Abstract
The primary DNA processing protein for conjugative mobilization of the plasmid R1162 is the transesterase MobA, which acts at a unique site on the plasmid, the origin of transfer (oriT). Both MobA and oriT are members of a large family of related elements that are widely distributed among bacteria. Each oriT consists of a highly conserved core and an adjacent region that is required for binding by its cognate MobA. The sequence of the adjacent region is important in determining the specificity of the interaction between the Mob protein and the oriT DNA. However, the R1162 MobA is active on the oriT of pSC101, another naturally occurring plasmid. We show here that MobA can recognize oriTs having different sequences in the adjacent region and, with varying frequencies, can cleave these oriTs at the correct position within the core. Along with the structure of the oriTs themselves, these characteristics suggest a model for the evolution of this group of transfer systems.
R1162 is a small, broad-host-range plasmid that can efficiently utilize the conjugative pore encoded by self-transmissible, IncP-1 plasmids, such as RK2 and R751 (20). Transfer requires a cis-acting site, the 38-bp origin of transfer (oriT) (Fig. 1), as well as three R1162-encoded proteins (7, 8, 11). The most important of these is MobA, which cleaves one of the DNA strands at nic within oriT (5). MobA and two accessory proteins assemble at oriT to form the relaxosome (18). In order for MobA to bind properly to oriT DNA, base pairing within the AT-rich region must be significantly disrupted, with adjacent DNA, making up the inner arm of the inverted repeat (Fig. 1), remaining in duplex form. Within the relaxosome, strand separation extending from the AT-rich region is initiated by the interaction of the DNA with the Mob proteins (23). The extension of this disruption through nic is required for strand cleavage, a transesterification in which MobA becomes covalently joined to the 5′ end of one strand by a tyrosyl phosphodiester linkage (18). After transfer of the cleaved strand, the circular plasmid is reformed and the protein is released by a second transesterification, in which the 3′-OH end of the DNA is the entering nucleophile. In this reaction, the AT-rich region and cleavage site are single stranded, so localized disruption of duplex DNA is not required, and base pairing between the arms of the inverted repeat restores sufficient duplex character to the adjacent DNA (6). Thus, the outer arm of the inverted repeat is required for termination of transfer, but not for initiation (14).
FIG. 1.
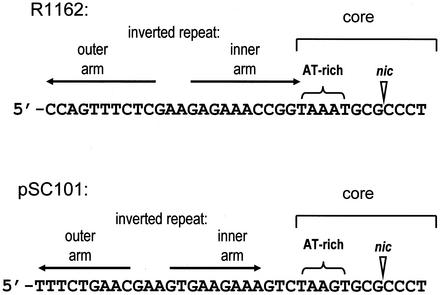
The oriTs of R1162 and pSC101, showing for each the location of the cleavage site (nic) (reference 8 and unpublished results), the core DNA, the AT-rich region, and the inverted repeats (horizontal arrows).
Overall, the plasmid pSC101 is unrelated to R1162, but the two encode very similar MobA proteins (15). The oriTs of these plasmids have a 12-bp region, containing nic and the AT-rich DNA, with almost identical base sequences (Fig. 1). Since this region is highly conserved in the oriTs of other plasmids as well (below), we call it the oriT core. In contrast, the sequence and size of the DNA making up the inverted repeat are significantly different. Despite these differences, the R1162 Mob proteins are active on the pSC101 oriT, whereas the pSC101 proteins are unable to process the R1162 oriT for transfer (15). These results indicate that, although the two relaxases are matched with their own oriTs, there is sufficient flexibility to allow an effective but nonreciprocal interaction between a Mob protein and a noncognate oriT. In particular, inverted repeat DNA with a different sequence is tolerated by the R1162 MobA. This observation has prompted us to ask whether there is relaxed sequence specificity in the binding of the R1162 MobA to the DNA making up the inverted repeat region of oriT. Our results, along with other observations about related proteins and their putative oriTs, suggest a model for the evolution of the R1162 family of mobilization systems.
MATERIALS AND METHODS
Plasmids, strains, and bacterial mating.
Plasmid DNA was isolated from MV10 (13), a derivative of the Escherichia coli K-12 strain C600, or DH5α (Invitrogen). R1162ΔmobA contains a 525-bp, in-frame deletion in mobA. It was derived by digesting R1162 DNA with PflMI. The plasmid R1162ΔoriT contains a 48-bp deletion that removes oriT but not the adjacent promoters (16).
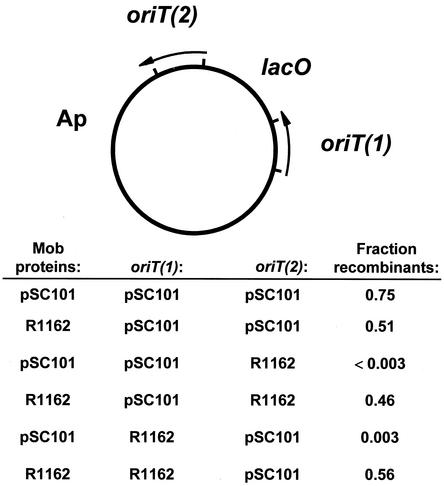
Bacteria were mated on semisolid medium as described previously (4). The recipient strain was a C600 derivative resistant to nalidixic acid. Transconjugant colonies were selected on medium containing nalidixic acid (25 μg/ml), ampicillin (100 μg/ml), and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (0.008%). In addition to the test plasmid (Fig. 2), donor strains contained the mobilizing plasmid R751 (20) and either R1162 or pUT1621 as a source of Mob proteins. R1162 is naturally occurring; pUT1621 consists of the mob genes of pSC101 cloned in the vector pACYC184 (15).
