As goes history, so goes research: this year, activity in areas of retrovirus research related only indirectly have provoked events that are notable when considered together. Last summer it was reported that a patient in one X-linked severe combined immunodeficiency retroviral vector gene therapy trial had developed leukemia. Now disquietingly, there has been a second such event, and a third patient is reported to have a vector insertion near the same gene (LMO2) as observed in the other two individuals (1). Meanwhile, in a basic research laboratory, experiments have moved us another step closer to understanding the mechanics of insertion specificity for retrovirus-type integrases (IN). As reported in this issue of PNAS, investigators have produced active retroviruslike elements with synthetic insertion specificities (2). Dan Voytas and colleagues at Iowa State University (Ames) study the Saccharomyces long terminal repeat (LTR)-retrotransposon Ty5, which targets heterochromatic regions (3). Now, in an elegant adaptation of the two-hybrid system, the 6-aa Ty5 targeting domain (TD) was exchanged for two heterologous domains shown to mediate interaction of their respective proteins with protein partners. When domains from those partners were produced fused to the LexA DNA-binding domain, targeting to LexA-binding sites was observed. Although integration specificity in the system was by no means absolute, these results are of interest to genetic engineers and future gene therapists.
Interest in the integration patterns of retroviruses is longstanding. Despite the potential danger of deleterious activating or even inactivating insertions, retroviruses present compelling advantages as therapy vectors (reviewed in ref. 4). Early investigations of oncogenic retrovirus insertion sites in transformed cells showed that insertions were linked to activation of flanking oncogenes or DNaseI hypersensitive sites, leading to the notion that insertion into open chromatin was favored (reviewed in ref. 5; see also refs. 6 and 7). The potential for deleterious retrovirus vector insertions fueled investigation into the mechanistic basis of insertion site selection. Development of PCR assays with which significant numbers of retrovirus integration sites could be mapped showed that genomes are broadly accessed by retroviruses, but that there are decidedly nonrandom patterns as well (8). More recently, large numbers of HIV type 1 (HIV-1) insertions have been mapped and compared with genomewide transcription patterns to globally probe the relationship between gene expression and retrovirus integration (9). These experiments showed that HIV-1 insertion favors transcribed regions. Nonetheless, the basis of the preference for transcribed regions has been elusive, and examination of at least one transcribed region for effects of transcriptional activity on integration activity have not shown a positive correlation (10).
At the heart of retroviral integration is the IN. It is a member of the D,D(35)E transposase/IN superfamily named after its conserved catalytic triad of amino acids. Because of its central role in the retrovirus lifecycle, the function and structure of this enzyme has been studied extensively (reviewed in refs. 11–15). Retroviral IN mediates a strand transfer of LTR DNA 3′ OH ends to staggered positions in the host DNA (16, 17). Combined evidence of many types shows a retroviral IN with three physically distinct domains. An N-terminal domain includes three α-helices and a zinc-binding motif. This domain has been implicated in dimerization and in binding the LTR ends. The central domain contains the conserved catalytic triad D,D(35)E. Members of this triad coordinate a divalent metal cation, probably Mg2+ in vivo (15) and are essential for catalytic activity. The C-terminal domain contributes to oligomerization, has nonspecific DNA-binding activity and is physically similar to the SH3 protein interaction domain. No full-length IN structure has yet been determined at high resolution.
Potential deleterious retrovirus insertions fueled investigation into the mechanism of insertion site selection.
In vivo a retroviral preintegration complex composed of IN bound to the ends of the full-length DNA mediates integration into host DNA. Isolation first of preintegration complexes from infected cells and then production of active, recombinant IN allowed examination of the effect of different target features on integration in vitro. A generalization that has emerged from studies conducted in several laboratories is that bending of DNA favors integration (18), as do hairpin structures (19). The former occurs in nucleosomes, which, contrary to expectations, were found to act as preferred targets over nonnucleosomal DNA, both in vitro and in vivo (20–22).
The relatively global distribution of retrovirus integration sites stands in interesting contrast to the distinctive insertion preferences of their LTR-retrotransposon cousins, the Pseudoviridiae (e.g., Ty1 and Ty5 copialike elements) (23) and the Metaviridae (e.g., Tf1 and Ty3 gypsylike elements) (24). IN proteins encoded by these elements have the zinc-binding motif, the highly conserved residues of the central domain and the poorly conserved C-terminal domain. The IN proteins of the Pseudoviridae and the Metaviridae differ from each other in the C-terminal domain where the Pseudoviridae have a conserved GKGY motif (23), and the Metaviridae have a conserved GPF/Y motif. Some members of the Metaviridae also have a chromodomain (24).
