Abstract
Photoactivation of the visual rhodopsin, a prototypical G protein-coupled receptor (GPCR), involves efficient conversion of the intrinsic inverse-agonist 11-cis-retinal to the all-trans agonist. This event leads to the rearrangement of the heptahelical transmembrane bundle, which is thought to be shared by hundreds of GPCRs. To examine this activation mechanism, we determined the x-ray crystallographic model of the photoreaction intermediate of rhodopsin, lumirhodopsin, which represents the conformational state having the nearly complete all-trans agonist form of the retinal. A difference electron density map clearly indicated that the distorted all-trans-retinal in the precedent intermediate bathorhodopsin relaxes by dislocation of the β-ionone ring in lumirhodopsin, along with significant peptide displacement in the middle of helix III, including approximately two helical turns. This local movement results in the breaking of the electrostatic interhelical restraints mediated by many of the conserved residues among rhodopsin-like GPCRs, with consequent acquisition of full activity.
Keywords: G protein-coupled receptor, membrane protein, visual pigment
G protein-coupled receptors (GPCRs) are heptahelical transmembrane proteins responsible for a variety of physiological functions mediated by cell membranes in eukaryotes (1, 2) and are therefore considered a major target for pharmacological intervention. The activity of GPCRs in catalyzing GDP/GTP exchange reaction in the α-subunit of heterotrimeric G protein strongly depends on the nature of the interaction between the receptor and ligand molecules. Even for a given GPCR, binding of structurally related ligand has different degrees of effect on activity. Effective drug design thus requires understanding of the structural consequences evoked in a receptor just upon its formation of a complex with a ligand. The retinal dim-light photoreceptor rhodopsin (RHO) is representative of hundreds of GPCRs in terms of the dramatic change in activity it undergoes before to after photoactivation, which involves isomeric structural change of the intrinsic ligand retinal (Vitamin A aldehyde) (3).
To date, RHO is the only 1 among 1,000 GPCRs for which a high-resolution experimental model is available. Its crystal structure was recently solved in the presence of intrinsic inverse-agonist 11-cis-retinal (4–6), which is covalently linked to the protein (ε-amino group of Lys-296 in the seventh transmembrane helix) via protonated Schiff base (SB). Because its photoexcitation leads to formation of a series of reaction intermediates having differently shaped retinal ligands and the protein moiety, it is thought that RHO crystal is an ideal material for investigation of the mechanism of activation of GPCRs, which share many key amino acids with RHO in the transmembrane helical bundle. Furthermore, beside previous studies on structural changes of membrane proteins, x-ray crystallography of RHO intermediates provides a previously undescribed example of an irreversible reaction, because the covalently bound retinal ligand is released from the protein after deprotonation of the SB.
These intermediates are trapped either by time-resolved technique from femtoseconds to milliseconds at physiological temperatures (7, 8) or steady-state experiments with low-temperature technique (9, 10). Although the observed sequences of spectrally distinct species are not completely identical for the two experimental methods, bathorhodopsin (BATHO) and lumirhodopsin (LUMI) are detected consistently regardless of such technical differences and are considered keys to understanding the mechanism of light-induced triggering RHO activation (see Fig. 5, which is published as supporting information on the PNAS web site).
The first crystallographic model of BATHO was recently proposed (11). This intermediate has been a focus of extensive experimental and theoretical studies, because it is the first stable product after the photoisomerization that is characterized by strict stereospecificity, high quantum efficiency, large photon energy storage, and ultrafast reaction time (12, 13). Although the resolution of our x-ray diffraction data was not very high, the model appeared to be accurate enough to explain the unique spectroscopic properties of this intermediate by quantum mechanical calculations. The results showed how the retinal of BATHO took a distorted shape during photoisomerization reaction in the confined protein-binding pocket.
To acquire full agonist activity, the distorted retinal should relax in some fashion, accompanying further rearrangement in the heptahelical transmembrane polypeptide region. LUMI, the next thermal product of BATHO, is key to connecting the photoisomerization process to the formation of fully activated metarhodopsin II, because previous time-resolved laser photolysis studies under physiological conditions have demonstrated that the decay of LUMI is directly linked to the deprotonation of the SB (8), which is known to be a prerequisite for the active form of RHO (14).
In this work, we show a crystallographic model of this thermal product, LUMI, in which the highly distorted retinal ligand in BATHO has changed to a more relaxed conformation. The resulting local peptide distortion in this reaction intermediate suggests the possible pathway to the fully active form of RHO.
