Abstract
Several reports have described an activity that modifies nitrotyrosine-containing proteins and their immunoreactivity to nitrotyrosine Abs. Without knowing the product of the reaction, this new activity has been called a “denitrase.” In those studies, some nonspecific proteins, which have multiple tyrosine residues, e.g., albumin, were used as a substrate. Therefore, the studies were based on an unknown mechanism of reaction and potentially a high background. To solve these problems, one of the most important things is to find a more suitable substrate for assay of the enzyme. We developed an assay strategy for determining the substrate for denitrase combining 2D-gel electrophoresis and an on-blot enzyme assay. The resulting substrate from RAW 264.7 cells was Histone H1.2, an isoform protein of linker histone. Histone H1.2 has only one tyrosine residue in the entire molecule, which ensures the exact position of the substrate to be involved. It has been reported that Histones are the most prominent nitrated proteins in cancer tissues. It was also demonstrated that tyrosine nitration of Histone H1 occurs in vivo. These findings lead us to the idea that Histone H1.2 might be an intrinsic substrate for denitrase. We nitrated recombinant and purified Histone H1.2 chemically and subjected it to an on-blot enzyme assay to characterize the activity. Denitrase activity behaved as an enzymatic activity because the reaction was time dependent and was destroyed by heat or trypsin treatment. The activity was shown to be specific for Histone H1.2, to differ from proteasome activity, and to require no additional cofactors.
Since the discovery of the first biological effects of nitric oxide (NO) more than two decades ago (1–3), NO has been identified as an important messenger molecule with diverse functions in many biological systems (4–8). Although many effects of NO are mediated through its direct interaction with guanylyl cyclase activation (1–8), its indirect action via secondary reactions with reactive oxygen species, forming reactive nitrogen species accounts for other effects. The reaction of NO and its reactive intermediates with protein bound tyrosine residues causes nitrotyrosine formation (9). Several mechanisms for nitrotyrosine formation have been suggested that involve peroxynitrite or myeloperoxidase (10).
Recently, a growing number of publications report the formation of nitrotyrosine-containing proteins in vivo. A long list of reports describes the increase in nitrotyrosine immunoreactivity in numerous proteins detected by immunohistochemical or immunoblot methods in various pathologic conditions (9, 10).
However, the possible role of tyrosine nitration in cellular function has not been fully explored. The nitration of proteins is reported to alter the conformation and structure of a protein, catalytic activity of enzymes, and susceptibility to proteolysis (11–18). It has also been shown that tyrosine nitration can diminish the effectiveness of a protein as a substrate for tyrosine kinases (14, 19, 20).
Thus, an important view is that protein nitration is associated with some inflammatory, toxic, or deleterious effects and with interruption of the cell-signaling processes. The hypothesis that protein tyrosine nitration renders the protein more susceptible to proteolysis has led to several reports, which describe that nitrated proteins are degraded by the proteasome more efficiently than native proteins (21, 22). On the other hand, some proteins are reported to be nitrated under physiological conditions (9, 10, 12). This phenomenon suggests that protein nitration may be, at least in some situations, reversible and contributing to regulatory processes.
Several reports have described denitrase as an activity that modifies nitrotyrosine-containing proteins (14, 23–26). In these studies, some nonspecific proteins like albumin, which has multiple tyrosine residues, were used as substrate for the activity. The denitrase activity was found to decrease the nitrotyrosine immunoreactivity or caused an increase in nitrate release into the media with incubation. The immunological methods used most commonly have the risk of artifacts by proteolysis and destruction of the epitope. To date, studies to examine denitrase were based on an unknown mechanism of reaction and potentially a high background and are subject to possible artifacts. To solve these problems, one of the most important things is to find a proper substrate to characterize and purify the denitrase activity.
The purpose of the present study was to establish a method with a specific substrate for denitrase and to reconstitute the reaction in vitro. Making use of 2D gel electrophoresis, we developed a solid support assay for denitrase to screen specific substrates. One substrate identified was Histone H1.2, an isoform protein of linker histone. This substrate allowed us to reconstitute and characterize the denitrase reaction.
Materials and Methods
Materials.
