HvnC is important during animal colonization. Interestingly, we have identified hvnC transcript from RNA extracted from colonized animals at 18 h postinoculation (data not shown), suggesting that hvnC is transcribed by bacterial cells at least within the light-organ environment.
In conclusion, we have identified a transcriptional regulator of flagellar synthesis in V. fischeri and have shown that it is required for the expression of motility. Furthermore, by comparing expression patterns of the wild-type and flrA mutant strains, we have discovered additional FlrA-regulated genes. Future studies will focus on determining the role of these proteins in light-organ colonization and symbiotic development by V. fischeri.
Acknowledgments
We thank K. Bidle and D. Bartlett for technical assistance with RAP-PCR, E. Stabb, J. McCann, and C. Lupp for helpful discussions, and A. Schaefer and C. Whistler for insightful comments on the manuscript. In its later stages, this work was aided by data provided by the Vibrio fischeri genome project (http://ergo.integratedgenomics.com/Genomes/VFI), with the support of the W. M. Keck Foundation.
This work was supported by National Institutes of Health grant RR12294 to E.G.R. and M. McFall-Ngai and National Science Foundation grant IBN9904601 to M. McFall-Ngai and E.G.R. D.S.M. was supported by a National Science Foundation Postdoctoral Fellowship in Microbial Biology.
REFERENCES
- 1.Aeckersberg, F., C. Lupp, B. Feliciano, and E. G. Ruby. 2001. Vibrio fischeri outer membrane protein OmpU plays a role in normal symbiotic colonization. J. Bacteriol. 183:6590-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Arora, S. K., B. W. Ritchings, E. C. Almira, S. Lory, and R. Ramphal. 1997. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J. Bacteriol. 179:5574-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auvrey, J. 2001. Flagellin polymerisation control by a cytosolic export chaperone. J. Mol. Biol. 308:221-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrios, H., B. Valderrama, and E. Morett. 1999. Compilation and analysis of σ54-dependent promoter sequences. Nucleic Acids Res. 27:4305-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bidle, K. A., and D. H. Bartlett. 2001. RNA arbitrarily primed PCR survey of genes regulated by ToxR in the deep-sea bacterium Photobacterium profundum strain SS9. J. Bacteriol. 183:1688-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boettcher, K. J., and E. G. Ruby. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 172:3701-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brahms, G., S. Brahms, and B. Magasanik. 1995. A sequence-induced superhelical DNA segment serves as transcriptional enhancer. J. Mol. Biol. 246:35-42. [DOI] [PubMed] [Google Scholar]
- 9.Carroll, P. A., G. Zhao, S. A. Boyko, M. E. Winkler, and S. B. Calderwood. 1997. Identification, sequencing, and enzymatic activity of the erythrose-4-phosphate dehydrogenase gene of Vibrio cholerae. J. Bacteriol. 179:293-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Correa, N., C. Lauriano, R. McGee, and K. Klose. 2000. Phosphorylation of the flagellar regulatory protein FlrC is necessary for Vibrio cholerae motility and enhanced colonization. Mol. Microbiol. 35:743-755. [DOI] [PubMed] [Google Scholar]
- 11.Cotter, P. A., and J. F. Miller. 1994. BvgAS-mediated signal transduction: analysis of phase-locked regulatory mutants of Bordetella bronchiseptica in a rabbit model. Infect. Immun. 62:3381-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeLoney, C. R., T. M. Bartley, and K. L. Visick. 2002. Role for phosphoglucomutase in Vibrio fischeri-Euprymna scolopes symbiosis. J. Bacteriol. 184:5121-5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eaton, K. A., D. R. Morgan, and S. Krakowka. 1992. Motility as a factor in the colonization of gnotobiotic piglets by Helicobacteri pylori. J. Med. Microbiol. 37:123-127. [DOI] [PubMed] [Google Scholar]
- 14.Foster, J. S., M. A. Apicella, and M. J. McFall-Ngai. 2000. Vibrio fischeri lipopolysaccharide induces developmental apoptosis, but not complete morphogenesis, of the Euprymna scolopes symbiotic light organ. Dev. Biol. 226:242-254. [DOI] [PubMed] [Google Scholar]
