Abstract
Bet3 is a component of the transport protein particle complex involved in vesicular trafficking to and through the Golgi complex. X-ray structural analysis of human and mouse Bet3 revealed a hydrophobic tunnel inside the protein, which is occupied by a fatty acid linked to cysteine-68. We show here that Bet3 has strong self-palmitoylating activity. Incubation of purified Bet3 with [3H]palmitoyl-CoA (Pal-CoA) leads to a rapid and stoichiometric attachment of fatty acids to cysteine-68. Bet3 has an intrinsic affinity for Pal-CoA, and the palmitoylation reaction occurs at physiological pH values and Pal-CoA concentrations. Moreover, Bet3 is also efficiently palmitoylated at cysteine-68 inside vertebrate cells. Palmitoylation can occur late after Bet3 synthesis, but once the fatty acids are bound they are not removed, not even by disassembly of the Golgi complex. Narrowing the hydrophobic tunnel by exchange of alanine-82 with bulkier amino acids inhibits palmitoylation, both in vitro and inside cells, indicating that the fatty acid must insert into the tunnel for stable attachment. Finally, we show that palmitoylation of Bet3 plays a structural role. CD spectroscopy reveals that chemically deacylated Bet3 has a reduced melting temperature. As a consequence of its structural defect nonacylated Bet3 does not bind to TPC6, a further subunit of the transport protein particle complex, and is degraded inside cells.
Keywords: Golgi, protein structure
Bet3p was described in a synthetic lethality screen in yeast to block endoplasmic reticulum (ER) to Golgi transport and to genetically interact with the SEC21 subunit of the coatomer, with the Rab-like GTPase YPT1, and with the SNARE proteins BOS1 and SEC22 (1). Bet3p was subsequently identified to be a component of the multisubunit transport protein particle (TRAPP) tethering complex that plays an important role for the recruitment of ER-derived vesicles to the Golgi apparatus (2–4). Two TRAPP complexes have been characterized that differ in size, localization within the Golgi, and function (3). Both TRAPP complexes facilitate nucleotide exchange on the small GTPase Ypt1 (5, 6). Bet3 is the best-conserved of all TRAPP subunits and is thought to have a central role within TRAPP. Yeast Bet3p is stably associated with the Golgi and does not continuously cycle through the ER (2). In contrast, mammalian Bet3 is present mainly in the cytosol, and lower amounts are membrane-bound. Cytosolic Bet3 was shown to be present in a low- and a high-molecular-weight form, the latter probably representing the TRAPP complex (7). However, it is not known whether the TRAPP complex assembles in the cytosol or on the Golgi membrane.
X-ray crystallography with Bet3 from mice and humans revealed a novel dimeric protein fold, which is also present in a second TRAPP subunit, TPC6 (8–12). Biochemical and crystallographic studies showed that Bet3 and TPC6 form heterodimers that are likely to be functional modules of TRAPP. The Bet3–TPC6 subcomplex is thought to represent the core of TRAPP and to be involved in anchoring the complex to the Golgi membrane.
Surprisingly, the crystals of Bet3 revealed a long-chain fatty acid covalently bound via a thioester linkage to the conserved residue cysteine-68, even if the recombinant protein used for crystallization was purified from Escherichia coli, which lack enzymes for acylation of proteins (8, 9). The carbon chain is not exposed at the surface of the molecule but is completely buried inside a hydrophobic tunnel. The homologous TPC6 does not contain a hydrophobic tunnel and is not acylated (10, 11). Genetic studies with yeast showed that cysteine-68 is not essential for membrane binding of Bet3 and for yeast survival, but narrowing the tunnel by exchange of alanine-82 with bulkier amino acids led to conditional lethality, mistargeting of Bet3, and general vesicular trafficking defects along the secretory pathway. It was therefore hypothesized that palmitoylation of Bet3 is an artifact of the E. coli expression system. Instead, an acyl chain linked to a Golgi-resident membrane protein was proposed to insert into the hydrophobic tunnel, thereby promoting membrane binding of Bet3. The same deleterious effects on yeast survival and vesicular trafficking were noticed if positively charged amino acids on a flat surface of Bet3 were substituted with negatively charged residues, suggesting that they also contribute to membrane binding of Bet3 (8). However, overexpression of Trs33 (yeast homolog of TPC6) was able to restore growth of these mutants (11).
Palmitoylation is the attachment of fatty acids to cysteine residues of proteins. In contrast to other hydrophobic modifications, i.e., myristoylation or isoprenylation, palmitoylation may be dynamic, with rapid cycles of acylation and deacylation. Palmitoylation controls targeting of the modified protein to membranes or membrane subdomains, affects protein–protein interactions, or influences the stability of proteins (13, 14). Palmitoylation of proteins is also involved in intracellular transport. Palmitoyl-CoA (Pal-CoA), the lipid donor in the palmitoylation reaction, stimulates the Golgi transport assay, supporting budding as well as fusion of vesicles. It was suggested that this stimulatory effect might be due to palmitoylation of a protein, which thereby gets activated to function in the transport reaction (15–17).
Pal-CoA is capable of spontaneously S-acylating cysteine residues of several proteins and especially of peptides at their natural palmitoylation sites (18–25). Most of these reactions are unlikely to operate inside cells. They are either too slow or require unphysiologically high pH values or Pal-CoA concentrations to account for palmitoylation in vivo.
The enzymology of protein palmitoylation is now beginning to be elucidated. The family of DHHC-CRD proteins (polytopic membrane proteins with the sequence DHHC and a cysteine-rich domain) has recently been identified as acyltransferases for a variety of substrate proteins in yeast and mammals (14, 26, 27). Ykt6, a member of the SNARE family of proteins, possesses an enzymatic, autocatalytic activity in vitro: the N-terminal domain binds Pal-CoA and transfers palmitate to cysteine residues located in its C terminus and also to at least one other protein (28–30).