FIG. 2.
Test for initiation and termination of transfer in the presence of the pSC101 or R1162 Mob proteins. The general structure of the test plasmid is shown at top, with the direction of transfer indicated by the arrows. Transconjugant colonies were scored for blue or white color on selective medium containing X-Gal. The white colonies contained recombinant plasmids due to initiation of transfer at oriT(1) and termination at oriT(2). In each case, the fraction of these was determined for two independent experiments (approximately 300 to 400 colonies scored for color), and the average value is reported.
Construction of a library of oriTs containing different sequences for the inner arm of the inverted-S repeat.
The partially degenerate oligonucleotide GCCCAAGCTTATGGAAGAA(N)10TAAATGCGCCCTGCCCTTTTGGCAATTGGGCCC (25% of each base in the degenerate region) was amplified with the primers GCCCAAGCTTATGGAA and GGGCCCAATTGCCAAAAG. The product was then digested with HindIII and MfeI and cloned into a pBR322 derivative (2) by replacement of a HindIII-MfeI lacO fragment. After transformation of MV10 containing either R1162ΔoriT or R1162ΔmobA, dilutions of the cells were plated on medium containing ampicillin and X-Gal to estimate the number of transformed cells and to determine the fraction of these which formed white colonies and were therefore likely to contain a recombinant plasmid with a cloned oriT. There were about 20,000 transformed cells for each recipient, and about 95% of these contained recombinant plasmids. In each case, the pooled transformants were grown to mid-log phase in 100 ml of 170 tryptone-0.5% yeast extract-0.5% NaCl containing 100 μg of ampicillin/ml and 25 μg of streptomycin/ml, and the plasmid DNA was isolated from cells by the cleared lysate method of Clewell and Helinski (9), modified as described previously (23). The DNA was dissolved in 10 μl of 50 mM Tris HCl, 5 mM EDTA, pH 8.
For tailing with T4 polydeoxynucleotidyl terminal transferase, 1 μl of the cleared lysate DNA was added to 39 μl of H2O, boiled for 3 min, and then quickly cooled in ice water. The reaction mixture consisted of this DNA made up to 50 μl of a solution (approximate pH 7.9) containing 20 mM Tris-acetate, 50 mM potassium acetate, 10 mM magnesium acetate, 1 mM dithiothreitol, 0.25 mM cobalt chloride, 0.4 mM dGTP, and 10 U of terminal transferase. Incubation was at 37°C for 30 min and then 70°C for 10 min. The DNA was precipitated and then amplified with the primers GGAAATGTTGAATACTCATACTCTTC (complementary to the vector) and CCCGAATTCCCCCCCCCC (partially complementary to the tailed DNA and introducing an EcoRI site). The amplified product was digested with HindIII and EcoRI and cloned into pUC19 (22). We transformed this DNA into E. coli strain DH5α; colonies containing recombinant molecules were identified by plating on medium containing 0.008% X-Gal and 0.16 mM isopropyl-β-d-thiogalactopyranoside. Plasmid DNA was then prepared from individual transformants for DNA sequencing by the facility at the University of Texas.
MobA protein-DNA binding assay.
The R1162 MobA protein is covalently joined at the carboxy-terminal end to the plasmid primase (19). The part of the protein required for mobilization was purified by affinity chromatography as previously described (3). The pSC101 MobA was prepared by the same method except that the entire protein was purified, since it is not naturally fused to another protein. The oligonucleotides used in the binding assay were TTCTGAACGAAGTGAAGAAAGTCGAAGTGCGCCCTGATTTTTGGGAATTC(top strand) and TTCTTCACTTCGTTCAGAAACGTGCGCCCTTCATTTTGGGAATTC (bottom strand). Samples consisted of 20 pmol of 5′-32P-end-labeled oligonucleotide and 23 pmol (pSC101 MobA) or 28 pmol (R1162 MobA) of purified protein together in 40 μl of buffer (50 mM Tris [pH 8], 50 mM NaCl, 0.5 mM EDTA, 15% glycerol). After several minutes at room temperature, the samples were loaded on a 10% polyacrylamide gel and the bands were visualized by autoradiography after electrophoresis.
Primer extension to detect nicking within the relaxosome.
DNA was prepared from cleared lysates as described above. One-tenth of this DNA was digested with AatII, which cleaves the DNA at a single site, downstream from the nick site in the direction of primer extension. The digested DNA was mixed with 20 pmol of primer and 10 nmol of each deoxynucleoside triphosphate in 20 μl of Qiagen PCR buffer. Primer DNA was extended by incubation with 2.5 U of Taq DNA polymerase for 35 thermal cycles (each cycle consisting of 1 min at 94, 58, and 72°C). The samples were then mixed with 5 μl of running dye containing 95% formamide and boiled for 5 min. Radioactive fragments were separated by electrophoresis through an 8% polyacrylamide-urea gel and were visualized by autoradiography.