Enhanced chemiluminescence (ECL) plus was from Amersham Pharmacia. Mouse anti-nitrotyrosine mAbs were from Upstate Biotechnology (Lake Placid, NY) and Zymed. Anti-Histone H1 Ab was also from Upstate Biotechnology. Protease inhibitor complete (PIC) was obtained from Roche (Gipf-Oberfrick, Switzerland). Lactacystin and purified recombinant human Histone H1.2 protein were obtained from Calbiochem.
Other materials including the constituents of protein inhibitors premix [(PIP) 10 μg/ml soybean trypsin inhibitor/10 μg/ml benzamadine/0.005 units/ml trypsin inhibitor/aprotinin/10 μg/ml leupeptin/10 μg/ml pepstatin A/5 μg/ml antipain/200 μM phenylmethanesulfonyl fluoride, respectively, at final concentration] and cofactors premix [(CoF) 0.1 mM NADPH/0.1 mM NADP/0.2 mM NADH/0.2 mM NAD/5 μM FAD/5 μM FMN/0.1 μM ATP/0.1 mM GTP/1 mM neutralized glutathione, respectively, at final concentration] were obtained from Sigma.
Cell Culture.
Murine macrophage-like RAW 264.7 cells were obtained from the American Type Culture Collection and cultured in RPMI medium 1640 supplemented with 10% FBS 0.2 mM glutamine. Cultures were maintained at 37°C in a humidified atmosphere of 5% CO2 and 95% air.
For the source of denitrase, cells were harvested at 90–95% confluence by gentle pipetting, resuspended in 20 mM Tris (pH 7.4), 2 × PIC, and lysed by sonication.
For stimulation by lipopolysaccharide (LPS), RAW 264.7 cells were grown in the same conditions described above. At 50% confluence, LPS was added to make the final concentration 20 mg/ml and incubated for 16 h.
Peroxynitrite Treatment of RAW 264.7 Cells.
RAW 264.7 cells were harvested by gentle pipetting and resuspended in PBS. Peroxynitrite was added to the suspension with continuous stirring to achieve 0.5 mM as the final concentration. Diluted HCl was added to maintain pH and osmolality for each addition of peroxynitrite. This combined treatment was repeated 1–10 times. The treated cells were washed with PBS twice, suspended in 20 mM Tris (7.4), 2× PIC and lysed by sonication. The soluble fraction was separated by ultracentrifugation (100,000 × g, 60 min, 4°C). The particulate fraction was washed with PBS/1× PIC for five times and extracted with 20 mM Tris (7.4)/2% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) to obtain “CHAPS solubilized fraction.”
2D Membrane Assay for Denitrase.
Proteins in the fractions of peroxynitrite-treated cells were precipitated in 80% acetone, washed with 80% acetone three times, and subjected to the 2D gels. For the first dimension, 250–800 μg of protein per strip was applied to ReadyStrip IPG strips 11 cm from Bio-Rad. To compare the results of final immunoblotting, two strips developed side by side were used as one set for each experiment.
For the second dimension, a pair of strips was developed on an ExcelGel SDS Gradient 8–18% from Amersham Pharmacia. The gel was transferred onto poly(vinylidene difluoride) membrane. Then the membrane was blocked with 5% skim milk/Tris-buffered saline with 0.1% Tween 20 for 1 h at room temperature, washed with PBS five times, cut into two pieces, and incubated for 1 h at 37°C with reaction solution (1× PBS/1× PIC/1× PIP/1× CoF/ribonuclease A 250 μg/ml), which contains (sample) or does not contain (control) cell lysate at final concentration of 0.4 mg/ml protein.
After incubation, the membranes were washed with Tris-buffered saline with 0.1% Tween 20 for three times, 1.5 M NaCl once, and Tris-buffered saline with 0.1% Tween 20 for five more times. Then they were incubated with 5% skim milk/Tris-buffered saline with 0.1% Tween 20 for 30 min at room temperature, followed by incubation with anti-nitrotyrosine mAb from Upstate Biotechnology for 16 h at 4°C. They were developed with ECL plus reagent according to the manufacture's instructions.
The 1D Membrane Assay for Denitrase.
Purified recombinant human Histone H1.2 was nitrated by either incubating with 0.5 mM peroxynitrite/PBS for 20 min at room temperature or by incubating with 20 mM Tris (7.4), 2 mM NaNO2, 1 mM H2O2, and 10 μM horseradish peroxidase for 16 h at room temperature. Then 2.5 μg of Histone H1.2 per lane was applied for a 12.5% SDS/PAGE and transferred to poly(vinylidene difluoride) membrane. Each lane was separated, and the pieces of membrane were subjected to the exactly same procedure as 2D membranes. The resulting x-ray film was scanned and analyzed densimetrically by using quantity one software from Bio-Rad. Each experiment was repeated at least three times.