Results and Discussion
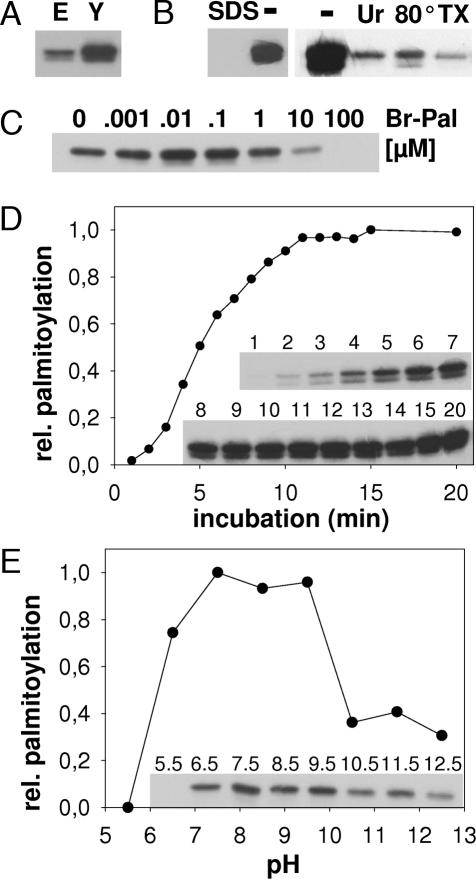
Human Bet3 purified from E. coli or from yeast was incubated with [3H]Pal-CoA, noncovalently bound lipids were extracted, and Bet3 was subjected to SDS/PAGE and fluorography (Fig. 1A). Bet3 from both sources shows strong [3H]palmitate incorporation, but palmitoylation is more effective with Bet3 from yeast, probably because more molecules are properly folded. Heating of Bet3 as well as inclusion of SDS, urea, and Triton X-100 in the reaction buffer inhibit palmitoylation, which is evidence for the specificity of the reaction (Fig. 1B). The reaction is inhibited by 2-bromo-palmitate (Br-Pal) at concentrations known to block protein palmitoylation inside cells (31), whereas low concentrations of Br-Pal stimulate Bet3 acylation (Fig. 1C). Although Br-Pal is frequently used to inhibit protein palmitoylation, its mechanism of action is not known. It may bind to the Pal-CoA binding site of Bet3 (see Fig. 2), but it cannot be covalently attached to the protein. Low Br-Pal concentrations might stabilize the Bet3 dimer by binding to only one subunit, and the other subunit might then perform its autocatalytic activity more efficiently. Furthermore, acylation proceeds very rapidly: 50% of total Bet3 acylation was completed after 5 min of labeling (Fig. 1D).
Fig. 1.
Purified Bet3 has self-palmitoylating activity. (A) Bet3 purified from E. coli (E; 19 μg) or from yeast (Y; 8 μg) was incubated with [3H]Pal-CoA for 30 min at 30°C. Samples were subjected to SDS/PAGE under nonreducing conditions and fluorography for 4 h. (B) Bet3 purified from yeast was incubated with [3H]Pal-CoA in the absence (−) or presence of SDS (0.1%), urea (Ur; 6.75 M), or Triton X-100 (TX; 0.1%) or heated to 80°C for 5 min before the assay was performed. (C) Increasing amounts of Br-Pal were added to the reaction with Bet3 purified from yeast. Palmitate alone had no effect on the reaction up to a concentration of 1 mM. (D) Bet3 purified from yeast was incubated with [3H]Pal-CoA for the times (in minutes) indicated. (Inset) The resulting fluorogram was quantified, and relative palmitoylation is plotted against time of incubation. (E) Bet3 purified from yeast was incubated with [3H]Pal-CoA at different pH values as indicated. (Inset) The resulting fluorogram was quantified, and relative palmitoylation is plotted against the pH.
Fig. 2.
Bet3 binds Pal-CoA at physiological concentrations. (A) His-Bet3-StrepII purified from E. coli was added to Pal-CoA agarose. Washed beads were eluted twice with 10 mM Pal-CoA and finally with SDS buffer. Comparable aliquots of input (IP), flow-through (FT), wash 1 (W1), wash 4 (W4), wash 9 (W9), and from eluates were subjected to SDS/PAGE and Coomassie staining. wt, wild type. (B) Bet3 purified from E. coli was incubated with Cibacron Blue Sepharose for 1 h. Beads were washed and incubated with increasing concentrations of Pal-CoA (Upper) or with CoA (Lower). Comparable aliquots of input (IP), flow-through (FT), and eluates were subjected to SDS/PAGE and Coomassie staining. (C) Bet3 C68S (CS) was incubated with Pal-CoA agarose (Upper) and Cibacron Blue Sepharose (Lower) and processed as described in A and B. (D) Soluble Cibacron Blue at the concentration indicated was added to the palmitoylation reaction with GST-Bet3 purified from E. coli. The resulting fluorogram is shown. (E) Bet3 from yeast (2 μM) was incubated with increasing concentrations of [3H]Pal-CoA for 5 min at 30°C. (Inset) The resulting fluorogram was quantified by densitometry, and the rate of palmitoylation in arbitrary units was plotted against the [3H]Pal-CoA concentration. The data points were fitted according to the Michaelis–Menten equation v = vmax × [S]/Km + [S], and the Km of Bet3 for Pal-CoA was calculated as 5.7 μM.
Acyl-chain transfer (either enzyme-mediated or chemical) occurs through nucleophilic attack of a cysteine in the substrate protein on the carbonyl of the C S bond in the Pal-CoA molecule (20). To act as good nucleophile the cysteine should be present in its deprotonated form. Spontaneous acylation is characterized by a linear increase in the palmitoylation efficiency when plotted against the pH (21). In contrast, palmitoylation of Bet3 has a broad pH optimum at neutral pH but is completely inhibited at acidic pH and reduced at basic pH (Fig. 1E). The residual labeling at basic pH may be due to acylation at irrelevant sites.