RESULTS AND DISCUSSION
The MobA of R1162 can form a functional relaxosome with oriTs having inner arms with many different base sequences.
The core sequences of the R1162 and pSC101 oriTs are the same except for a single base difference (Fig. 1). Indeed, the core is identical for pSC101 and RSF1010, a plasmid virtually identical to R1162 (19). It is therefore the remaining oriT DNA, which makes up the inverted repeat, that determines the specificity of the interaction with the Mob proteins. Since the R1162 mobilization proteins are functional with the pSC101 oriT, they can recognize the inverted repeat DNA of this oriT. One possibility is that the structure of the (imperfect) hairpin loop formed by the inverted repeat determines specificity, because it is required for termination of a round of transfer. The structure has some importance, because in a MobA-based phage recombination assay single base mutations in one arm of the inverted repeat are suppressed by second-site mutations restoring base complementarity within the hairpin loop (4). To test this possibility, we examined transfer-dependent recombination for plasmids containing two directly repeated copies of oriT. Transfer of the intact plasmid results in the formation of blue transconjugant colonies on media containing X-Gal, due to titration of the lac repressor in the recipient (4). However, when transfer is initiated from oriT(1) and terminated at oriT(2), the colonies are white. No recombinants were formed in the presence of the pSC101 Mob proteins when the R1162 oriT was present at either the initiating or terminating position (Fig. 2). Thus, the sequence of the inner arm DNA, either as duplex or as formed by the inverted repeat, is in itself an important factor in determining MobA specificity.
How much variation in the sequence of the oriT inner arm is tolerated by the R1162 MobA? We amplified by PCR an oligonucleotide containing the oriT core sequence but degenerate for the inner arm of the inverted repeat (see Materials and Methods). We constructed a collection of plasmids having oriTs with different base sequences for the inner arm by cloning the PCR product into a derivative of pBR322. We could not simply test the resulting plasmids for mobilization, since the oriTs in general would not form an inverted repeat and thus would not terminate correctly. Instead, we identified those plasmids with oriTs that could be cleaved at nic in the relaxosome.
The plasmid library was used to transform the E. coli strains MV10 (R1162ΔoriT) and MV10 (R1162ΔmobA). We then pooled the transformants in each case, cultured these cells, and prepared plasmid DNA by a method that allows the isolation of both nicked and supercoiled molecules (23). The DNA was denatured, tailed with terminal deoxynucleotidyl transferase and dGTP, and amplified by PCR (Fig. 3). Molecules cleaved at the nick site will result in an amplified fragment of approximately 300 bp, depending on the size of the tail. In fact, the fragments will all be nearly the same size, since the repeated cycles of synthesis can result in shortening but not elongation of the poly(dG) extension. Randomly located nicks will also be tailed, but after PCR these will result in fragments of different sizes rather than a discrete band.
FIG. 3.
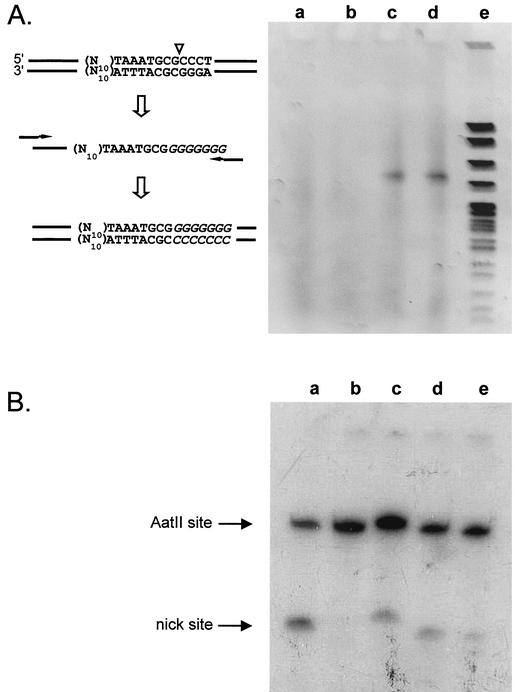
(A) Cloned oriT DNA tailed with T4 polydeoxynucleotidyl transferase, amplified by PCR, and displayed on a 5% polyacrylamide gel. The strategy for amplification and cloning is shown on the left. N10 designates the region with variable base sequence. Plasmid DNA for tailing was isolated from pooled transformants of cells containing R1162 ΔmobA (lanes a and b) or R1162 ΔoriT (lanes c and d). Marker (lane e) is 0.5 μg of MspI-digested pBR322 DNA. (B) Primer extension to nick site for plasmids containing cloned oriT DNA with the inner arm base sequences (see Table 1) for wild type (a), 16-1A (b), 2B-13 (c), 2A-17 (d), and 2B-20 (e). The bands in panel c were delayed in entering the gel and, thus, appear to migrate more slowly, because urea in the well was not removed completely before loading the sample.