Amino Acid Sequencing.
The 2D gel was stained with Coomassie brilliant blue. The spot of interest was excised and sequenced by the Protein Core Laboratory at Baylor College of Medicine by using matrix-assisted laser desorption ionization mass spectrometric analysis.
Results
Strategy to Search for a Substrate of Denitrase.
RAW 264.7 is a mouse cell line, which has many similarities with macrophages. LPS treatment will activate these cells, changing the morphology as well as their biological properties in a manner similar to macrophages. Among the features acquired, the induction of inducible NOS (NOS-2) activity and myeloperoxydase activity combined with increased oxidative stress, these cells provide favorable conditions for nitrotyrosine formation, which is thought to interfere with the cellular functions. This situation raises a question as to why macrophage or RAW 264.7 cells can survive and function under such stressful circumstances. We hypothesized that these cells have mechanisms, which can reduce or remove protein nitration products and allow the cells to survive. According to this hypothesis, both denitrase activity and its substrates should exist in these activated RAW 264.7 cells.
As is illustrated in Fig. 1, a strategy was developed to search for a specific substrate for denitrase in RAW 264.7 cells. To search for any putative substrate for denitrase, whole RAW 264.7 cells were treated with peroxynitrite. They were washed with PBS, lysed by sonication, and fractionated by using the ultracentrifuge. The fractions were subjected to 2D gel electrophoresis and blotted onto poly(vinylidene difluoride) membranes. A pair of membranes were prepared under similar conditions and blocked with 5% skim milk. One membrane was incubated with cell lysate from LPS-treated RAW 264.7 cells, as a source of denitrase, whereas the other membrane was incubated with control solution. After washing, the membranes were treated with anti-nitrotyrosine Ab. Comparison of the pair of membranes permitted is to identify proteins as possible substrates for denitrase.
Figure 1.
Strategy for searching for substrates for denitrase. To obtain a library of nitrotyrosine-containing proteins, RAW 264.7 cells were treated with peroxynitrite. Sequential extraction according to solubility was performed and each fraction was subjected to 2D membrane assay for denitrase. A pair of samples was applied for 2D electrophoresis and transferred onto poly(vinylidene difluoride) membranes. The membranes were blocked with milk and cut into two pieces of which one was incubated with lysate from LPS-treated RAW 264.7 cells and the other with control solution. The membranes were washed with 1.5 M NaCl, blocked again, and treated with anti-nitrotyrosine Ab. The resulting immunoreactive spots were compared between these membranes, and the spots that showed a decrease of Ab staining intensity after incubation with lysate were assumed to be denitrase substrates and were subjected to peptide sequencing.
2D Membrane Assay for Denitrase.
Soluble fractions or CHAPS solubilized fractions of peroxynitrite-treated RAW 264.7 cells were subjected to the 2D membrane assay for denitrase (Fig. 2). Several spots (Fig. 2, arrowhead and asterisk) showed a decrease in nitrotyrosine immunoreactivity, whereas some spots (double asterisk) showed an increase. The spot from the CHAPS solubilized fraction [pI = 8.5, Mr 45 kDa (Fig. 2, arrowhead)] was the most prominent to lose nitrotyrosine immunoreactivity. This spot was excised from the 2D gel stained with Coomassie brilliant blue and sequenced by the peptide mass fingerprint method by using matrix-assisted laser desorption ionization–time-of-flight mass spectrometric analysis. This protein was found out to be mouse Histone H1.2, which has a high degree of homology between species. This protein has only one tyrosine residue in its amino acid sequence.
Figure 2.
A 2D membrane assay for denitrase. The soluble fraction (Upper) and the CHAPS solubilized fraction (Lower) from peroxynitrite-treated RAW 264.7 cells were subjected to 2D membrane assay for denitrase. A spot corresponding pI = 8.5, Mr of 45 kDa (arrow head) was found to disappear after incubation with cell lysate. This spot was excised and found out to be mouse Histone H1.2.
The 1D Membrane Assay for Denitrase.