S bond in the Pal-CoA molecule (20). To act as good nucleophile the cysteine should be present in its deprotonated form. Spontaneous acylation is characterized by a linear increase in the palmitoylation efficiency when plotted against the pH (21). In contrast, palmitoylation of Bet3 has a broad pH optimum at neutral pH but is completely inhibited at acidic pH and reduced at basic pH (Fig. 1E). The residual labeling at basic pH may be due to acylation at irrelevant sites.
Assuming an enzymatic mechanism, Bet3 should have an intrinsic affinity for Pal-CoA. Bet3 shows chromatographic properties that are consistent with this assumption. Bet3 binds almost quantitatively to Pal-CoA agarose (Fig. 2A) and to Cibacron Blue Sepharose (Fig. 2B), a medium for enzymes with an affinity for adenyl-containing cofactors such as CoA or Pal-CoA. It can be eluted from Cibacron Blue with Pal-CoA (Fig. 2B Upper), but not with CoA (Fig. 2B Lower), suggesting that Bet3 has a higher affinity for Pal-CoA compared with CoA and that Pal-CoA competes with Cibacron Blue for the same binding site on Bet3. We performed the same binding experiments with the nonacylated C68S mutant of Bet3 (see next page). Bet3 C68S also binds to Pal-CoA agarose and to Cibacron Blue Sepharose, although elution with Pal-CoA is somewhat less efficient, probably because less protein is properly folded (Fig. 2C). Moreover, [3H]palmitoylation of wild-type Bet3 is completely inhibited by adding cold Pal-CoA (data not shown) or soluble Cibacron Blue at a concentration of 250 μM (Fig. 2D), indicating that binding of Pal-CoA is a prerequisite for the self-palmitoylating activity of Bet3.
To calculate the Pal-CoA concentration at which the rate of Bet3 palmitoylation is most efficient we incubated Bet3 for 5 min with increasing amounts of [3H]Pal-CoA (Fig. 2E). Quantification of the fluorogram shows that the reaction begins to saturate at 10 μM Pal-CoA. Analysis of these data according to Michaelis–Menten yielded an apparent Km of Bet3 for Pal-CoA of 5.7 μM. Although the Michaels–Menten equation might not be accurate for a reaction were the enzyme is used up, the data nevertheless indicate that palmitoylation of Bet3 occurs in the range reported for Pal-CoA concentrations inside cells (32).
From this experiment we analyzed the stoichiometry of Bet3 palmitoylation. Liquid scintillation counting of excised [3H]palmitoylated Bet3 at saturating conditions (20 μM Pal-CoA) revealed that ≈50 pmol of palmitate were transferred to 200 pmol of protein. Thus, 25% of all Bet3 molecules were acylated in this reaction. Mass spectrometry analysis revealed that approximately two-thirds of purified Bet3 is already palmitoylated (data not shown). Thus, only one-third of Bet3's palmitoylation sites are available in the assay. Self-palmitoylation of Bet is 2-fold more efficient compared with the glyceraldehyde phosphate dehydrogenase (data not shown), which is known to contain one molecule of palmitate per tetramer (33). In summary, circumstantial evidence shows that palmitoylation of Bet3 in vitro occurs at an almost stoichiometric ratio.
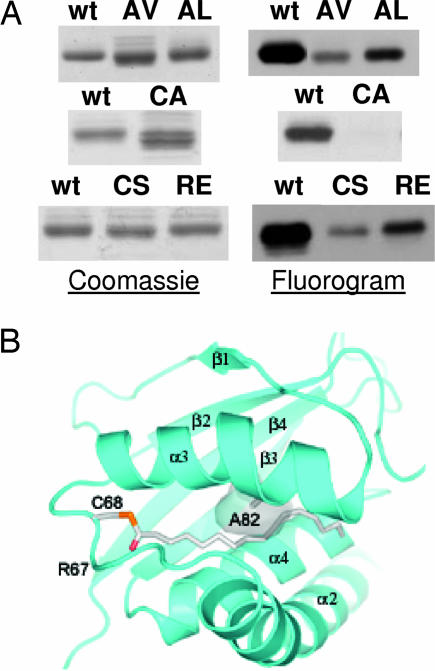
[3H]Palmitate labeling of Bet3 can be cleaved with hydroxylamine and with mercaptoethanol, indicating a thioester fatty acid linkage (data not shown). Exchange of cysteine-68 by serine or by alanine almost completely inhibited [3H]palmitate labeling of Bet3 (Fig. 3A). Thus, self-palmitoylation of Bet3 occurs at cysteine-68. Does acylation of Bet3 in vitro require an open hydrophobic tunnel? Modeling a replacement of alanine-82 in the middle of the tunnel by a bulkier amino acid such as valine narrows the pocket such that it can no longer accommodate a hydrocarbon chain (Fig. 3B). Acylation of Bet3 A82V and of A82L is marginal (Fig. 3A), indicating that the fatty acid must insert into the tunnel to become stably attached. Multiple sequence alignment of Bet3 sequences showed adjacent to the palmitoylated cysteine an arginine, which is completely conserved from yeast to man (8, 9). Basic amino acids in the vicinity of palmitoylated cysteines often affect the acylation reaction, probably by decreasing the pKa of the cysteine's -sulfhydryl group (19, 20). Replacing arginine-67 by glutamic acid almost completely blocked palmitoylation of Bet3 (Fig. 3A).
Fig. 3.
Palmitoylation occurs on cysteine-68, requires arginine-67, and is inhibited by channel-blocker mutations. (A) Similar amounts of GST-Bet3 wild-type (wt), C68S, C68A, A82V, A82L, and R67E were incubated with [3H]Pal-CoA. The Coomassie-stained gels (Left) and the fluorograms (Right) are shown. Similar results were obtained with Bet3 without GST tag. (B) View of the hydrophobic tunnel of Bet3 with the buried palmitoyl group. Secondary structure elements and cysteine-68 carrying the covalent modification are labeled. Alanine-82 is shown in stick representation, and the modeled channel-blocking mutation to valine is drawn as a semitransparent surface.