A PCR product consisting of approximately 300-bp fragments was obtained when plasmids from MV10 (R1162ΔoriT) were used as template in the reaction (Fig. 3A, lanes c and d). No distinct bands were observed for plasmids from the MV10 (R1162ΔmobA) strain (Fig. 3B, lanes a and b). Thus, the amplified band is the result of nicking at oriT. We cloned the PCR product and then sequenced the oriT DNA in several of the recombinant molecules. We could determine unambiguously from the sequence whether the poly(dG) tail originated from nic or from some other site in the DNA. Although about a third of the molecules had poly(dG) tails originating at unexpected positions within the oligonucleotide, probably the result of extension from randomly nicked strands, most were located at the nick site. Moreover, the sequence of the inner arm DNA varied significantly for different clones (Table 1). The variation was not the same at each position in the arm; in particular, the bases at positions 4, 6, 9, and 10 were predominantly A, A, G, and G, respectively. We also determined the sequence of the cloned DNA for eight, randomly chosen colonies from the original library, prior to enrichment of the nicked population by tailing and PCR (Table 1). These sequences showed variation in the bases at positions 4, 6, 9, and 10. Thus, the conserved bases in the tailed population reflect the base sequences that allow nicking within the relaxosome. Interestingly, positions 4 and 6 are within the sequence AGAAA that is conserved for both the pSC101 and R1162 oriTs. The bases adjacent to the core, at positions 9 and 10, also seem to be important, but on the other hand they are not the same for the two oriTs.
TABLE 1.
Inner arm base sequences of a cloned, partially degenerate oriT oligonucleotide
| Strain | Sequence in degenerate region at position:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| Tailed at nic site | ||||||||||
| wt | G | A | G | A | A | A | C | C | G | G |
| 16-1A | C | T | A | A | T | A | C | G | A | G |
| 2B-6 | T | C | C | G | T | A | G | G | G | G |
| 1B-7 | C | A | A | A | A | A | G | G | G | G |
| 1B-16 | T | G | A | A | A | A | C | A | G | G |
| 2A-17 | T | G | G | A | T | A | C | C | G | G |
| 2B-13 | G | A | G | A | A | A | G | G | G | G |
| 2B-20 | G | T | C | A | A | A | G | C | G | G |
| Chosen randomly without tailing | ||||||||||
| CN-1 | G | C | A | A | G | G | A | T | C | G |
| CN-2 | C | A | C | A | G | T | T | G | T | A |
| CN-3 | A | T | T | T | G | G | T | G | G | A |
| CN-4 | C | G | G | A | C | A | A | A | C | G |
| CN-5 | T | T | G | A | C | G | G | A | C | G |
| CN-6 | G | G | A | G | C | A | G | A | T | A |
| CN-7 | T | G | C | C | A | T | C | C | T | G |
| CN-8 | T | T | A | A | T | G | G | A | C | C |
Although these results do not indicate the efficiency of nicking in each case, they do suggest that oriTs with a variety of different sequences for the inner arm DNA are potential targets for nicking by the relaxosome. To show that these sequences in fact support an active relaxosome, we cloned oriT DNA fragments similar to those in the degenerate library but having one of the putative active sequences for the inner arm. Cleared lysates were made as before, and then nicking was assayed by primer extension. Nicking was detected in all but one case (Fig. 3B), although as expected the efficiency of this cleavage varied. We conclude that different sequences for the inner arm can support cleavage within oriT.
The oriTs in the R1162 mobilization family have a highly conserved core, but very different inverted repeats.
Naturally occurring plasmids encoding a protein similar to MobA, and also having a putative oriT similar to that of R1162, have been obtained from different bacteria. A sample derived from a search of GenBank and selected to indicate the diversity and range of the group is shown in Table 2 . The core sequence within the oriT-like DNA is highly conserved, but the inverted repeat next to the core, while always identifiable, varies considerably in sequence, size, and potential folding structure. In addition, the distance between these putative oriTs and the gene encoding the MobA-like protein varies from 49 to 264 bp. While some of the inverted repeats are obviously related (for example, those of pDN1 and RSF1010), it seemed likely that others had originated independently, despite the high degree of conservation of the core DNA. We supposed that the inverted repeat region formed on several different occasions in unrelated DNA by duplication and inversion of DNA containing the core. For some of these plasmids, a closer examination of the sequences adjacent to the oriTs supports this idea.
TABLE 2.