Further investigation of denitrase activity was done by using purified recombinant human Histone H1.2, which is commercially available. Human Histone H1.2 was treated with peroxynitrite and subjected to the 1D membrane assay (Fig. 3A). With either Upstate Biotechnology or Zymed Abs against nitrotyrosine, nitration of Histone H1.2 decreased after incubation with cell lysate from LPS-treated RAW 264.7 cells. This also was confirmed when the resulting bands for assay were quantified by densimetric analysis (Fig. 3B).
Figure 3.
A 1D membrane assay for denitrase. Purified recombinant human Histone H1.2 was nitrated and subjected to 1D membrane assay for denitrase. (A) CTL, membrane treated with control solution; L/R, membrane treated with cell lysate from LPS-treated RAW 264.7 cells; (Z), membrane detected by anti-nitrotyrosine from Zymed; (U), membrane detected by anti-nitrotyrosine from Upstate Biotechnology. Nitrotyrosine immunoreactivity was detected with two different anti-nitrotyrosine mAbs on the control membrane at the Mr of Histone H1.2. These signals of nitration were almost abolished after incubation of the membranes with cell lysate from LPS-treated RAW 264.7 cells. (B) The signals of 1D membrane assay for denitrase were quantified densitometrically. (C) Normalization of signals by Histone H1 Ab. The membrane incubated with LPS-treated RAW 267.4 cell lysate showed less intensity of nitrotyrosine immunoreactivity but similar intensity for anti-Histone H1 Ab.
Further confirmation by normalization of Histone H1 immunoreactivity was performed (Fig. 3C). Whereas immunoreactivity for nitrotyrosine decreased after the membrane was incubated with RAW 264.7 cell lysate, the immunoreactivity for Histone H1 was not reduced. This result indicated that the reduction of nitrotyrosine immunoreactivity is not due to the nonspecific loss of Histone H1.2 protein.
Characterization of Denitrase Activity.
The time course of the denitrase reaction was assessed by incubating pieces of the membrane with cell lysate for various times. The immunoreactivity for nitrotyrosine was reduced according to the incubation time (Fig. 4).
Figure 4.
Time course analysis of denitrase activity. (A) Each piece of membrane containing nitrated histone H1.2 was incubated with cell lysate from LPS-treated RAW 264.7 cells for various lengths of time. The signals for the 1D membrane assay are shown. (B) The signals were quantified densitometrically and plotted according to the incubation time. Note that the y axis is logarithmic. A decrease of signal in accordance with the incubation time was seen.
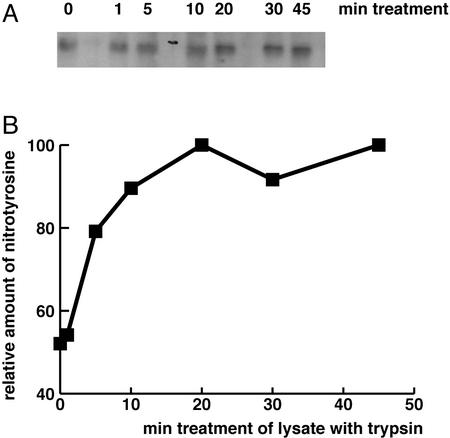
To confirm that this activity is due to the function of an enzymatic protein, trypsin treatment of the cell lysate was performed. After dialysis, the cell lysate mixture was aliquoted and incubated with trypsin (20 μg/ml) for various lengths of time (0–60 min). Then, using these aliquots, the 1D membrane assay was performed. The denitrase activity was destroyed after incubation with trypsin in a time-dependent manner (Fig. 5).
Figure 5.
Time-dependent effect of trypsin on the denitrase activity. The cell lysate from LPS-treated RAW 264.7 cells was incubated with trypsin for various lengths of time. (A) Resulting signals of the 1D membrane assay for denitrase are shown. (B) The signals were densitometrically quantified and plotted for the incubation time. A 20-min incubation with trypsin destroyed denitrase activity almost completely.
To confirm that this phenomenon is due to the effect of the specific decrease in nitrated Histone H1.2 instead of nitrotyrosine-specific degradation by proteasome (23, 24), lactacystin was used to inhibit proteasome activity. Lactacystin did not inhibit denitrase activity, suggesting that the effect is not due to proteasome (Fig. 6).
Figure 6.