Bet3 has self-palmitoylating activity in vitro, which is likely to be of physiological significance. Bet3 is the only known eukaryotic protein containing covalently bound fatty acids when purified from E. coli, which lack enzymes for palmitoylation (8). Thus, even in cells acylation of Bet3 does not require an exogenous enzyme. If self-palmitoylation of Bet3 were not of physiological relevance, it is unclear how it is suppressed. Several spontaneous palmitoylation reactions are inhibited by the acyl-CoA binding protein (ACBP) (34, 35), but human Bet3 is also palmitoylated in yeasts (9) that contain functional ACBP (32).
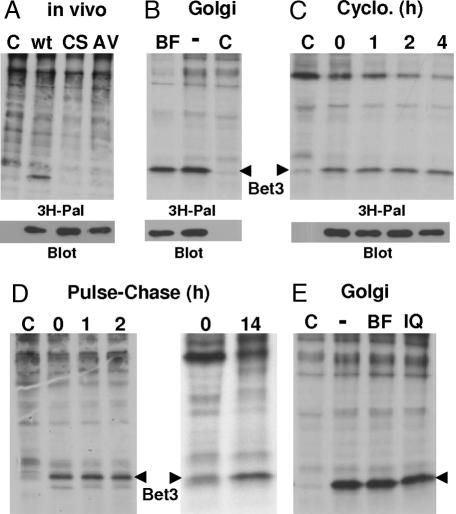
Is palmitoylation of mammalian Bet3 an artifact of the E. coli or yeast expression systems or detectable only with purified protein? BHK cells transfected with the human Bet3 gene were long-term labeled with [3H]palmitate. SDS/PAGE of total cellular proteins and fluorography showed that Bet3 is the major palmitoylated protein under steady-state conditions (Fig. 4A). In contrast, Bet3 mutants C68S and A82V are not labeled with [3H]palmitate, although both proteins are expressed at similar levels compared with the wild-type protein.
Fig. 4.
Wild-type Bet3, but not C68S and A82V, is palmitoylated inside cells. (A) BHK cells were mock-transfected (C) or transfected with wild-type myc-Bet3 (wt), C68S myc-Bet3 (CS), or A82V myc-Bet3 (AV) and were labeled for 19 h with [3H]palmitate. Cells were extracted with chloroform/methanol, and precipitated proteins were subjected to SDS/PAGE under nonreducing conditions and fluorography ([3H]palmitate) (Upper) or Western blotting with anti-myc antibodies (Lower). (B) Mock-transfected (C) or myc-Bet3-transfected BHK cells were treated with BFA (BF) for 30 min before labeling with [3H]palmitate for 3 h in the presence of BFA. (C) Protein synthesis was blocked in mock-transfected (C) or myc-Bet3-transfected BHK cells by the addition of cycloheximide (Cyclo.) for 0, 1, 2, or 4 h before labeling with [3H]palmitate for 2 h. (D Left) Mock-transfected (C) or myc-Bet3-transfected BHK cells were labeled 24 h later with [3H]palmitate for 19 h. The labeling medium was removed, and cells were processed immediately (lane 0) or chased in the presence of cycloheximide for 1 h (lane 1) or 2 h (lane 2). (D Right) BHK cells were labeled for 2 h and chased for 0 or 14 h. (E) Mock-transfected (C) or myc-Bet3-transfected BHK cells were labeled with [3H]palmitate for 3 h. Cells were chased in the presence of cycloheximide in the absence (−) or presence of BFA (BF) or ilimaquinone (IQ) for 1 h.
Brefeldin A (BFA) is known to inhibit palmitoylation of several proteins that are in transit to the plasma membrane. These proteins (including SNAP-25 and GAP-43) are palmitoylated in a compartment distal from the block in intracellular transport exerted by the drug, i.e., in the Golgi. In contrast, palmitoylation of other proteins, such as N-ras and H-ras, is not inhibited by BFA because their palmitoylating enzyme is located in a more proximal compartment, i.e., in the late ER or in the intermediate compartment (13). Bet3 does not cycle between the ER and the Golgi, and its palmitoylation should therefore be blocked by BFA if it requires a Golgi-located enzyme. We preincubated cells with BFA before labeling with [3H]palmitate, but palmitoylation of Bet3 remains unimpaired (Fig. 4B).
Palmitoylation is a posttranslational modification, but initial fatty acid attachment to most proteins begins a few minutes after their synthesis and is completed after 1 h (36). Protein synthesis in BHK cells transfected with the Bet3 gene was blocked by the addition of cycloheximide for 1, 2, or 4 h before labeling with [3H]palmitate. This treatment continuously reduced labeling of most cellular proteins, but palmitoylation of Bet3 was only marginally affected (Fig. 4C). Thus, synthesis of Bet3 and its palmitoylation are not strictly coupled, and there is a pool of nonacylated Bet3 with an open hydrophobic tunnel, which can be used to bind Bet3 to the Golgi membrane (8).
Protein palmitoylation might be reversible, and half-times for depalmitoylation as fast as 15 min have been estimated from pulse–chase experiments (13, 37). To analyze whether the fatty acids bound to Bet3 turn over rapidly, a pulse–chase experiment with [3H]palmitate was performed. However, no cleavage of fatty acids from [3H]palmitoylated Bet3 was obvious; even after 14 h of chase the fatty acids were stably attached (Fig. 4D). There is even an increase in the labeling intensity of Bet3 detectable after prolonged chase, which might be due to cleavage of [3H]palmitate from lipids and its reutilization after activation as lipid donor in the palmitoylation reaction (38).
We finally asked whether disassembly of the Golgi apparatus induced by BFA or by ilimaquinone might promote cleavage of the fatty acids from Bet3 (39). BHK cells were transfected with the Bet3 gene, labeled with [3H]palmitate, and chased in the presence of BFA or ilimaquinone for 1 h, which is sufficient time to cause complete dispersal of the Golgi. However, cleavage of fatty acids from Bet3 was not detectable (Fig. 4E). Thus, the fatty acids are stably attached to Bet3, even in the absence of a functional Golgi apparatus.