Known and putative oriTs
| Organismc | Plasmid | Inverted repeat | Core | Similarity to R1162 MobA (E value)b | Core-mobA separation (bp) |
|---|---|---|---|---|---|
| Plasmids with known or putative oriTs and associated relaxase | |||||
| E. coli | R1162a | CCAGTTTCTCGAAGAGAAACCGG | TAAATGCGCCCT | 102 | |
| S. enterica serovar Panama | pSC101 | TTTCTGAACGAAGTGAAGAAAGTC | TAAGTGCGCCCT | 2e − 35 | 52 |
| D. nodosus | pDN1 | GTTTCTTGAAGAGAAACCGG | TAAGTGCGCCCT | 2e − 127 | 85 |
| Unknown | pIE1115 | GTTTCTTGAAGAGAAACCGG | TAAGTGCGCCCT | 2e − 128 | 84 |
| Unknown | pIE1130 | CCAGTTTCTCGAAGAGAAACCGG | TAAGTGCGCCCT | 2e − 143 | 85 |
| A. ferrooxidans | pTF1 | GGGTAATCTGAAGAGATTACTC | TAAGTGCGCCCT | 4e − 12 | 71 |
| S. aureus | pSK41 | GCGAACGGAACGTTCGCA | TAAGTGCGCCCT | 5e − 10 | 99 |
| B. longum | pKJ36 | ATGCAACCTCCGGTTGCATG | TAAGTGCGCCCT | 5e − 11 | 196 |
| B. longum | pKJ50 | ATGTTACCACCGGTAACATG | TAAGTGCGCCCT | 4e − 08 | 264 |
| N. meningitidis | pAB6 | CAGTTTCTCGAAGAGAAACTGA | TAAGTGGGCATT | 5e − 49 | 65 |
| L. lactis | pMRC01 | ACACCACCCAATTTTGGAGTGGTGTG | TAAGTGCGCATT | 3e − 20 | 108 |
| X. fastidiosa | pXF5823 | ACCTTTCGTAGAAAGGTA | TAAGTGCGCCCT | 3e − 36 | 49 |
| S. enteriditis | Pp | GTTTCTCGACGAAGGAGAGAAACGT | TAAGTGCGCATC | 1e − 39 | 55 |
| E. faecalis | pRE25(A) | TACGAAGTAACGAAGTTACTGCGTA | TAAGTGCGCCCT | 1e − 11 | 141 |
| Plasmids with putative oriT but no identifiable relaxase | |||||
| S. ruminantium | pONE429 | GTGTCTTAAATTGCAAATTTGAGACACTCA | TAAGTGCGCCCT | ||
| L. lactis | pCRL291.1(A) | CACCAAAATATTTTGGTGGGTG | TAAGTGCGCCCT | ||
| (B) | AATTTTTCGTAAGAAAAATTTGGTGGGTA | TAAGTGCGCCCT | |||
| (C) | TAAATAAAATTTATGGGTA | TAAGTGCGCCCT | |||
| L. lactis | pHP003 | TATACCATGATTTTTCATGGTATA | TAAGTGCGCCCT | ||
| L. fermentum | pKC5B | CACTAAATGAAATTTAGGGAGTA | TAAGTGCGCCCT | ||
| A. pasteurianus | pAP12875(A) | CACGAACGAAAGTTTGTG | TAAGTGCGCCCT | ||
| (B) | CAAACGGGAGTTTGTA | TAAGTGCGCTTC | |||
| E. faecalis | pRE25(B) | ATATACAAGATTGAAAATCGTGTA | TAAGTGCGCCCT | ||
| (C) | TACATCAATTTTTTAAAATGATGTA | TAAGTGCGCCCT | |||
| S. aureus | p21kb | ATGTCGATTTATCCGACGTA | TAAGTGCGCCCT | ||
| E. faecium | pEFR(A) | CACCAAAATATTTTGGTGGGTG | TAAGTGCGCCCT | ||
| (B) | AAAAAATTCGGTAGATTTTTTTACTGCGTA | TAAGTGCGCCCT | |||
| L. helveticus | pLJ1 | ACAACTGATTTATCTTGTTGTGTG | TAAGTGCGCCCT |
RSF1010 has the core sequence TAAGTGCGCCCT but is otherwise essentially identical.
Probability of the amino acid sequence occurring by chance (GenBank).
Reference organisms: E. coli, Salmonella enterica serovar Panama, Dichelobacter nodosus, Acidithiobacillus ferrooxidans, Staphylococcus aureus, Bifidobacterium longum, Neisseria meningitidis, L. lactis, Xylella fastidiosa, Salmonella enterica serovar Enteriditis, Enterococcus faecalis, Selenomonas ruminantium, Lactobacillus fermentum, Acetobacter pasteurianus, Enterococcus faecium, Lactobacillus helveticus.
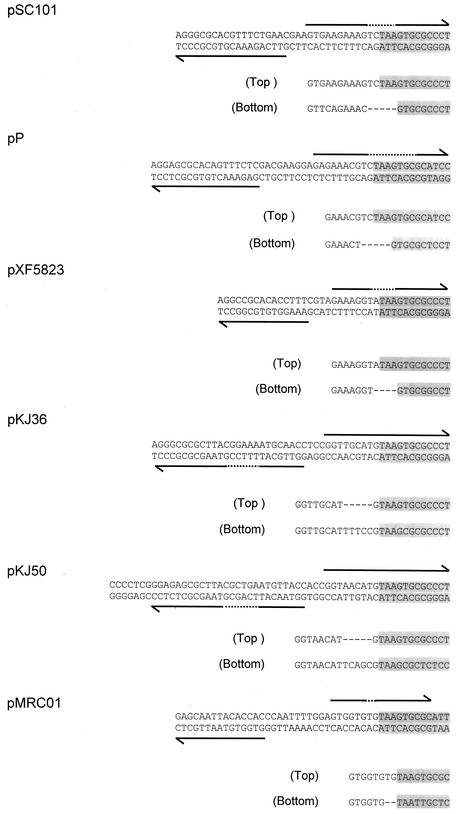
If the inverted repeat is formed by inversion, then in some cases part or all of the highly conserved core sequence would also be duplicated. Evidence for this duplication is shown in Fig. 4. In the plasmids pSC101, pP, and pXF5823, part of the core, GTGCGCCC, is present in the inverted orientation. Plasmids pKJ36 and pK50, both derived from Bifidobacterium longum, have oriTs with inverted repeats that are clearly different from those in the first group of plasmids, but again both show evidence for ancestral duplication and inversion of the core. In these plasmids, the duplicated core is intact, but there is a deletion in the remaining DNA in one of the two arms. The plasmids pKJ36 and pKJ50 show similarities in the sequences of their oriTs and might have been derived from a plasmid with the same initial inversion. A final example is pMRC01, from Lactobacillus lactis. Here there are several point mutations in one of the core sequences and a small deletion in the adjacent DNA.