As a group, the yeast LTR retrotransposons have notable insertion preferences. The specificity of Ty5 for heterochromatin is discussed further below. In budding yeast, the Pseudoviridae Ty1, 2, and 4 reside mostly within 750 bp of the 5′ ends of tRNA genes (25, 26). In vivo insertions fall along a gradient beginning at about −80 bp from the 5′ coding end of the tRNA gene and extending upstream. Integration appears to rise and fall in a pattern which could correlate with some feature of the nucleosome (27). The pattern of integration of the Metaviridae element Ty3 is even more restricted. The gene-proximal strand transfer in this case occurs within one or two nucleotides of tRNA gene transcription initiation sites. In vivo it is likely that transcription factors TFIIIB and TFIIIC are essential for Ty3 targeting (28–30). Furthermore, it has been shown that yeast elements Ty1–4 target other genes transcribed by RNA polymerase III with similar patterns to those observed flanking tRNA genes (27, 30). In vitro, Ty3 targeting to the U6 gene requires only TATA-binding protein and Brf1 (29).
Observation of highly specific integration in yeast helped to motivate a series of experiments to confer novel insertion specificities on retrovirus IN proteins (reviewed in refs. 31 and 32). Recombinant retroviral IN has been expressed as a fusion with relatively compact DNA-binding domains including lambda repressor (33), LexA DNA-binding domain (34, 35), and the DNA-binding domain of Zif268 (36). Recombinant proteins have been shown to target in vitro integration to the respective DNA-binding sites of the fusion proteins. Disappointingly, these chimeric IN species, appear to be incompatible with high levels of infectious virus. Presumably this is caused by some failure to structurally accommodate the heterologous domain. To circumvent some of these problems, a strategy involving trans expression of IN has been used. In this variation, a fusion of HIV-1 structural protein p6 to an IN-LexA targeting domain directs IN to the virion and complements catalytically defective IN contributed from Gag-Pol (37, 38). However, there are no naturally occurring LexA-binding sites in mammalian cells, and targeting to synthetic sites has not yet been reported.
Ty5 is distinct among the yeast elements. Originally identified as a degenerate element at the ends of Saccharomyces cerevisiae chromosomes (39), the Voytas laboratory recovered an active copy from Saccharomyces paradoxus and transferred it into S. cerevisiae (3). In this context, they showed that Ty5 inserted into heterochromatic DNA (40). Mutations in Sir3p or Sir4p that disrupted silencing of telomeric DNA also resulted in loss of targeting to silenced regions (41). The pieces of the puzzle fell quickly into place. A targeting domain of 6 aa (TD), virtually at the C-terminus of Ty5 IN, was mapped, which was required for targeting (42) and which mediated interactions with a large C-terminal portion of Sir4p (43).
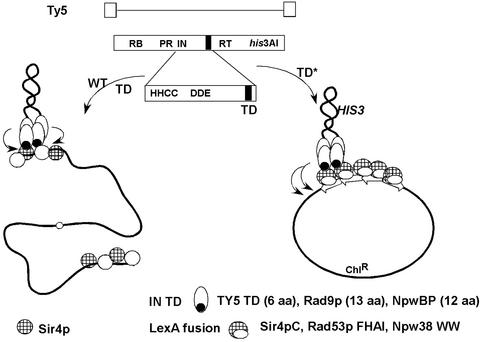
In the current article (2), the Voytas laboratory accomplishes design-based integration. The strategy is outlined in Fig. 1. They fused the LexA DNA-binding domain to one of several TD-interacting domains: first the C-terminal domain of Sir4p (Sir4pC). Next the 6-aa IN TD and the Sir4pC fusion domains were swapped with two pairs of heterologous partner domains. Such domains were carefully chosen to minimize disruption of IN. A 13-aa sequence in Rad9p mediates its interaction with a forkhead-associated domain (FHA1) in another DNA repair protein, Rad53p. A 12-aa domain in NpwBP mediates interaction with the WW domain of another nuclear protein Npw38. The Rad9p and NpwBP domains were substituted for the natural Ty5 TD. The partner interacting domains (i.e., FHA1 from Rad53p and WW from Npw38) were expressed fused to the LexA DNA-binding domain. Yeast were transformed with the synthetic Ty5 TD elements, constructs from which fusion DNA-binding domains were expressed, and a target plasmid containing LexA-binding sites embedded in Arabidopsis DNA. Target plasmids were recovered in Escherichia coli for analysis. For Ty5-TD and Ty5-Rad9p targeting, 26 integrant joints were sequenced and shown to be within 120 bp of LexA-binding sites, and of 18 further analyzed, all had the direct flanking repeats characteristic of bona fide integrants. In the case of targeting to Sir4p-, Rad53p FHA1-, and Npw38 WW-LexA fusions and target plasmids with four copies of the LexA operator, about one-sixth of transposition was into the target.