We found that Bet3 mutant proteins lacking efficient self-palmitoylation activity were harder to express, and recombinant proteins were unstable after cleavage of the GST tag. It is also striking that in all crystal structures determined Bet3 was found to be palmitoylated, indicating that the fatty acids have a beneficial effect on the protein's structure (8–11). We quantitatively determined the difference in protein stability between acylated Bet3 and Bet3 gently depalmitoylated with hydroxylamine. Temperature melting curves of both samples were recorded with CD spectroscopy. Both proteins revealed similar CD spectra in the beginning (data not shown), indicating that hydroxylamine had no effect on the secondary structure of the protein. However, unfolding of deacylated Bet3 starts earlier (Fig. 5A). The melting temperatures differ by 10°C, proving a significant decrease in protein stability upon loss of palmitoylation. The lack of a ligand in the hydrophobic tunnel might lead to a partial misfolding that leads to the destabilization of the protein. At high temperatures both acylated and nonacylated Bet3 are not completely unfolded but form β-aggregates, as seen in the absence of a plateau in the melting curves and characteristic CD spectra of heated samples.
Fig. 5.
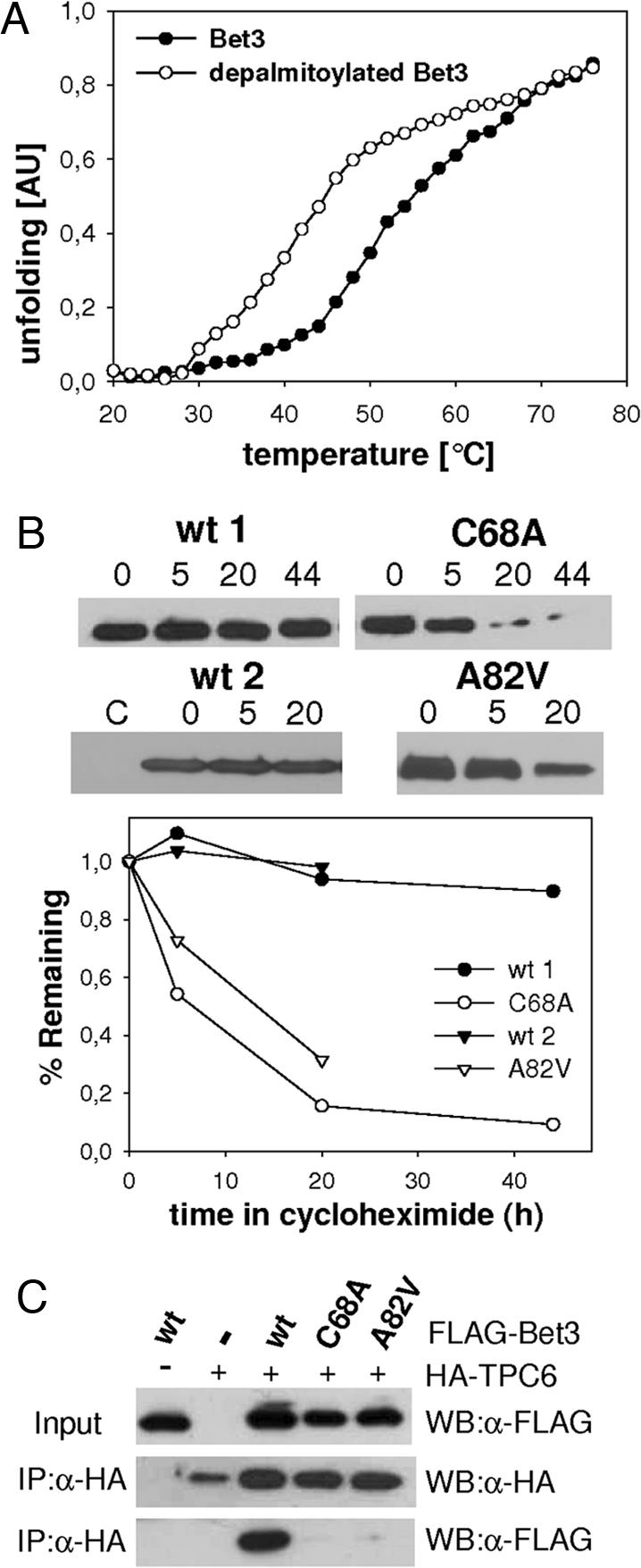
Palmitoylation of Bet3 increases the stability of the molecule in vitro and in vivo. (A) Melting curve as determined by CD spectroscopy of wild-type Bet3 purified from yeast, either mock-deacylated (Bet3) or depalmitoylated with hydroxylamine. (B) BHK cells were mock-transfected or transfected with FLAG-Bet3 wild type (wt), FLAG-Bet3 C68A, or FLAG-Bet3 A82V. Twenty hours later, protein synthesis was blocked with cycloheximide, and cells were further incubated for 0, 5, 20, or 44 h. Cellular proteins were precipitated and probed for Bet3 expression with anti-FLAG antibodies, and band intensities were quantified. (C) HEK293 cells were transfected with FLAG-Bet3 wild type (wt), FLAG-Bet 3 C68A, or FLAG-Bet 3 A82V and HA–TPC6. Immunoprecipitations with anti-HA affinity matrix (IP:α-HA) from cleared cell lysates were probed with anti-HA and anti-FLAG antibodies (WB). (Top) Blot without prior immunoprecipitation.
Does palmitoylation also affect the stability of Bet3 in vivo? BHK cells were transfected with plasmids encoding acylated and nonacylated Bet3 and incubated for 24 h to allow expression of the protein. Protein synthesis was then blocked, and cells were incubated further for 5, 20, and 44 h before analyzing the remaining amount of Bet3 in cellular extracts by blotting. Whereas palmitoylated Bet3 is stable for at least 44 h, the amount of the nonacylated mutants A82V and C68A decreased rapidly after 5 h of incubation (Fig. 5B). Degradation in vivo has been reported before for other proteins, notably for SNARE proteins, upon removal of their palmitoylation sites (40).