FIG. 4.
Evidence for the inversion of core and adjacent DNA. Core DNA is shaded and the repeated and inverted DNA is indicated by the horizontal arrows, with the dotted part indicating the region missing in one arm. In each case the duplicated regions are also aligned, with the deleted region indicated by dashes.
Model for evolution of the R1162 mobilization family.
Our observations lead us to propose that a family of related conjugative transfer systems was created by the acquisition, on several separate occasions, of a transesterase and its recognition sequence, consisting of core and adjacent duplex DNA (Fig. 5). These elements might have been involved in transfer of the chromosome or in some other function requiring strand cleavage, such as site-specific recombination. Possibly the core segment and adjacent DNA were acquired first, with the relaxase protein active in trans. Initially, these elements would result in inefficient plasmid transfer, since there would be no mechanism for stabilizing the plasmid in the recipient except by recombination with the chromosome or with other preexisting plasmids. Inversion of the DNA adjacent to the core would result in a substrate suitable for recircularization of the plasmid after transfer, and those inversions most able to form a hairpin loop would be selected. The relaxase would become adapted by selection to the particular sequence determined by the inverted repeat, resulting in the gain of some degree of specificity of the protein for its oriT.
FIG. 5.
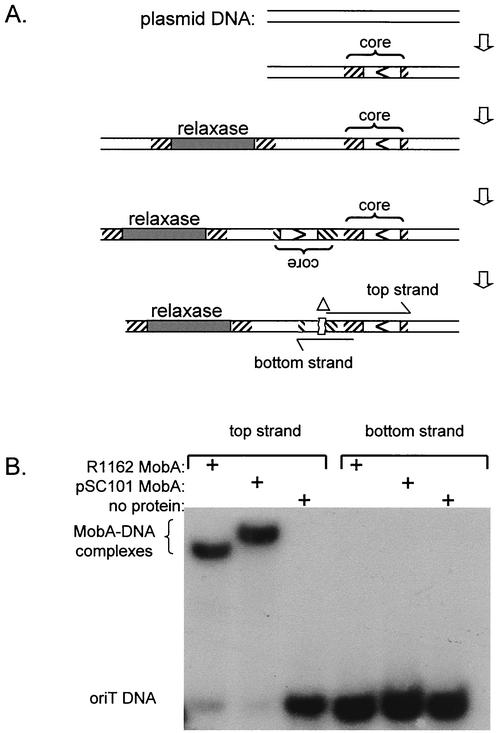
(A) Model for evolution of oriT. The double horizontal line indicates plasmid DNA, the box with an arrow is core DNA, and the filled segment represents the relaxase gene. The slashed segment is DNA derived from a foreign source by recombination and containing either core or the relaxase gene. Following duplication and inversion of DNA containing the core, a deletion (Δ) inactivates one of the nascent oriTs. The extent of this deletion is different for each plasmid and is illustrated here for pSC101 (compare with Fig. 4). (B) Gel retardation assay for single-stranded oligonucleotides mixed with purified R1162 or pSC101 MobA. The location of the pSC101 DNA present in each oligonucleotide is shown in panel A.
Since in many cases duplication and inversion of the DNA would involve the core itself, both strands of the plasmid DNA might become susceptible to cleavage by the cognate MobA. As the system becomes more efficient, the probability of simultaneous cleavage of both strands would increase and this could destabilize the plasmid, for example, by impairing processes such as plasmid replication. Therefore, mutations resulting in inactivation of one of the sites would be selected. In pSC101, five base pairs have been deleted from one of the duplications, and this results in the failure of either the pSC101 or R1162 MobA to bind the DNA (Fig. 5).
In R1162 and pSC101, oriT is oriented so that the relaxase gene is transferred last (14), and this also seems to be true for most of the plasmids listed in (Table 2). In the case of pMRC01, pKJ36, and pKJ50, it is not possible to infer the direction of transfer, since the core is conserved in both orientations. The relative orientations of oriT and mobA in R1162 are required neither by the system of mobilization nor by other aspects of the biology of the plasmid. We have successfully inverted the R1162 oriT, or placed it at a new location, without any significant effect on the frequency of transfer (reference 16 and unpublished data). However, the R1162 system for mobilization is already very efficient. This would probably not be the case, at least at the termination step, for a system where the oriT had been newly created by inversion. When the nascent inverted repeat and the relaxase gene are closely linked in the direction of transfer, they will be better able to coevolve toward greater efficiency because they will be coinherited most often. Because the inverted repeat part of the core is transferred last, selection will also place the relaxase gene at the end of transfer.