Figure 1.
Strategy for retargeting Ty5 integration. Top, schematic of Ty5 single ORF encoding RNA binding (RB), protease (PR), integrase (IN), reverse transcriptase (RT), and marker gene (his3AI) (open box). View of IN is expanded to show conserved residues and targeting domain (TD) (solid). Lower left, preintegration complex showing wild-type IN bound to ends of Ty5 DNA (thick line) and integrating into telomeric heterochromatin, mediated by Sir4p (hatched). Lower right, same as left except that the natural TD is replaced with heterologous domains (TD*) from Rad9p and NpwBP (solid). LexA DNA-binding domain (open) is expressed fused to Sir4pC, Rad53p FHA1, and Npw38 WW domains (hatched). Integration occurs proximal to LexA-binding sites (open arrows) in plasmid target (closed circle).
Many questions remain. For example, how does Ty5 access the DNA after docking at Sir4p? What is the distribution of the majority of (nontarget plasmid) Ty5 integrations? Do nonplasmid insertions default to random, to native Rad53p direction in the case of the Rad9p-based TD, or do natural, as yet unidentified, functions continue to operate on the Ty5 IN? Is it possible to generate integration that is more highly restricted, perhaps through the use of phage panning or slightly larger domains?
The experiments by Voytas suggest many new avenues for genome exploration. The occurrence of a compact and independent interaction domain in a retroviral-type IN of course poses the question of whether other such domains exist. In the case of Ty3, interactions between the N-terminal domain and TFIIIC subunit Tfc1p have been documented in vitro and are consistent with in vivo results (44). Ty3 also has a relatively extended C-terminal domain that could interact with targeting proteins including TFIIIB subunits, but this has not been demonstrated. It seems likely that the S. cerevisiae Pseudoviridae element Ty1 will be targeted by some feature of chromatin which distinguishes regions directly upstream of tRNA genes (27). An alignment of Metaviridae element IN C-terminal domains recently resulted in the identification of a chromodomain motif (24). Tf1, a Schizosaccharomyces pombe element of this class has been shown to insert in inter-ORF spaces, apparently with preference for the region within 100–300 bp from the ORF initiation codon (45, 46). Results of recent experiments suggest that Tf1 integration is actually targeted through interaction of the chromodomain with histone H3 methylated at K4 (H. Levin, National Institutes of Health, Bethesda, personal communication). These observations are exciting because they not only hint at the subtlety and diversity of integration specificity, but suggest that integration can be used to learn about chromatin structure as well as to manipulate the genome.
It is not clear to what extent retroviral proteins will be shown to interact with specific proteins for targeting in the manner observed for the yeast LTR retrotransposons. The C-terminal domain of characterized retroviral IN proteins has an SH3 structure and the SH3 motif mediates a wide variety of protein interactions albeit mostly having to do with signal transduction (47). In addition, it has been shown that several chromatin-related proteins enhance retroviral integration in vitro and potentially in vivo; one such case is INI1 (48), and another is LEDGF/p75 (49). The recent findings in yeast are likely to encourage further exploration for proteins that contribute to the loosely defined preference of at least some retroviruses for insertion into transcriptionally active regions and into particular hotspots.
What are the lessons that could be applied to better laboratory retrovirus vectors, or even make safer therapeutic vectors? One observation, so obvious it can hardly be considered a lesson, is that relatively subtle changes are likely to be better tolerated by the virion. A second point is that the known structure of the C-terminal domain of retroviral IN might be used to identify positions actually within the IN, which are compatible with replacements or insertions of small TD cassettes. The Ty5 study underscores the findings from in vitro targeting studies with retroviral IN, namely that the C-terminal domain can deliver active IN to the integration site. Finally, although protein–protein mediation of IN docking does not have the reassuring simplicity of an IN that binds unique DNA sequences, it offers the rich combinatorial complexity of the natural proteome.