Substrate specificity and the effect of proteasome inhibitor. (A) Nitrated Cu/Zn SOD was subjected to the 1D membrane assay and compared to Histone H1.2. The nitrotyrosine immunoreactivity of Histone H1.2 (upper arrow) was abolished by incubation with cell lysate from LPS-treated RAW 264.7 cells, whereas the immunoreactivity of nitrated Cu/Zn SOD (lower arrow) was not. In addition, a proteasome inhibitor, lactacystin (L) did not suppress this effect of cell lysate. CTL, membrane treated with control solution; L/R, membrane treated with cell lysate from LPS-treated RAW 264.7 cells. (B) The signals were quantified densitometrically.
To study the specificity of substrate, nitrated copper/zinc superoxide dismutase (Cu/Zn SOD) was subjected to the 1D membrane assay and compared to Histone H1.2. In contrast to Histone H1.2, Cu/Zn SOD, which also has a single tyrosine residue, showed no significant decrease in nitrotyrosine immunoreactivity after incubation with cell lysate (Fig. 6). Similar results were obtained when nitrated BSA or phosphoglycerate kinase was used as substrate. (data not shown). These results suggest sequence specificity of denitrase activity, which was also observed in the 2D membrane assay.
The induction of denitrase activity was assessed by comparing LPS-treated and nontreated RAW 264.7 cells (Fig. 7A). Nonstimulated cell showed denitrase activity, but LPS treatment increased the activity. Boiling of cell lysate destroyed denitrase activity (Fig. 7B). This heat lability also suggests that denitrase activity is catalyzed by an enzymatic protein.
Figure 7.
Characterization of denitrase activity. (A) Resulting immunoreactivity for nitrotyrosine with the 1D membrane assay (Upper) and its quantification (Lower) are shown. CTL, membrane treated with control solution; R, membrane treated with nontreated RAW 264.7 cell lysate; L/R, membrane treated with cell lysate from LPS-treated RAW 264.7 cells. LPS-treated and nontreated RAW 264.7 cell lysates were analyzed for denitrase activity. Both lysates showed a significant amount of denitrase activity. (B) Cell lysate from LPS-treated RAW 264.7 cells was boiled and analyzed. The boiling procedure abolished denitrase activity in the lysate. (C) Effect of DNase, dialysis of the lysate to remove intrinsic cofactors and removal of the additional cofactors mixture were analyzed. These treatments did not change denitrase activity significantly.
To test whether the denitrase is a cofactor requiring enzyme, several experiments were performed (Fig. 7C). Addition of EDTA or DTT did not affect denitrase activity. Elimination of the cofactor mixture including NADPH, NADP, NADH, NAD, FAD, FMN, ATP, GTP, and glutathione did not affect the activity. Furthermore, denitrase activity was retained after dialysis. These experiments indicated that denitrase did not seem to require any additional cofactors or that the required cofactors, if any, are associated with the enzyme.
The dialysis membrane has a 3.5-kDa Mr cutoff. Furthermore, an ultrafiltration experiment by using a YM30 membrane, which has a 30-kDa Mr cutoff, revealed that the size of denitrase activity was >30 kDa (data not shown).
Discussion
We developed an assay for a putative denitrase activity and found Histone H1.2 as a substrate for denitrase. Histone H1.2 has only one tyrosine residue in the molecule, which ensures the exact position of the nitrotyrosine substrate. Recombinant and purified Histone H1.2 was chemically nitrated and subjected to the denitrase assay to characterize the enzyme activity. Denitrase activity behaved as an enzymatic activity because the reaction was time dependent and was destroyed by heat or trypsin. The activity was shown to be specific for some substrates including Histone H1.2, was different from proteasome activity, and required no additional cofactors. These findings demonstrate the existence of an activity and describe a simple assay for further characterization and purification of the enzyme.