Results and Discussion
Crystallography and Microspectrophotometry.
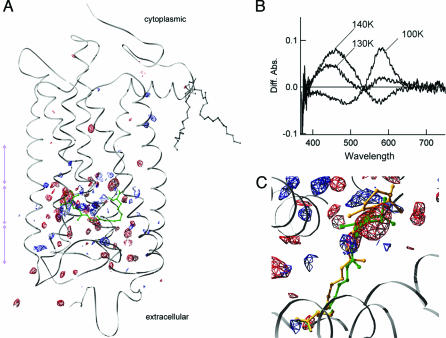
In Fig. 1A, the overall difference electron densities reproducibly observed from cryogenic x-ray diffraction data for LUMI are shown on the ground state polypeptide backbone. The temperature for trapping LUMI was chosen based on microspectrophotometry of the crystals of RHO (16). As shown in Fig. 1B, formation of BATHO at 100 K by illumination with visible light and successive warming of the crystal results in gradual conversion from BATHO to LUMI. From these observations, the difference densities obtained at 160 K can be considered as that of LUMI minus RHO. The pattern of difference densities is found to be consistent for the two RHO molecules in the crystallographic asymmetric unit, and the details of the x-ray data statistics are summarized in Table 2, which is published as supporting information on the PNAS web site. To minimize possible radiation damage (17), the intensity of x-ray was attenuated to less than one-third of that used for BATHO (11).
Fig. 1.
Conversion from RHO to LUMI in a 3D crystal. (A) Difference electron densities calculated from x-ray diffraction data at 160 K. Three purple double arrows indicate the slab sections shown in Fig. 3 A–C. (B) Spectral changes observed upon conversion from RHO to BATHO (100 K) and to LUMI (130 K, 140 K) in a 3D crystal. These spectra were obtained after subtraction of the ground-state spectrum. (C) Expanded view of the difference electron densities around the retinal. In A and C, positive and negative electron densities are shown in blue and red, respectively, on the ground-state α-carbon polypeptide chain of RHO. The maps are contoured to 3.5σ level for one of the two molecules in the crystallographic asymmetric unit. The chromophores (11-cis-retinal + Lys-296) of RHO and LUMI are in green and orange, respectively. Figs. 1 and 3 were prepared with SPDBV (15).
In contrast to the very limited differences between BATHO and RHO (11), which represents only the process of photoisomerization reaction of the retinal chromophore, current data for LUMI clearly demonstrate the propagation of structural changes after the initial photoreaction in RHO. The structural refinement of LUMI was performed by using the x-ray data to 2.8-Å resolution. Whereas the two data sets in Table 2 gave almost identical models, the results shown below are based on data set 1 (Table 1) because it contains a smaller amount of merohedral twinning, which was found to be better in terms of the quality of difference electron density map. Because the intermediate structure models contain large domains that remained identical to that in RHO, they were unambiguously overlaid in the following figures to present the differences correctly.
Table 1.
Refinement statistics
| Alternate conformation refinement | Value |
|---|---|
| Resolution range, Å | 50–2.8 |
| Twin fraction | 0.123 |
| Protein atom | 3,630 |
| Retinal atom | 40 |
| Rcryst, % (outer shell) | 21.8 (30.5) |
| Rfree, % (outer shell) | 23.8 (32.0) |
| rmsd of bonds, Å | 0.012 |
| rmsd of angles, ° | 1.41 |
The final refinements included dual conformations of the selected residues as described in the text. Rcryst = Σhkl|Fo − Fc|/Σhkl|Fo|. Rfree values were calculated from a set of 5% randomly selected reflections that were omitted from refinement. This set was the same as that used for the dark state.
Movement of the Retinal Chromophore.
The positional/conformational change of the β-ionone ring moiety of the retinal chromophore during the RHO activation has been controversial (18–20). One of the important findings in our difference map is the appearance of a positive electron density just beside the β-ionone ring of the retinal (Fig. 1C; also see Fig. 3B and Fig. 6, which is published as supporting information on the PNAS web site). This result is obviously different from the finding obtained for BATHO and is well interpreted as the result of displacement of the ring in the direction toward helices III and IV. It is noteworthy that helix IV contains Ala-169, which was suggested to get closer to the β-ionone ring at the later stages of RHO activation (18).
Fig. 3.