Clearly, much work remains to explore the mechanisms, implications, and applications of targeted retroviral integration. Integration by design in a model organism from the Voytas laboratory hints at the possibilities.
Footnotes
See companion article on page 5891.
References
- 1.Kaiser J. Science. 2003;299:991. doi: 10.1126/science.299.5609.991b. [DOI] [PubMed] [Google Scholar]
- 2.Zhu Y, Dai J, Fuerst P G, Voytas D F. Proc Natl Acad Sci USA. 2003;100:5891–5895. doi: 10.1073/pnas.1036705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zou S, Ke N, Kim J M, Voytas D F. Genes Dev. 1996;10:634–645. doi: 10.1101/gad.10.5.634. [DOI] [PubMed] [Google Scholar]
- 4.Galimi F, Verma I M. Curr Top Microbiol Immunol. 2002;261:245–254. doi: 10.1007/978-3-642-56114-6_13. [DOI] [PubMed] [Google Scholar]
- 5.Sandmeyer S B, Hansen L J, Chalker D L. Annu Rev Genet. 1990;24:491–518. doi: 10.1146/annurev.ge.24.120190.002423. [DOI] [PubMed] [Google Scholar]
- 6.Scherdin U, Rhodes K, Breindl M. J Virol. 1990;64:907–912. doi: 10.1128/jvi.64.2.907-912.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mooslehner K, Karls U, Harbers K. J Virol. 1990;64:3056–3058. doi: 10.1128/jvi.64.6.3056-3058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Withers-Ward E S, Kitamura Y, Barnes J P, Coffin J M. Genes Dev. 1994;8:1473–1487. doi: 10.1101/gad.8.12.1473. [DOI] [PubMed] [Google Scholar]
- 9.Schroder A R, Shinn P, Chen H, Berry C, Ecker J R, Bushman F. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 10.Weidhaas J B, Angelichio E L, Fenner S, Coffin J M. J Virol. 2000;74:8382–8389. doi: 10.1128/jvi.74.18.8382-8389.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haren L, Ton-Hoang B, Chandler M. Annu Rev Microbiol. 1999;53:245–281. doi: 10.1146/annurev.micro.53.1.245. [DOI] [PubMed] [Google Scholar]
- 12.Hindmarsh P, Leis J. Microbiol Mol Biol Rev. 1999;63:836–843. doi: 10.1128/mmbr.63.4.836-843.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wlodawer A. Adv Virus Res. 1999;52:335–350. doi: 10.1016/s0065-3527(08)60305-x. [DOI] [PubMed] [Google Scholar]
- 14.Craigie R. J Biol Chem. 2001;276:23213–23216. doi: 10.1074/jbc.R100027200. [DOI] [PubMed] [Google Scholar]
- 15.Rice P A, Baker T A. Nat Struct Biol. 2001;8:302–307. doi: 10.1038/86166. [DOI] [PubMed] [Google Scholar]
- 16.Fujiwara T, Mizuuchi K. Cell. 1988;54:497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- 17.Craigie R, Mizuuchi K. Cell. 1985;41:867–876. doi: 10.1016/s0092-8674(85)80067-2. [DOI] [PubMed] [Google Scholar]
- 18.Müller H-P, Varmus H E. EMBO J. 1994;13:4704–4714. doi: 10.1002/j.1460-2075.1994.tb06794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katz R A, Gravuer K, Skalka A M. J Biol Chem. 1998;273:24190–24195. doi: 10.1074/jbc.273.37.24190. [DOI] [PubMed] [Google Scholar]
- 20.Pryciak P M, Müller H-P, Varmus H E. Proc Natl Acad Sci USA. 1992;89:9237–9241. doi: 10.1073/pnas.89.19.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pryciak P M, Varmus H E. Cell. 1992;69:769–780. doi: 10.1016/0092-8674(92)90289-o. [DOI] [PubMed] [Google Scholar]
- 22.Pruss D, Bushman F D, Wolffe A P. Proc Natl Acad Sci USA. 1994;91:5913–5917. doi: 10.1073/pnas.91.13.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peterson-Burch B D, Voytas D F. Mol Biol Evol. 2002;19:1832–1845. doi: 10.1093/oxfordjournals.molbev.a004008. [DOI] [PubMed] [Google Scholar]