If nonpalmitoylated Bet3 adopts a tertiary structure different from that of the palmitoylated form in vivo one would expect that protein–protein interactions of Bet3 are impaired. We thus tested the binding of Bet3 A82V and C68A to TPC6 (10, 11). In coimmunoprecipitation experiments with transfected HEK293 cells, binding of Bet3 mutants to TPC6 was almost completely abolished (Fig. 5C). Because the mutation sites of unpalmitoylated Bet3 (A82V and C68A) are spatially far apart from its interaction interface with TPC6, a profound change in the tertiary arrangement must have occurred.
Why then does mutating cysteine-68 of Bet3, in contrast to exchange of alanine-82 by bulkier amino acids, not cause conditional lethality in yeast (8)? Nonacylated Bet3 might be stabilized by the postulated insertion of an acyl chain linked to a Golgi-resident membrane protein into the hydrophobic tunnel (8), which is not possible if the tunnel is too narrow. As a consequence, Bet3 C68S is recruited to the Golgi, ensuring proper trafficking through the secretory pathway and cell viability.
However, yeast and human Bet3 might have different requirements for palmitoylation. Most of mammalian Bet3 is not membrane-anchored (7), and we speculate that this pool of Bet3 might serve a different function beyond acting as a tethering factor in the framework of the TRAPP complex. Although increased palmitoylation of proteins upon overexpression of Bet3 was not detected in cell lysate (Fig. 4), it is intriguing to speculate that Bet3 acylates proteins of lower abundance or lipids, and a function as carrier of activated fatty acids is also conceivable.
Very recently the palmitoyl proteome of Saccharomyces cerevisiae was characterized, and the effect of deletion of DHHC proteins on palmitoylation of individual proteins was determined (41). Only Bet3 shows a palmitoylation that was undiminished in any of the DHHC mutant strains, strongly arguing in favor of an autocatalytic mode for its acylation.
Methods
Recombinant Protein Expression.
Human Bet3 with an N-terminal His tag and C-terminal StrepII tag was expressed and purified from S. cerevisiae as described (9). For expression of GST fusion protein in E. coli, human Bet3 cDNA was cloned into the bacterial expression vector pGEX-4T1 (GE Healthcare, Munich, Germany). Site-directed mutagenesis was performed according to a QuikChange mutagenesis protocol (42). E. coli BL21 (DE3) cells were transformed with the resulting plasmids. An overnight culture was inoculated in LB medium containing 100 μg/ml ampicillin. The preculture was diluted 20-fold in the same medium, grown at 37°C to an OD600 of ≈0.7, and induced with 1 mM isopropyl-1-thio-β-d-galactopyranoside. Growth was continued for 4 h, cells were harvested by centrifugation, and pellets were stored at −80°C. Cells expressing GST-Bet3 were lysed by using a French press (SLM-Aminco, Schwäbisch Gmünd, Germany) in PBS supplemented with 1 mM PMSF, 1 mM DTT, and DNase I (Roche, Mannheim, Germany). A cleared lysate obtained by centrifugation was loaded on 2 ml of glutathione-Sepharose (GE Healthcare) in a polypropylene column. The column was washed with PBS, protein was eluted with elution buffer (50 mM Tris, pH 8/10 mM GSH), and elution fractions were dialyzed against 25 mM Tris (pH 8.4)/120 mM NaCl.
Palmitoylation Assay.
[3H]Pal-CoA, prepared as described (21), was dried in a SpeedVac and dissolved in 20 mM Tris·HCl (pH 7.4)/120 mM NaCl/0.05% Triton X-100 to yield a final concentration of 100,000 cpm/μl. For most experiments [3H]Pal-CoA was diluted 1:1 with 10 μM cold Pal-CoA. The standard palmitoylation reaction contains purified Bet3 (5–10 μl, 8 μg, 3.2 μM), 5 μl of [3H]Pal-CoA (500 nM, 250,000 cpm), and 1 mM DTT in a final volume of 100 μl of Tris·NaCl buffer (20 mM Tris·HCl, pH 8.4/120 mM NaCl) and was incubated for 30 min at 30°C. One milliliter of chloroform/methanol (1:1) was added to the samples, and precipitated proteins were pelleted (15 min, 14,000 × g), washed, resuspended in 20 μl of nonreducing SDS/PAGE sample buffer, and boiled for 5 min. Samples were subjected to SDS/PAGE, staining with Coomassie, and fluorography for 5 h or overnight. For the pH experiment, the following buffers were used: 20 mM Pipes/KOH at pH 5.5, 6.5, and 7.5; 20 mM Tris·HCl at pH 8.5 or 20 mM glycine/NaOH at pH 9.5, 10.5, 11.5, or 12.5. Br-Pal (Acros, Geel, Belgium) and Cibacron Blue were added from a 100× stock solution in ethanol and Tris buffer, respectively. For the Pal-CoA titration experiment, [3H]Pal-CoA was diluted 1:1 with 100 μM unlabeled Pal-CoA and incubated with Bet3 for 5 min at 30°C. To deacylate Bet3, 10 μg of purified protein was incubated with 50 μl of 1 M hydroxylamine (pH 7) or 1 M Tris·HCl for 1 h at 4°C. The reaction mix was then desalted with a spin column (Zeba Desalt mini column; Perbio, Bonn, Germany).
Affinity Chromatography.