Differences between RHO and LUMI. (A–C) Projection views of the three regions containing the difference electron densities between RHO and LUMI. Each image is a 10-Å slab section from the extracellular (A) to cytoplasmic (C) side of the transmembrane helical domain. Positive (blue) and negative (red) electron densities contoured to 3.5σ level are shown on the α-carbon traces of the seven helices. (D) Superposition of the crystallographic models of RHO (green) and LUMI (orange). Only the α-carbon traces and the retinal chromophore are shown.
The structure of the retinal in LUMI modeled according to the x-ray data takes a nearly complete all-trans conformation, although the C9 C10 and C11
C10 and C11 C12 double bonds are still twisted to some extent (Fig. 2 C and D). Compared with the retinal in BATHO (Fig. 2 A and B), it is evident that relaxation of the curved polyene chain has taken place, without involving flip-over of the ring moiety (Fig. 2D). We suppose this step to be a key part of conversion of the stored photon energy to the surrounding protein moiety through conversion of 11-cis inverse-agonist to all-trans agonist in RHO. Whereas the retinal does not completely escape from the ground-state binding pocket, keeping the position of the protonated SB, interactions with some nearby amino acids are substantially altered (see Fig. 4A). Two bulky hydrophobic residues, Phe-212 and Trp-265 (5.47 and 6.48, respectively, in GPCR numbering according to ref. 23), and Glu-113, the counterion to the protonated SB, are especially notable in the consistent but larger displacement of the side chains compared with that found in BATHO. The change of Glu-113 position and possible movement of nearby waters, which remain unresolved in this study, need to be supported by further experimental or theoretical studies to understand the spectroscopic and functional properties of LUMI (24, 25).
C12 double bonds are still twisted to some extent (Fig. 2 C and D). Compared with the retinal in BATHO (Fig. 2 A and B), it is evident that relaxation of the curved polyene chain has taken place, without involving flip-over of the ring moiety (Fig. 2D). We suppose this step to be a key part of conversion of the stored photon energy to the surrounding protein moiety through conversion of 11-cis inverse-agonist to all-trans agonist in RHO. Whereas the retinal does not completely escape from the ground-state binding pocket, keeping the position of the protonated SB, interactions with some nearby amino acids are substantially altered (see Fig. 4A). Two bulky hydrophobic residues, Phe-212 and Trp-265 (5.47 and 6.48, respectively, in GPCR numbering according to ref. 23), and Glu-113, the counterion to the protonated SB, are especially notable in the consistent but larger displacement of the side chains compared with that found in BATHO. The change of Glu-113 position and possible movement of nearby waters, which remain unresolved in this study, need to be supported by further experimental or theoretical studies to understand the spectroscopic and functional properties of LUMI (24, 25).
Fig. 2.
Structural changes of the retinal chromophore after photoexcitation of the RHO crystal. (A and B) Conversion from RHO (green) to BATHO (red) is derived from ref. 11. (C and D) Conversion from BATHO (red) to LUMI (orange). B and D are different views from A and C, respectively, rotated ≈90° around the horizontal axis. Nitrogen atoms are in blue. Figs. 2 and 4 were prepared with MolScript (21) and Raster3D (22).
Fig. 4.

Crystallographic models of the three states of RHO. (A) Projection view around the retinal with some amino acid residues. Shown are the three models: RHO (green), BATHO (red), and LUMI (orange). (B) Structural changes between RHO (green) and LUMI (orange) around the middle of helix III. The four bound water molecules in this site are shown as small light blue spheres, which are fixed in the positions found in the ground-state structure.
Distortion in the Middle of Helix III.
The most remarkable rearrangement in the protein is seen in the middle of helix III, where a strong shift of electron densities is evident (Fig. 3B). This pattern is a clear indication of outward movement of the polypeptide backbone (Fig. 3D), which has not been demonstrated before. On both the extracellular (Fig. 3A) and the cytoplasmic (Fig. 3C) sides of this region, the difference electron density signals are much weaker. We therefore conclude that the protein structural changes in LUMI at 160 K are still limited and do not propagate to the cytoplasmic surface where G protein activation is mediated after full activation. As for the second extracellular loop, we suppose that the structure might become slightly disordered from dark state to LUMI, as indicated by some negative densities (Fig. 3A). Also, a pair of densities at the S–S bond can be seen as detected in the BATHO minus RHO map (11).