Our interpretation does not account for the accessory proteins MobB and MobC, also part of the R1162 relaxosome. Even for similar MobA proteins, the presence or absence of accessory proteins is a variable and unpredictable feature of each relaxase. For example, despite the similarity of the pSC101 and R1162 MobA proteins, there is no MobC homolog present in the pSC101 relaxosome (15). In contrast, a MobC-like protein is encoded by the mobilization system of plasmid pTF1, but a protein similar to MobB has not been identified for this plasmid (12, 17). We believe that these accessory proteins became secondarily associated with the relaxase because they enhanced the interaction of MobA with the newly generated oriT, whereas in other cases mutations in the MobA gene itself accomplished the same purpose. However, it is possible that the accessory proteins allow the R1162 MobA to recognize a broader range of oriTs, as indicated by our results. By assisting in the melting of oriT, MobC could permit a looser fit between MobA and its DNA target.
We might expect mobA-like genes on the chromosome and putative oriTs on plasmids lacking the relaxase gene. A protein similar to the R1162 MobA (E = 9e−10) is encoded by Xanthomonas axonopodis pv. citri (10). The gene is designated mobL, and while this organism has two plasmids, mobL is located on the chromosome. Although there at least two core-like sequences in the X. axonopodis chromosome, neither is adjacent to mobL and both lack an adjacent inverted repeat. Similarly, the linear chromosome of Agrobacterium tumefaciens strain C58 encodes a MobA-like protein (21). In this case, there is an adjacent core, separated from the gene by 56 bp, but no inverted repeat. We would not expect the selection of an inversion, since the parent element is linear.
We can also identify putative oriTs on plasmids that do not appear to encode a relaxase. Some of these plasmids are listed in Table 2. It is likely that at least some of these elements are involved in mobilization. The plasmids pSK41 and p21kb have identical putative oriTs, located within a 41-bp segment that has nearly 100% sequence identity, but only pSK41 encodes a recognizable relaxase. The two plasmids are not related by simple deletion; although they have several large regions of nearly identical sequence, approximately 200 bp on either side of the 41-bp oriT-containing segment, p21kb shows little relatedness to pSK41 DNA. The conservation of oriT as a “patch” suggests that it has been selected during the evolution of the plasmid.
Among the putative oriTs listed in Table 2, subfamilies made up of very similar members can be identified. For example, the oriTs of R1162, pDN1, pIE1115, pIE1130, and pAB6 have inverted repeats with very similar sequences, and it is reasonable to assume that they were derived by horizontal gene transfer and recombination. As a group, however, the elements in Table 2 do not have a consensus base sequence apart from the core. This indicates that plasmids acquired the ancestral nicking elements on different occasions. Either the DNA adjacent to the core in these elements does not need to have a specific sequence, or there is a family of these elements, each with a different sequence for this adjacent DNA. The relaxed specificity of the R1162 MobA and the imperfect hairpin loops for the oriTs in Table 2 favor the first possibility.
Several of the plasmids in Table 2 have more than one putative oriT, arranged either in direct or inverted orientation. Multiple oriTs could arise by duplication of the element, or by the acquisition of two unrelated oriTs. It is likely that both mechanisms have occurred. For example, the two directly repeated oriTs of pEFR show no sequence similarity apart from the core, whereas the oriTs of pCRL291.1 are all surrounded by tracts of DNA having similar sequences. When mobilization is inefficient, molecules containing multiple oriTs will be selected. If the oriTs are cleaved by different relaxases, then the probability of transfer, and possibly the host range of the plasmid, will be increased. Even when the oriTs are functionally the same, the benefits of more than one oriT could outweigh the difficulties of simultaneous cleavage of both strands, or termination at the wrong oriT, possible problems when transfer is efficient.
Recently, the relationship between conjugative transfer and type IV secretion has been emphasized (1). However, there has been little effort to understand the origin of the proteins involved in the DNA processing required for mobilization. We propose here that the oriTs of the R1162/RSF1010 mobilization family were derived by duplication and inversion of a conserved recognition sequence for a protein involved in single-strand cleavage. The protein could be part of a mechanism for horizontal gene transfer of the chromosome, with rescue of the incoming DNA by recombination, or it could have been derived from some other DNA processing activity in the cell. Once the duplication became fixed on the plasmid, it became part of a potent mechanism for transfer of plasmid DNA and became disseminated among other species of bacteria.
Acknowledgments
This work was supported by Public Health Service grant GM-37462 from the National Institutes of Health.