Combining 2D gel electrophoresis with on-blot enzyme assay (27), we developed an assay strategy for identifying the substrates for denitrase. In the assay described previously (23, 25, 26), the substrate was incubated with the enzyme in solution. Then, SDS/PAGE was done followed by immunoblotting with anti-nitrotyrosine Ab for assessment of denitrase activity. These studies suggested that the nitrotyrosine immunoreactivity was reduced, whereas the amount of substrate protein was not altered. In some cases, the amount of substrate by using Coomassie brilliant blue staining was carried out (23). However, in the assay, nitrotyrosine immunoreactivity and protein staining must be in a linear range. In contrast, the membrane assay for denitrase used here is more resistant against artifacts from proteolysis because the substrate is bound to the membrane. This assures that the immunoreactive group will stay at the same position on the membrane even if the substrate is digested by a protease, provided that the nitrated epitope remains on the membrane. Blocking membrane with 5% milk will coat the surface of the membrane with protein and also reduce the chance of nonspecific proteolysis. Taken together, nonspecific proteolysis should be negligible in this membrane assay. On the other hand, there might be a possibility that something from the lysate binds to the Histone H1.2 protein on the membrane and blocks the epitope of immunoreactivity. To solve this problem, the membrane was washed with 1.5 M NaCl, and possible binding molecules should be washed away. This concentration of salt is sufficient to destroy protein–protein interactions as well as protein–nucleic acid interactions (28). Among the spots that showed a decrease in nitrotyrosine immunoreactivity, the spot from the CHAPS solubilized fraction corresponding to a spot (pI = 8.5, Mr = 45 kDa) was excised and sequenced because it was the most prominent to lose nitrotyrosine immunoreactivity. The CHAPS solubilized fraction contained much less spots in the Coomassie-stained gel. This protein turned out to be Histone H1.2, which has only one tyrosine residue in its amino acid sequence. With the soluble fraction, some spots gained immunoreactivity after incubation with LPS-treated RAW 264.7 cell lysate (Fig. 2, double asterisk). This might be caused by nitration during the membrane incubation because LPS treatment is known to increase nitrotyrosine immunoreactivity in macrophages (29).
In the present study, Histone H1.2 was artificially nitrated, which raises the question whether Histone H1.2 is a natural substrate for denitrase. Some reports describe chemical nitration of histones (30–32), whereas others demonstrate that nuclear staining for nitrotyrosine by immunohistochemistry is often observed in tissues and in NO-exposed cell cultures (33–35). Recently, Haqqani et al. (36) described that histones are the most prominent nitrated proteins in the Mutatect tumor tissue as well as in the cultured Mutatect cells that are exposed to NO. They also suggest tyrosine nitration of Histone H1 in vivo, which support the view that Histone H1.2 might be an intrinsic substrate for denitrase. Actually, in the NO exposed Mutatect cells, they observed a discrepancy in the time course of nitration between histone and other proteins. They speculate that the delay in histone nitration may be due to relative inaccessibility of nuclei to the nitrating reactive nitrogen oxide species. But it may be due to the existence of mechanisms that reduce nitrated histone, namely denitrase.
Although both of the nitration reactions mediated by peroxynitrite and horseradish peroxidase are already established (26, 37), they may involve different mechanisms of nitration (5), possibly resulting in different tyrosine residues to be nitrated. But in the specific cases of Histone H1.2 or Cu/Zn SOD, this is not a problem, because these proteins have only one tyrosine residue in the entire protein molecule. When nitration was mediated by horseradish peroxidase, the horseradish peroxidase itself was also nitrated and caused a strong signal. But this signal appears in the area corresponding to a greater Mr than Histone H1.2, which can be distinguished easily.
Antinitrotyrosine Abs are not as well characterized as antiphosphotyrosine Abs. We used two different mAbs, and both of them recognized nitrated Histone H1.2, which assured us that the immunoreactivity is specific for nitrotyrosine. In other experiments (data not shown), Western immunoblot staining of nitrotyrosine proteins can be blocked with pretreatment of these Abs with nitrotyrosine.
Some of the nitrotyrosine-containing proteins are reported to be degraded by proteasome compared with the native protein (21, 22). All of these activities thus far are inhibited by lactacystin. But, the denitrase activity we report here is not inhibited by lactacystin (Fig. 6), which indicates that denitrase activity is distinguishable from the effect of proteasome.
Quantification of nitrotyrosine immunoreactivity was performed by scanning the resulting x-ray films from immunoblotting. By this method, the comparison of band intensity within one film should be reliable, whereas the comparison between different films is not.
Furthermore, every preparation of nitrated Histone H1.2 may have different degrees of nitration. This makes it difficult to compare activities between films or to obtain absolute values of activity. The nature of the denitrase reaction is not known; nor it is known if a single or multiple enzymes participation in the alteration of the epitope. These studies are in progress.