The largest displacement of the α-carbon atoms is detected in the Gly–Gly sequence at 120 and 121 (3.35 and 3.36), which is highly conserved among vertebrate visual pigments and is very close to the known ligand-binding site (3.32) in many of the catecholamine receptors (1, 2). This finding of structural change appears to be consistent with the constitutive activity of the mutants at Gly-121 in RHO (26). The magnitude of movement at Gly-120, 1.16 and 0.99 Å for the two molecules in the asymmetric unit, and that of nearby residues is large enough to be detected with the resolution of our data (see Fig. 7, which is published as supporting information on the PNAS web site, for comparison with other helices). We also observed a reproducible shift of the side chain of Thr-118 (3.33) in BATHO (Fig. 4A), and the difference densities around this side chain become even larger in LUMI (data not shown). On the basis of these observations, we can explain the sequential structural perturbations evoked in helix III upon photoexcitation of RHO. In addition to the interaction change between the retinal and Thr-118, the side chain of Glu-122 (3.37) is found to be affected by the dislocation of the β-ionone ring. Although the outward movement of helix III observed at 160 K in the 3D crystal is not very large, it also should be consistent with the previous notion that a small increase of the protein volume occurs during the conversion from ground-state RHO to LUMI under physiological conditions (27).
In the archaeal retinal protein bacteriorhodopsin, which has a heptahelical transmembrane bundle differently arranged from that of RHO, a structural model of the L-intermediate has been shown to involve polypeptide displacement in the middle of helix C (III) (28). This change was considered critical for the proton-pumping function of this membrane protein. Notably, however, the direction of movement is almost opposite that seen here in LUMI. Therefore, the functional difference between RHO and bacteriorhodopsin correlates well with the nature of the structural changes occurring during the photoreaction cascade.
Effects on Other Helices.
The outward movement in the middle of helix III in LUMI directly affects the nearby region in helix IV, where the highly conserved tryptophan in RHO-like GPCRs, Trp-161 (4.50), is located (Fig. 4B). This residue and the preceding Thr-160 (4.49) are hydrogen-bonded with Asn-78 (2.45) in helix II in the inactive ground state. These electrostatic restraints must be important for keeping the inactive state, because they help to define the highly tilted angle of helix III. Partly because of these direct interactions between helices II and IV, the cytoplasmic half of helix III is occluded inside of the helical bundle (see Fig. 8, which is published as supporting information on the PNAS web site). Upon formation of LUMI, the distances between these residues increase substantially (Fig. 4B), preparing for further breakage of restraints in this region. This effect is presumably due to the slight movement of helix IV, although it is unclear in the difference map of LUMI whether this movement is translational or rotational.
The distortion in the middle of helix III can also change the interactions with helix II around the highly conserved aspartic acid Asp-83 (2.50). The side chain of this residue is bridged to the peptide carbonyl of Gly-120 (3.35) via a water molecule in the ground-state structure. This region has been assigned as the largest water-binding site containing four molecules in RHO (5, 6, 16) and is also thought to overlap with the putative metal-binding site in certain families of other RHO-like GPCRs. Our findings are therefore consistent with much previous evidence of regulation of receptor activity in this region. Because Asp-83 (2.50) is also involved in mediating the other electrostatic interhelical restraints with Asn-55 (1.50) in helix I and Ala-299 (7.46) in helix VII, the consequences of the initial perturbation around the Gly–Gly sequence of helix III should extend even more than those in the current model of LUMI at the later stage of activation. This suggestion is also supported by the previous finding that the interhelical space around the cytoplasmic side of helix VII from Val-304 (7.51) to Lys-311 (7.58) becomes exposed upon formation of the active state (29).
The significant displacement of the β-ionone ring of retinal directly causes perturbations in some surrounding residues. These structural changes observed in LUMI are even larger and mostly consistent with that found in BATHO (11). The largest perturbations occur in the middle of helix V, where a small helical bulge toward the interior of the RHO molecule is observed in the ground-state structure of RHO. The general importance of this region is evident from the identification of ligand-binding residues in the receptors for catecholamines (1, 2, 30). In RHO, Met-207 (5.42), His-211 (5.46), and Phe-212 (5.47) form a part of the retinal-binding site. Along with the movement of the β-ionone ring, not only the side chains but also the peptide backbone of these residues undergo substantial rearrangement (Fig. 4A). It should be noted that the bulge is presumably due to the presence of the nearby highly conserved proline, Pro-215 (5.50). Because the regular helical structure is disrupted even in the ground state as such, the perturbation due to movement of retinal can easily cause outward shift of the backbone in this region.