A total of 100 μl of immobilized Cibacron Blue F3GA (Perbio) or Pal-CoA-agarose (Sigma, Munich) was equilibrated with 20 mM Tris·HCl (pH 8.4)/120 mM NaCl. The beads were incubated with 40 μg of Bet3 for 1 h at 4°C. Beads were pelleted (3 min, 500 × g), and the supernatant was removed. Beads were then resuspended in 20 mM Tris·HCl (pH 8.4)/120 mM NaCl and again pelleted, and the supernatant was removed (wash fraction). Washing of the beads was repeated eight times. Pelleted beads were then incubated with different CoA compounds as indicated for 15 min at 4°C, beads were again pelleted, and the eluate was removed. Elution with CoA compounds was repeated once. Comparable aliquots of input, flow-through, several wash fractions, and the eluates were analyzed by SDS/PAGE and Coomassie staining.
CD.
For CD spectroscopy, protein samples of human Bet3 purified from yeast and chemically depalmitoylated Bet3 were diluted to ≈0.1 mg/ml, and buffer was exchanged to PBS. Measurements were carried out with a J720 spectropolarimeter (Jasco) with temperature control at 222 nm with a 0.2-cm-path-length cuvette. Unfolding was monitored after the decrease of ellipticity during heating of the sample 0.5°C min−1 from 20°C to 80°C.
Transfection and Labeling of Cells.
BHK cells were transfected with plasmids containing human myc-Bet3 (pRK5) or FLAG-Bet3 (pcDNA3) by using either Superfect (Qiagen, Hilden, Germany) or the Nucleofector technology (Amaxa, Cologne, Germany). Eighteen hours (Nucleofector) or 42 h (Superfect) later, cells were labeled with 300 μCi/ml [3H]palmitate (1 Ci = 37 GBq) for the times indicated. For the pulse–chase experiments, the radioactive medium was removed, and cells were washed and further incubated in medium containing cycloheximide. BFA (Sigma), cycloheximide, and ilimaquinone (Sigma) were added to final concentrations of 10 μg/ml, 50 μg/ml, and 30 μM, respectively. Cells were extracted with chloroform/methanol (1:2). Precipitated proteins were pelleted (15 min, 14,000 × g), washed with methanol and subjected to SDS/PAGE under nonreducing conditions, staining with Coomassie, and fluorography. An aliquot was probed by blotting with anti-myc antibody 9E10 (sc-40; Santa Cruz Biotechnology, Santa Cruz, CA) or anti-FLAG antiserum (sc-807; Santa Cruz Biotechnology) by using the ECL system (Amersham Pharmacia). HEK293 cells were transfected with the calcium precipitation method and lysed 24 h later with 150 mM NaCl/50 mM Hepes (pH 7.5)/0.2% Nonidet P-40/1 mM glycerol/1 mM DTT/protease inhibitors. Immunoprecipitation from cleared lysate with anti-HA–affinity matrix (Roche) was performed overnight, and beads were washed, subjected to SDS/PAGE, and probed by blotting with anti-FLAG M2 (Sigma) or anti-HA Y11 (Santa Cruz Biotechnology) antibodies.
Acknowledgments
This work was supported by the German Federal Ministry for Education and Research through National Genome Network Grant FZK 01GR0471, by Deutsche Forschungsgemeinschaft Grant Ve 141/6, and by the Fonds der Chemischen Industrie.
Abbreviations
- TRAPP
transport protein particle
- Pal-CoA
palmitoyl-CoA
- BFA
brefeldin A
- ER
endoplasmic reticulum
- Br-Pal
2-bromo-palmitate
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Rossi G., Kolstad K., Stone S., Palluault F., Ferro-Novick S. Mol. Biol. Cell. 1995;6:1769–1780. doi: 10.1091/mbc.6.12.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrowman J., Sacher M., Ferro-Novick S. EMBO J. 2000;19:862–869. doi: 10.1093/emboj/19.5.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sacher M., Barrowman J., Wang W., Horecka J., Zhang Y., Pypaert M., Ferro-Novick S. Mol. Cell. 2001;7:433–442. doi: 10.1016/s1097-2765(01)00190-3. [DOI] [PubMed] [Google Scholar]
- 4.Sacher M., Jiang Y., Barrowman J., Scarpa A., Burston J., Zhang L., Schieltz D., Yates J. R., III, Abeliovich H., Ferro-Novick S. EMBO J. 1998;17:2494–2503. doi: 10.1093/emboj/17.9.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang W., Sacher M., Ferro-Novick S. J. Cell Biol. 2000;151:289–296. doi: 10.1083/jcb.151.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones S., Newman C., Liu F., Segev N. Mol. Biol. Cell. 2000;11:4403–4411. doi: 10.1091/mbc.11.12.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loh E., Peter F., Subramaniam V. N., Hong W. J. Cell Sci. 2005;118:1209–1222. doi: 10.1242/jcs.01723. [DOI] [PubMed] [Google Scholar]
- 8.Kim Y. G., Sohn E. J., Seo J., Lee K. J., Lee H. S., Hwang I., Whiteway M., Sacher M., Oh B. H. Nat. Struct. Mol. Biol. 2005;12:38–45. doi: 10.1038/nsmb871. [DOI] [PubMed] [Google Scholar]
- 9.Turnbull A. P., Kummel D., Prinz B., Holz C., Schultchen J., Lang C., Niesen F. H., Hofmann K. P., Delbruck H., Behlke J., et al. EMBO J. 2005;24:875–884. doi: 10.1038/sj.emboj.7600565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kummel D., Muller J. J., Roske Y., Misselwitz R., Bussow K., Heinemann U. EMBO Rep. 2005;6:787–793. doi: 10.1038/sj.embor.7400463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim M. S., Yi M. J., Lee K. H., Wagner J., Munger C., Kim Y. G., Whiteway M., Cygler M., Oh B. H., Sacher M. Traffic. 2005;6:1183–1195. doi: 10.1111/j.1600-0854.2005.00352.x. [DOI] [PubMed] [Google Scholar]