- 15.Ghelardi, E., F. Celandroni, S. Salvetti, D. J. Beecher, M. Gominet, D. Lereclus, A. C. L. Wong, and S. Senesi. 2002. Requirement of flhA for swarming differentiation, flagellin export, and secretion of virulence-associated proteins in Bacillus thuringiensis. J. Bacteriol. 184:6424-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Givaudan, A., and A. Lanois. 2000. flhDC, the flagellar master operon of Xenorhabdus nematophilus: requirement for motility, lipolysis, extracellular hemolysis, and full virulence in insects. J. Bacteriol. 182:107-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Givskov, M., L. Eberl, G. Christiansen, M. J. Benedik, and S. Molin. 1995. Induction of phospholipase and flagellar synthesis in Serratia liquefaciens is controlled by expression of the flagellar master operon flhD. Mol. Microbiol. 15:445-454. [DOI] [PubMed] [Google Scholar]
- 18.Goodier, R. I., and B. M. Ahmer. 2001. SirA orthologs affect both motility and virulence. J. Bacteriol. 183:2249-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graf, J., P. V. Dunlap, and E. G. Ruby. 1994. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J. Bacteriol. 176:6986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graf, J., and E. G. Ruby. 1998. Host-derived amino acids support the proliferation of symbiotic bacteria. Proc. Natl. Acad. Sci. USA 95:1818-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graf, J., and E. G. Ruby. 2000. Novel effects of a transposon insertion in the Vibrio fischeri glnD gene: defects in iron uptake and symbiotic persistence in addition to nitrogen utilization. Mol. Microbiol. 37:168-179. [DOI] [PubMed] [Google Scholar]
- 22.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 23.Hawley, D. K., and W. R. McClure. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 11:2237-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragol, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helmann, J. D., and M. J. Chamberlin. 1987. DNA sequence analysis suggests that expression of flagellar and chemotaxis genes in Escherichia coli and Salmonella typhimurium is controlled by an alternative sigma factor. Proc. Natl. Acad. Sci. USA 84:6422-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoch, J. A., and T. J. Silhavy. 1995. Two-component signal transduction. ASM Press, Washington, D.C.
- 28.Josenhans, C., and S. Suerbaum. 2002. The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 291:605-614. [DOI] [PubMed] [Google Scholar]
- 29.Klose, K. E., and J. J. Mekalanos. 1998. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol. Microbiol. 28:501-520. [DOI] [PubMed] [Google Scholar]
- 30.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 31.Liu, X., D. L. Brutlag, and J. S. Liu. 2001. BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac. Symp. Biocomput. 2001:127-138. [PubMed] [Google Scholar]
- 32.Macnab, R. M. 1996. Flagella and motility, p.123-145. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 33.McCarter, L. L. 2001. Polar flagellar motility of the Vibrionaceae. Microbiol. Mol. Biol. Rev. 65:445-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McFall-Ngai, M. J. 1999. Consequences of evolving with bacterial symbionts: insights from the squid-vibrio associations. Annu. Rev. Ecol. Syst. 30:235-256. [Google Scholar]
- 35.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Millikan, D. S., and E. G. Ruby. 2002. Alterations in Vibrio fischeri motility correlate with a delay in symbiosis initiation and are associated with additional symbiotic colonization defects. Appl. Environ. Microbiol. 68:2519-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montgomery, M., and M. McFall-Ngai. 1994. Bacterial symbionts induce host organ morphogenesis during early post embryonic development of the squid Euprymna scolopes. Development 120:1719-1729. [DOI] [PubMed] [Google Scholar]
- 38.Montgomery, M. K., and M. J. McFall-Ngai. 1993. Embryonic development of the light organ of the sepiolid squid Euprymna scolopes Berry. Biol. Bull. 184:296-308. [DOI] [PubMed] [Google Scholar]
- 39.Nyholm, S. V., B. Deplancke, H. R. Gaskins, M. A. Apicella, and M. J. McFall-Ngai. 2002. Roles of Vibrio fischeri and nonsymbiotic bacteria in the dynamics of mucus secretion during symbiont colonization of the Euprymna scolopes light organ. Appl. Environ. Microbiol. 68:5113-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nyholm, S. V., E. V. Stabb, E. G. Ruby, and M. J. McFall-Ngai. 2000. Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc. Natl. Acad. Sci. USA 97:10231-10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ottemann, K. M., and J. F. Miller. 1997. Roles for motility in bacterial-host interactions. Mol. Microbiol. 24:1109-1117. [DOI] [PubMed] [Google Scholar]