Possible Activation Mechanism.
It has been proposed that the activation of RHO-like GPCRs involves substantial movement of helix VI (31, 32). However, the electron density differences around this helix in LUMI are not as large as expected. This result is consistent with a recent electron microscopic study on metarhodopsin I, for which no difference in the peptide backbone from the ground state was demonstrated (33). In our x-ray data, a few positive densities on the extracellular side of the highly conserved Pro-267 (6.50) suggest a slight outward displacement (Fig. 3A). It is possible that this change further results in a rotational motion of the cytoplasmic side by using Pro-267 as a hinge, because the two major hydrophobic interactions between this helix and the retinal at Phe-261 (6.44) and Trp-265 (6.48) are mostly lost in LUMI. In any case, an important conclusion to be drawn from our results is that the possibility of movement of helix VI must be the consequence of the displacement of other structural elements in the transmembrane region, including the retinal ligand and helices II–V. We also speculate that such activation mechanism should be relevant for a number of RHO-like GPCRs accommodating a vast variety of external diffusible ligands.
The present experimental observations of the differences between the inactive ground-state RHO and LUMI are quite useful in elucidating possible structural sequences of activation in many of RHO-like GPCRs in more details. In the ground-state structure of bovine RHO, the intramolecular restraints in the transmembrane heptahelical domain are dominated by electrostatic interactions on the extracellular side and by the hydrophobic interactions on the cytoplasmic side (34). We now understand that the major consequence of the ligand conversion from an inverse-agonist (11-cis-retinal) to an agonist (all-trans-retinal) is to break and/or weaken most of the electrostatic restraints between the helices. The affected hydrogen-bonding sites include many highly conserved residues/motifs such as the N (1.50)–D (2.50) pair (Asn-55–Asp-83 in RHO), Asn-78 (2.45), Trp-161 (4.50), and Asn-302 (7.49) in the so-called NPxxY motif. The region of irregular helix on the extracellular side of Pro-215 (5.50) is also the site of initial perturbation. This protein rearrangement would evoke further dynamic motion of helix III, then of helix VI (31, 32) and possibly VII (19) with a concomitant change of the environment of the SB (35), leading to the conformation of the fully active form, metarhodopsin II. Because the two regions identified in this study as sites of light-induced perturbation, the middle regions of helices III and V, overlap with the ligand-binding residues in many of the receptors for catecholamines, the proposed scheme of activation may be of general importance.
Conclusions
In a series of recent x-ray crystallographic studies including this work, we have shown three different states of bovine RHO: ground state, BATHO, and LUMI. These models represent the ligand–protein interaction changes during isomeric conversion of the retinal from inverse-agonist (11-cis) to agonist (all-trans). Further improvement of the data for these states and crystallographic and computational studies on the later intermediates will unveil the complete picture of the molecular mechanism of RHO activation. More importantly, structural studies on the photoreaction intermediates of RHO would be valuable for understanding the multiple conformational states (36, 37) observed upon ligand binding to other RHO-like GPCRs.
Materials and Methods
Crystallization.
3D crystals of RHO from bovine retina were grown by hanging-drop vapor-diffusion method at 283 K as recently reported (5). Briefly, RHO (6–7 mg/ml) purified in heptylthioglucoside micelles was mixed with a crystallization solution containing 7–10 mM 2-mercaptoethanol, 0.2–0.5% heptylthioglucoside, 0.6–0.7 M ammonium sulfate, and 0.6–1.0% PEG 600. Some of the crystals were grown in the presence of low-concentration (0.01–0.05%) sodium silicate–acetate mixture, which has been found to promote the crystallization process. Five microliters of the mixed sample was dispensed on a coverslip siliconized with hexamethyldisilazane, which was then fixed with paraffin oil on a well of a culture plate (Iwaki, Tokyo, Japan). The reservoir solution for vapor diffusion contained 20–30 mM Mes (pH 5.9–6.1) and various concentrations of ammonium sulfate (2.6–2.9 M). The course of crystallization was examined with a microscope (Olympus, Tokyo, Japan) by using dim red light (>650 nm filter; Fuji, Tokyo, Japan) to prevent photoreaction of samples. After adding 9 μl of cryoprotectant solution containing 15–20% trehalose directly to the hanging drop, crystals were mounted in the nylon loops (Hampton, San Diego, CA) and immediately flash-frozen in liquid nitrogen.