- 12.Kummel D., Müller J. J., Roske Y., Henke N., Heinemann U. J. Mol. Biol., 2006 doi: 10.1016/j.jmb.2006.06.012. in press. [DOI] [PubMed] [Google Scholar]
- 13.Bijlmakers M. J., Marsh M. Trends Cell Biol. 2003;13:32–42. doi: 10.1016/s0962-8924(02)00008-9. [DOI] [PubMed] [Google Scholar]
- 14.Smotrys J. E., Linder M. E. Annu. Rev. Biochem. 2004;73:559–587. doi: 10.1146/annurev.biochem.73.011303.073954. [DOI] [PubMed] [Google Scholar]
- 15.Pfanner N., Glick B. S., Arden S. R., Rothman J. E. J. Cell Biol. 1990;110:955–961. doi: 10.1083/jcb.110.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfanner N., Orci L., Glick B. S., Amherdt M., Arden S. R., Malhotra V., Rothman J. E. Cell. 1989;59:95–102. doi: 10.1016/0092-8674(89)90872-6. [DOI] [PubMed] [Google Scholar]
- 17.Glick B. S., Rothman J. E. Nature. 1987;326:309–312. doi: 10.1038/326309a0. [DOI] [PubMed] [Google Scholar]
- 18.Quesnel S., Silvius J. R. Biochemistry. 1994;33:13340–13348. doi: 10.1021/bi00249a021. [DOI] [PubMed] [Google Scholar]
- 19.Belanger C., Ansanay H., Qanbar R., Bouvier M. FEBS Lett. 2001;499:59–64. doi: 10.1016/s0014-5793(01)02513-3. [DOI] [PubMed] [Google Scholar]
- 20.Bizzozero O. A., Bixler H. A., Pastuszyn A. Biochim. Biophys. Acta. 2001;1545:278–288. doi: 10.1016/s0167-4838(00)00291-0. [DOI] [PubMed] [Google Scholar]
- 21.Duncan J. A., Gilman A. G. J. Biol. Chem. 1996;271:23594–23600. doi: 10.1074/jbc.271.38.23594. [DOI] [PubMed] [Google Scholar]
- 22.Veit M. Biochem. J. 2000;345:145–151. [PMC free article] [PubMed] [Google Scholar]
- 23.Veit M., Sachs K., Heckelmann M., Maretzki D., Hofmann K. P., Schmidt M. F. Biochim. Biophys. Acta. 1998;1394:90–98. doi: 10.1016/s0005-2760(98)00097-6. [DOI] [PubMed] [Google Scholar]
- 24.Bano M. C., Jackson C. S., Magee A. I. Biochem. J. 1998;330:723–731. doi: 10.1042/bj3300723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bizzozero O. A., McGarry J. F., Lees M. B. J. Biol. Chem. 1987;262:13550–13557. [PubMed] [Google Scholar]
- 26.Lobo S., Greentree W. K., Linder M. E., Deschenes R. J. J. Biol. Chem. 2002;277:41268–41273. doi: 10.1074/jbc.M206573200. [DOI] [PubMed] [Google Scholar]
- 27.Roth A. F., Feng Y., Chen L., Davis N. G. J. Cell Biol. 2002;159:23–28. doi: 10.1083/jcb.200206120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dietrich L. E., Peplowska K., LaGrassa T. J., Hou H., Rohde J., Ungermann C. EMBO Rep. 2005;6:245–250. doi: 10.1038/sj.embor.7400350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fukasawa M., Varlamov O., Eng W. S., Sollner T. H., Rothman J. E. Proc. Natl. Acad. Sci. USA. 2004;101:4815–4820. doi: 10.1073/pnas.0401183101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veit M. Biochem. J. 2004;384:233–237. doi: 10.1042/BJ20041474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Webb Y., Hermida-Matsumoto L., Resh M. D. J. Biol. Chem. 2000;275:261–270. doi: 10.1074/jbc.275.1.261. [DOI] [PubMed] [Google Scholar]
- 32.Faergeman N. J., Knudsen J. Biochem. J. 1997;323:1–12. doi: 10.1042/bj3230001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J., Gibson B., Snider J., Jenkins C. M., Han X., Gross R. W. Biochemistry. 2005;44:11903–11912. doi: 10.1021/bi0508082. [DOI] [PubMed] [Google Scholar]
- 34.Leventis R., Juel G., Knudsen J. K., Silvius J. R. Biochemistry. 1997;36:5546–5553. doi: 10.1021/bi963029h. [DOI] [PubMed] [Google Scholar]
- 35.Dunphy J. T., Schroeder H., Leventis R., Greentree W. K., Knudsen J. K., Silvius J. R., Linder M. E. Biochim. Biophys. Acta. 2000;1485:185–198. doi: 10.1016/s1388-1981(00)00060-3. [DOI] [PubMed] [Google Scholar]
- 36.van’t Hof W., Resh M. D. J. Cell Biol. 1997;136:1023–1035. doi: 10.1083/jcb.136.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wedegaertner P. B., Bourne H. R. Cell. 1994;77:1063–1070. doi: 10.1016/0092-8674(94)90445-6. [DOI] [PubMed] [Google Scholar]
- 38.Qanbar R., Bouvier M. Biochemistry. 2004;43:12275–12288. doi: 10.1021/bi049176u. [DOI] [PubMed] [Google Scholar]
- 39.Takizawa P. A., Yucel J. K., Veit B., Faulkner D. J., Deerinck T., Soto G., Ellisman M., Malhotra V. Cell. 1993;73:1079–1090. doi: 10.1016/0092-8674(93)90638-7. [DOI] [PubMed] [Google Scholar]
- 40.Valdez-Taubas J., Pelham H. EMBO J. 2005;24:2524–2532. doi: 10.1038/sj.emboj.7600724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roth A. F., Wan J., Bailey A. O., Sun B., Kuchar J. A., Green W. N., Phinney B. S., Yates J. R., III, Davis N. G. Cell. 2006;125:1003–1013. doi: 10.1016/j.cell.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang W., Malcolm B. A. BioTechniques. 1999;26:680–682. doi: 10.2144/99264st03. [DOI] [PubMed] [Google Scholar]