REFERENCES
- 1.Baron, C., D. O'Callaghan, and E. Lanka. 2002. Bacterial secrets of secretion: EuroConference on the biology of type IV secretion processes. Mol. Microbiol. 43:1359-1365. [DOI] [PubMed] [Google Scholar]
- 2.Becker, E., and R. Meyer. 2000. Recognition of oriT for DNA processing at termination of a round of conjugal transfer. J. Mol. Biol. 300:1067-1077. [DOI] [PubMed] [Google Scholar]
- 3.Becker, E. C., and R. Meyer. 2002. MobA, the DNA strand transferase of plasmid R1162. The minimal domain required for DNA processing at the origin of transfer. J. Biol. Chem. 277:14575-14580. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharjee, M., X. M. Rao, and R. J. Meyer. 1992. Role of the origin of transfer in termination of strand transfer during bacterial conjugation. J. Bacteriol. 174:6659-6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhattacharjee, M. K., and R. J. Meyer. 1991. A segment of a plasmid gene required for conjugal transfer encodes a site-specific, single-strand DNA endonuclease and ligase. Nucleic Acids Res. 19:1129-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhattacharjee, M. K., and R. J. Meyer. 1993. Specific binding of MobA, a plasmid-encoded protein involved in the initiation and termination of conjugal DNA transfer, to single-stranded oriT DNA. Nucleic Acids Res. 21:4563-4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brasch, M. A., and R. J. Meyer. 1986. Genetic organization of plasmid R1162 DNA involved in conjugative mobilization. J. Bacteriol. 167:703-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brasch, M. A., and R. J. Meyer. 1987. A 38 base-pair segment of DNA is required in cis for conjugative mobilization of broad host-range plasmid R1162. J. Mol. Biol. 198:361-369. [DOI] [PubMed] [Google Scholar]
- 9.Clewell, D. B., and D. R. Helinski. 1969. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an open circular DNA form. Proc. Natl. Acad. Sci. USA 62:1159-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.da Silva, A., J. Ferro, F. Reinach, C. Farah, L. Furlan, R. Quaggio, C. Monteiro-Vitorello, M. Van Sluys, N. Almeida, L. Alves, A. do Amaral, M. Bertolini, L. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. Ciapina, R. Cicarelli, L. Coutinho, J. Cursino-Santos, H. El-Dorry, J. Faria, A. Ferreira, R. Ferreira, M. Ferro, E. Formighieri, M. Franco, C. Greggio, A. Gruber, A. Katsuyama, L. Kishi, R. Leite, E. Lemos, M. Lemos, E. Locali, M. Machado, A. Madeira, N. Martinez-Rossi, E. Martins, J. Meidanis, C. Menck, C. Miyaki, D. Moon, L. Moreira, M. Novo, V. Okura, M. Oliveira, V. Oliveira, H. Pereira, A. Rossi, J. Sena, C. Silva, R. de Souza, L. A. Spinola, M. Takita, R. Tamura, E. Teixeira, R. Tezza, M. Trindade dos Santos, D. Truffi, S. Tsai, F. White, J. Setubal, and J. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 11.Derbyshire, K., G. Hatfull, and N. Willetts. 1987. Mobilization of the non-conjugative plasmid RSF1010: a genetic and DNA sequence analysis of the mobilization region. Mol. Gen. Genet. 206:161-168. [DOI] [PubMed] [Google Scholar]
- 12.Drolet, M., P. Zanga, and P. C. K. Lau. 1990. The mobilization and origin of transfer regions of a Thiobacillus ferrooxidans plasmid: relatedness to plasmids RSF1010 and pSC101. Mol. Microbiol. 4:1381-1391. [DOI] [PubMed] [Google Scholar]
- 13.Hershfield, V., H. W. Boyer, C. Yanofsky, M. A. Lovett, and D. R. Helinski. 1974. Plasmid ColE1 as a molecular vehicle for cloning and amplification of DNA. Proc. Natl. Acad. Sci. USA 71:3455-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, K., and R. J. Meyer. 1989. Unidirectional transfer of broad host-range plasmid R1162 during conjugative mobilization. Evidence for genetically distinct events at oriT. J. Mol. Biol. 208:501-505. [DOI] [PubMed] [Google Scholar]
- 15.Meyer, R. 2000. Identification of the mob genes of plasmid pSC101 and characterization of a hybrid pSC101-R1162 system for conjugal mobilization. J. Bacteriol. 182:4875-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perwez, T., and R. Meyer. 1996. MobB protein stimulates nicking at the R1162 origin of transfer by increasing the proportion of complexed plasmid DNA. J. Bacteriol. 178:5762-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rawlings, D., and E. Tietze. 2001. Comparative biology of IncQ and IncQ-like plasmids. Microbiol. Mol. Biol. Rev. 65:481-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scherzinger, E., R. Lurz, S. Otto, and B. Dobrinski. 1992. In vitro cleavage of double- and single-stranded DNA by plasmid RSF1010-encoded mobilization proteins. Nucleic Acids Res. 20:41-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scholz, P., V. Haring, B. Wittmann-Liebold, K. Ashman, M. Bagdasarian, and E. Scherzinger. 1989. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene 75:271-288. [DOI] [PubMed] [Google Scholar]
- 20.Willetts, N., and C. Crowther. 1981. Mobilization of the nonconjugative IncQ plasmid RSF1010. Genet. Res. 37:311-316. [DOI] [PubMed] [Google Scholar]
- 21.Wood, D. W., J. C. Setubal, R. Kaul, D. Monks, L. Chen, G. E. Wood, Y. Chen, L. Woo, J. P. Kitajima, V. K. Okura, N. F. Almeida, Jr., Y. Zhou, D. Bovee, Sr., P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, D. Guenthner, T. Kutyavin, R. Levy, M. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. Wu, D. Gordon, J. A. Eisen, I. Paulsen, P. Karp, P. Romero, S. Zhang, H. Yoo, Y. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z. Zhao, M. Dolan, S. V. Tingey, J. Tomb, M. P. Gordon, M. V. Olson, and E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317-2323. [DOI] [PubMed] [Google Scholar]
- 22.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 23.Zhang, S., and R. J. Meyer. 1995. Localized denaturation of oriT DNA within relaxosomes of the broad host-range plasmid R1162. Mol. Microbiol. 17:727-735. [DOI] [PubMed] [Google Scholar]