Single-Crystal Microspectrophotometry.
To confirm the formation of LUMI, visible absorption spectral changes of the 3D crystals of bovine RHO were recorded by a system based on a microspectrophotometer (4DX, Uppsala, Sweden) with a CCD detector (Andor, Tokyo, Japan). A crystal mounted on the goniometer head was illuminated under a stream of cold nitrogen gas by tunable argon laser light (Melles Griot, Irvine, CA), which was depolarized and aligned approximately along the long axis of the rod-shaped crystals to maximize the extent of photoreaction. The temperature at the crystal position was recorded by a thermocouple (Rigaku, Tokyo, Japan) manufactured specifically for the goniometer head.
Data Collection.
All x-ray diffraction data sets were collected on a MAR165 CCD detector (MAR Research, Hamburg, Germany) at BL41XU of SPring-8 (Harima, Japan), with a beam wavelength of 1.0000 Å. We carefully adjusted the intensity of the x-ray by using a series of attenuators so that two successive data sets collected from a dark state crystal did not indicate significant damage. The temperature of cold nitrogen gas at the position of the crystal was kept at 160 K during data collection. The temperature of 160 K, which is still low enough to prevent the decay of LUMI, was chosen to ensure that BATHO had completely decayed in the crystal. For illumination of crystals, depolarized argon laser light (514 nm, 2–7 mW) was aligned in a manner similar to that for the offline microspectroscopic studies and brought to the crystal during several minutes of data collection. X-ray data sets were processed with HKL2000 (38).
Model Building and Refinement.
For refinement of the LUMI structures, the ground-state structural model determined at 2.2 Å (5) was used as a starting model. By using phases of this structure and after rigid-body refinement, Fo − Fc maps were calculated for a number of ground-state (Fdark) and illuminated (Flight) data sets with different laser light intensities and temperatures to obtain reproducible structural differences between RHO and LUMI. The structure factors were used after appropriate treatment of the twin fractions of the data sets determined by the method described by Yeates (39). The refinement procedures used for LUMI basically followed those described in the recent report on BATHO (11) and the photoreaction intermediates of archaeal retinal proteins (28). The occupancy of RHO (α) in an illuminated crystal was estimated for each of the data sets by performing alternate conformation refinement of the structure of intermediate with its occupancy of 1α, while the rest (RHO) were fixed. The amino acids considered in refinement are Trp-35 → His-65, Thr-70 → Glu-134, Met-155 → Phe-220, and Val-250 → Asn-315, excluding some regions having high temperature factors in the ground state model. The value of α was changed from 0.4 to 0.75 in steps of 0.05, and the resulting R, Rfree, and 2Fo − Fc maps were compared. Finally, occupancy refinement of the two conformations converged to yield a value of α = 0.6 with CNS (40). We also carried out the selected-atom structure refinement of the intermediate by using extrapolated structure factor amplitudes, (Flight − α Fdark)/(1 − α), and found that the results, including the optimal value of α, were qualitatively the same as those for the alternate conformation refinement. Our recent crystallographic analysis on 9-cis-RHO at 2.9 Å (H.N. and T.O., unpublished data) supports an assumption that a possible two-photon reaction cannot contribute to the differences found at 160 K.
Supplementary Material
Acknowledgments
We thank M. Kawamoto and H. Sakai for excellent conditioning of the beamline BL41XU at SPring-8. This work was supported in part by grants from the Japanese Ministry of Education, Culture, Sports, Science, and Technology and by the New Energy and Industrial Technology Development Organization.
Abbreviations
- BATHO
bathorhodopsin
- GPCR
G protein-coupled receptor
- LUMI
lumirhodopsin
- RHO
rhodopsin
- SB
Schiff base
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2HPY).