In conclusion, this study provides an important approach for investigation of denitrase to further characterize and purify this activity. Purification of denitrase has been planned since its existence was first discussed (23), because it is attractive to assume that tyrosine nitration is reversible and contributes to signal transduction. The present study is also important in this context, because if denitrase does contribute to signal transduction, its substrates should be specific proteins rather than a wide range of nonspecific proteins.
Acknowledgments
These studies were supported by awards from the National Institutes of Health, the J. S. Dunn, G. Harold and Leila Y. Mathers, and Welch Foundations, and the U.S. Department of Defense.
Abbreviations
- LPS
lipopolysaccharide
- CHAPS
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
- PIC
protease inhibitor complete
- Cu/Zn SOD
copper/zinc superoxide dismutase
References
- 1.Katsuki S, Arnold W, Mittal C K, Murad F. J Cyclic Nucleotide Res. 1977;3:23–35. [PubMed] [Google Scholar]
- 2.Arnold W P, Mittal C K, Katsuki S, Murad F. Proc Natl Acad Sci USA. 1977;74:3203–3207. doi: 10.1073/pnas.74.8.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murad F, Mittal C K, Arnold W P, Katsuki S, Kimura H. Adv Cyclic Nucleotide Res. 1978;9:145–158. [PubMed] [Google Scholar]
- 4.Murad F. Braz J Med Biol Res. 1999;32:1317–1327. doi: 10.1590/s0100-879x1999001100001. [DOI] [PubMed] [Google Scholar]
- 5.Hanafy K A, Krumenacker J S, Murad F. Med Sci Monit. 2001;7:801–819. [PubMed] [Google Scholar]
- 6.Ignarro L, Murad F. In: Advances in Pharmacology. Ignarro L, Murad F, editors. Vol. 34. San Diego: Academic; 1995. pp. 1–516. [Google Scholar]
- 7.Murad F. J Am Med Assoc. 1996;276:1189–1192. [Google Scholar]
- 8.Murad F. Recent Prog Horm Res. 1998;53:43–60. [PubMed] [Google Scholar]
- 9.Davis K L, Martin E, Turko I V, Murad F. Annu Rev Pharmacol Toxicol. 2001;41:203–236. doi: 10.1146/annurev.pharmtox.41.1.203. [DOI] [PubMed] [Google Scholar]
- 10.Greenacre S A, Ischiropoulos H. Free Radical Res. 2001;34:541–581. doi: 10.1080/10715760100300471. [DOI] [PubMed] [Google Scholar]
- 11.Castro L, Rodriguez M, Radi R. J Biol Chem. 1994;269:29409–29415. [PubMed] [Google Scholar]
- 12.Marcondes S, Turko I V, Murad F. Proc Natl Acad Sci USA. 2001;98:7146–7151. doi: 10.1073/pnas.141222598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berlett B S, Friguet B, Yim M B, Chock P B, Stadtman E R. Proc Natl Acad Sci USA. 1996;93:1776–1780. doi: 10.1073/pnas.93.5.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gow A J, Duran D, Malcolm S, Ischiropoulos H. FEBS Lett. 1996;385:63–66. doi: 10.1016/0014-5793(96)00347-x. [DOI] [PubMed] [Google Scholar]
- 15.Greis K D, Zhu S, Matalon S. Arch Biochem Biophys. 1996;335:396–402. doi: 10.1006/abbi.1996.0522. [DOI] [PubMed] [Google Scholar]
- 16.Cassina A M, Hodara R, Souza J M, Thomson L, Castro L, Ischiropoulos H, Freeman B A, Radi R. J Biol Chem. 2000;275:21409–21415. doi: 10.1074/jbc.M909978199. [DOI] [PubMed] [Google Scholar]
- 17.Zhu S, Basiouny K F, Crow J P, Matalon S. Am J Physiol. 2000;278:L1025–L1031. doi: 10.1152/ajplung.2000.278.5.L1025. [DOI] [PubMed] [Google Scholar]