- 24.Malik H S, Eickbush T H. J Virol. 1999;73:5186–5190. doi: 10.1128/jvi.73.6.5186-5190.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji H, Moore D P, Blomberg M A, Braiterman L T, Voytas D F, Natsoulis G, Boeke J D. Cell. 1993;73:1007–1018. doi: 10.1016/0092-8674(93)90278-x. [DOI] [PubMed] [Google Scholar]
- 26.Kim J M, Vanguri S, Boeke J D, Gabriel A, Voytas D F. Genome Res. 1998;8:464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- 27.Devine S E, Boeke J D. Genes Dev. 1996;10:620–633. doi: 10.1101/gad.10.5.620. [DOI] [PubMed] [Google Scholar]
- 28.Kirchner J, Connolly C M, Sandmeyer S B. Science. 1995;267:1488–1491. doi: 10.1126/science.7878467. [DOI] [PubMed] [Google Scholar]
- 29.Yieh L, Kassavetis G, Geiduschek E P, Sandmeyer S B. J Biol Chem. 2000;275:29800–29807. doi: 10.1074/jbc.M003149200. [DOI] [PubMed] [Google Scholar]
- 30.Chalker D L, Sandmeyer S B. Genes Dev. 1992;6:117–128. doi: 10.1101/gad.6.1.117. [DOI] [PubMed] [Google Scholar]
- 31.Bushman F D. Curr Top Microbiol Immunol. 2002;261:165–177. doi: 10.1007/978-3-642-56114-6_8. [DOI] [PubMed] [Google Scholar]
- 32.Holmes-Son M L, Appa R S, Chow S A. Adv Genet. 2001;43:33–69. doi: 10.1016/s0065-2660(01)43003-3. [DOI] [PubMed] [Google Scholar]
- 33.Bushman F. Proc Natl Acad Sci USA. 1994;91:9233–9237. doi: 10.1073/pnas.91.20.9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goulaouic H, Chow S A. J Virol. 1996;70:37–46. doi: 10.1128/jvi.70.1.37-46.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katz R A, Merkel G, Skalka A M. Virology. 1996;217:178–190. doi: 10.1006/viro.1996.0105. [DOI] [PubMed] [Google Scholar]
- 36.Bushman F D, Miller M D. J Virol. 1997;71:458–464. doi: 10.1128/jvi.71.1.458-464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holmes-Son M L, Chow S A. J Virol. 2000;74:11548–11556. doi: 10.1128/jvi.74.24.11548-11556.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holmes-Son M L, Chow S A. Mol Ther. 2002;5:360–370. doi: 10.1006/mthe.2002.0559. [DOI] [PubMed] [Google Scholar]
- 39.Voytas D F, Boeke J D. Nature. 1992;358:717. doi: 10.1038/358717a0. [DOI] [PubMed] [Google Scholar]
- 40.Zou S, Voytas D F. Proc Natl Acad Sci USA. 1997;94:7412–7416. doi: 10.1073/pnas.94.14.7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu Y, Zou S, Wright D A, Voytas D F. Genes Dev. 1999;13:2738–2749. doi: 10.1101/gad.13.20.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gai X, Voytas D F. Mol Cell. 1998;1:1051–1055. doi: 10.1016/s1097-2765(00)80105-7. [DOI] [PubMed] [Google Scholar]
- 43.Xie W, Gai X, Zhu Y, Zappulla D C, Sternglanz R, Voytas D F. Mol Cell Biol. 2001;21:6606–6614. doi: 10.1128/MCB.21.19.6606-6614.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aye M, Dildine S L, Claypool J A, Jourdain S, Sandmeyer S B. Mol Cell Biol. 2001;21:7839–7851. doi: 10.1128/MCB.21.22.7839-7851.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Behrens R, Hayles J, Nurse P. Nucleic Acids Res. 2000;28:4709–4716. doi: 10.1093/nar/28.23.4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singleton T L, Levin H L. Eukaryot Cell. 2002;1:44–55. doi: 10.1128/EC.01.1.44-55.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mayer B J. J Cell Sci. 2001;114:1253–1263. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- 48.Kalpana G V, Marmon S, Wang W, Crabtree G R, Goff S P. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 49.Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. J Biol Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]