- 42.Prouty, M. G., N. E. Correa, and K. E. Klose. 2001. The novel σ54- and σ28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 39:1595-1609. [DOI] [PubMed] [Google Scholar]
- 43.Reich, K. A., T. Beigel, and G. K. Schoolnik. 1997. The light organ symbiont Vibrio fischeri possesses two distinct secreted ADP-ribosyltransferases. J. Bacteriol. 179:1591-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reich, K. A., and G. K. Schoolnik. 1996. Halovibrin, secreted from the light organ symbiont Vibrio fischeri, is a member of a new class of ADP-ribosyltransferases. J. Bacteriol. 178:209-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruby, E. G., and L. M. Asato. 1993. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch. Microbiol. 159:160-167. [DOI] [PubMed] [Google Scholar]
- 46.Schmiel, D. H., G. M. Young, and V. L. Miller. 2000. The Yersinia enterocolitica phospholipase gene yplA is part of the flagellar regulon. J. Bacteriol. 182:2314-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmitt, C. K., S. C. Darnell, V. L. Tesh, B. A. Stocker, and A. D. O'Brien. 1994. Mutation of flgM attenuates virulence of Salmonella typhimurium, and mutation of fliA represses the attenuated phenotype. J. Bacteriol. 176:368-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spohn, G., and V. Scarlato. 1999. Motility of Helicobacter pylori is coordinately regulated by the transcriptional activator FlgR, an NtrC homolog. J. Bacteriol. 181:593-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stabb, E., K. Visick, D. S. Millikan, A. A. Corcoran, L. Gilson, S. V. Nyholm, M. McFall-Ngai, and E. Ruby. 2001. The Vibrio fischeri-Euprymna scolopes symbiosis: a model marine animal-bacteria interaction, p. 269-277. In N. Saxena (ed.), Recent advances in marine science and technology. Pacon International, Honolulu, Hawaii.
- 50.Stabb, E. V., K. A. Reich, and E. G. Ruby. 2001. Vibrio fischeri genes hvnA and hvnB encode secreted NAD+-glycohydrolases. J. Bacteriol. 183:309-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stabb, E. V., and E. G. Ruby. 2003. New RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 358:413-426. [DOI] [PubMed] [Google Scholar]
- 52.Stewart, B. J., and L. L. McCarter. 1996. Vibrio parahaemolyticus FlaJ, a homologue of FliS, is required for production of a flagellin. Mol. Microbiol. 20:137-149. [DOI] [PubMed] [Google Scholar]
- 53.Suerbaum, S., T. Brauer-Steppkes, A. Labigne, B. Cameron, and K. Drlica. 1998. Topoisomerase I of Helicobacter pylori: juxtaposition with a flagellin gene (flaB) and functional requirement of a fourth zinc finger motif. Gene 210:151-161. [DOI] [PubMed] [Google Scholar]
- 54.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Visick, K. L., J. Foster, J. Doino, M. McFall-Ngai, and E. G. Ruby. 2000. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J. Bacteriol. 182:4578-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Visick, K. L., and E. G. Ruby. 1997. New genetic tools for use in the marine bioluminescent bacterium Vibrio fischeri, p. 119-122. In J. W. Hastings, L. J. Kricka, and P. E. Stanley (ed.), Bioluminescence and chemiluminescence. World Scientific Publishing, River Edge, N.J.
- 57.Visick, K. L., and E. G. Ruby. 1998. The periplasmic, group III catalase of Vibrio fischeri is required for normal symbiotic competence and is induced both by oxidative stress and by approach to stationary phase. J. Bacteriol. 180:2087-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Visick, K. L., and E. G. Ruby. 1998. Temporal control of lux gene expression in the symbiosis between Vibrio fischeri and its squid host, p. 277-279. In Y. Le Gal and H. O. Halvorson (ed.), New developments in marine biotechnology. Plenum Press, New York, N.Y.
- 59.Visick, K. L., and L. M. Skoufos. 2001. Two-component sensor required for normal symbiotic colonization of Euprymna scolopes by Vibrio fischeri. J. Bacteriol. 183:835-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu, J., and A. Newton. 1997. Regulation of the Caulobacter flagellar gene hierarchy, not just for motility. Mol. Microbiol. 24:233-239. [DOI] [PubMed] [Google Scholar]
- 61.Young, G. M., D. H. Schmiel, and V. L. Miller. 1999. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc. Natl. Acad. Sci. USA 96:6456-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]