- 18.Eiserich J P, Estevez A G, Bamberg T V, Ye Y Z, Chumley P H, Beckman J S, Freeman B A. Proc Natl Acad Sci USA. 1999;96:6365–6370. doi: 10.1073/pnas.96.11.6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong S K, Yim M B, Stadtman E R. Proc Natl Acad Sci USA. 1996;93:3377–3382. doi: 10.1073/pnas.93.8.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin B L, Wu D, Jakes S, Graves D J. J Biol Chem. 1990;265:7108–7111. [PubMed] [Google Scholar]
- 21.Grune T, Blasig I E, Sitte N, Roloff B, Haseloff R, Davies K J. J Biol Chem. 1998;273:10857–10862. doi: 10.1074/jbc.273.18.10857. [DOI] [PubMed] [Google Scholar]
- 22.Souza J M, Choi I, Chen Q, Weisse M, Daikhin E, Yudkoff M, Obin M, Ara J, Horwitz J, Ischiropoulos H. Arch Biochem Biophys. 2000;380:360–366. doi: 10.1006/abbi.2000.1940. [DOI] [PubMed] [Google Scholar]
- 23.Kamisaki Y, Wada K, Bian K, Balabanli B, Davis K, Martin E, Behbod F, Lee Y C, Murad F. Proc Natl Acad Sci USA. 1998;95:11584–11589. doi: 10.1073/pnas.95.20.11584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenacre S A, Evans P, Halliwell B, Brain S D. Biochem Biophys Res Commun. 1999;262:781–786. doi: 10.1006/bbrc.1999.1309. [DOI] [PubMed] [Google Scholar]
- 25.Kuo W N, Kanadia R N, Shanbhag V P. Biochem Mol Biol Int. 1999;47:1061–1067. doi: 10.1080/15216549900202183. [DOI] [PubMed] [Google Scholar]
- 26.Kuo W N, Kanadia R N, Shanbhag V P, Toro R. Mol Cell Biochem. 1999;201:11–16. doi: 10.1023/a:1007024126947. [DOI] [PubMed] [Google Scholar]
- 27.Tamura M, Gu J, Matsumoto K, Aota S, Parsons R, Yamada K M. Science. 1998;280:1614–1617. doi: 10.1126/science.280.5369.1614. [DOI] [PubMed] [Google Scholar]
- 28.Burgess R R. Methods Enzymol. 1991;208:3–10. doi: 10.1016/0076-6879(91)08003-z. [DOI] [PubMed] [Google Scholar]
- 29.Ding J W, Dickie J, O'Brodovich H, Shintani Y, Rafii B, Hackam D, Marunaka Y, Rotstein O D. Am J Physiol. 1998;274:L378–L387. doi: 10.1152/ajplung.1998.274.3.L378. [DOI] [PubMed] [Google Scholar]
- 30.Bustin M. Biochim Biophys Acta. 1971;251:172–180. doi: 10.1016/0005-2795(71)90100-0. [DOI] [PubMed] [Google Scholar]
- 31.Ptitsyn L A, Chepyzheva M A, Kolomiitseva G I, Senchenkov E P. Biokhimiia. 1978;43:1823–1829. [PubMed] [Google Scholar]
- 32.Prutz W A, Monig H, Butler J, Land E J. Arch Biochem Biophys. 1985;243:125–134. doi: 10.1016/0003-9861(85)90780-5. [DOI] [PubMed] [Google Scholar]
- 33.Beckmann J S, Ye Y Z, Anderson P G, Chen J, Accavitti M A, Tarpey M M, White R. Biol Chem Hoppe-Seyler. 1994;375:81–88. doi: 10.1515/bchm3.1994.375.2.81. [DOI] [PubMed] [Google Scholar]
- 34.Sandhu J K, Privora H F, Wenckebach G, Birnboim H C. Am J Pathol. 2000;156:509–518. doi: 10.1016/S0002-9440(10)64755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Behar-Cohen F F, Heydolph S, Faure V, Droy-Lefaix M T, Courtois Y, Goureau O. Biochem Biophys Res Commun. 1996;226:842–849. doi: 10.1006/bbrc.1996.1438. [DOI] [PubMed] [Google Scholar]
- 36.Haqqani A S, Kelly J F, Birnboim H C. J Biol Chem. 2002;277:3614–3621. doi: 10.1074/jbc.M105730200. [DOI] [PubMed] [Google Scholar]
- 37.Sampson J B, Ye Y, Rosen H, Beckman J S. Arch Biochem Biophys. 1998;356:207–213. doi: 10.1006/abbi.1998.0772. [DOI] [PubMed] [Google Scholar]