References
- 1.Kristiansen K. Pharmacol. Ther. 2004;103:21–80. doi: 10.1016/j.pharmthera.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 2.Gether U. Endocr. Rev. 2000;21:90–113. doi: 10.1210/edrv.21.1.0390. [DOI] [PubMed] [Google Scholar]
- 3.Hubbell W. L., Altenbach C., Hubbell C. M., Khorana H. G. Adv. Protein Chem. 2003;63:243–290. doi: 10.1016/s0065-3233(03)63010-x. [DOI] [PubMed] [Google Scholar]
- 4.Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., et al. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 5.Okada T., Sugihara M., Bondar A.-N., Elstner M., Entel P., Buss V. J. Mol. Biol. 2004;342:571–581. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 6.Li J., Edwards P. C., Burghammer M., Villa C., Schertler G. F. J. Mol. Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 7.Kukura P., McCamant D. W., Yoon S., Wandschneider D. B., Mathies R. A. Science. 2005;310:1006–1009. doi: 10.1126/science.1118379. [DOI] [PubMed] [Google Scholar]
- 8.Lewis J. W., Kliger D. S. Methods Enzymol. 2000;315:164–178. doi: 10.1016/s0076-6879(00)15842-2. [DOI] [PubMed] [Google Scholar]
- 9.Yoshizawa T., Wald G. Nature. 1963;197:1279–1286. doi: 10.1038/1971279a0. [DOI] [PubMed] [Google Scholar]
- 10.Rothschild K. J., Cantore W. A., Marrero H. Science. 1983;219:1333–1335. doi: 10.1126/science.6828860. [DOI] [PubMed] [Google Scholar]
- 11.Nakamichi H., Okada T. Angew. Chem. Int. Ed. 2006;45:4270–4273. doi: 10.1002/anie.200600595. [DOI] [PubMed] [Google Scholar]
- 12.Birge R. R. Biochim. Biophys. Acta. 1990;1016:293–327. doi: 10.1016/0005-2728(90)90163-x. [DOI] [PubMed] [Google Scholar]
- 13.Kandori H., Shichida Y., Yoshizawa T. Biochemistry (Moscow) 2001;66:1197–1209. doi: 10.1023/a:1013123016803. [DOI] [PubMed] [Google Scholar]
- 14.Kim J. M., Altenbach C., Kono M., Oprian D. D., Hubbell W. L., Khorana H. G. Proc. Natl. Acad. Sci. USA. 2004;101:12508–12513. doi: 10.1073/pnas.0404519101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guex N., Peitsch M. C. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 16.Okada T., Fuiyoshi Y., Silow M., Navarro J., Landau E., Shichida Y. Proc. Natl. Acad. Sci. USA. 2002;99:5982–5987. doi: 10.1073/pnas.082666399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burmeister W. P. Acta. Crystallogr. D. 2000;56:328–341. doi: 10.1107/s0907444999016261. [DOI] [PubMed] [Google Scholar]
- 18.Borhan B., Souto M. L., Imai H., Shichida Y., Nakanishi K. Science. 2000;288:2209–2212. doi: 10.1126/science.288.5474.2209. [DOI] [PubMed] [Google Scholar]
- 19.Patel A. B., Crocker E., Reeves P. J., Getmanova E. V., Eilers M., Khorana H. G., Smith S. O. Proc. Natl. Acad. Sci. USA. 2004;101:10048–10053. doi: 10.1073/pnas.0402848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ebrey T. G., Kumauchi M. Biophys. J. 2005;88:L41–L42. doi: 10.1529/biophysj.105.060269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kraulis P. J. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 22.Merritt E. A., Murphy M. E. P. Acta. Crystallogr. D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 23.Ballesteros J. A., Weinstein H. Methods Neurosci. 1995;25:366–425. [Google Scholar]
- 24.Pan D. H., Mathies R. A. Biochemistry. 2001;40:7929–7936. doi: 10.1021/bi010670x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganter U. M., Gartner W., Siebert F. Biochemistry. 1988;27:7480–7488. doi: 10.1021/bi00419a046. [DOI] [PubMed] [Google Scholar]
- 26.Han M., Lou J., Nakanishi K., Sakmar T. P., Smith S. O. J. Biol. Chem. 1997;272:23081–23085. doi: 10.1074/jbc.272.37.23081. [DOI] [PubMed] [Google Scholar]
- 27.Marr K., Peters K. S. Biochemistry. 1991;30:1254–1258. doi: 10.1021/bi00219a013. [DOI] [PubMed] [Google Scholar]
- 28.Edman K., Royant A., Larsson G., Jacobson F., Taylor T., van der Spoel D., Landau E. M., Pebay-Peyroula E., Neutze R. J. Biol. Chem. 2004;279:2147–2158. doi: 10.1074/jbc.M300709200. [DOI] [PubMed] [Google Scholar]
- 29.Abdulaev N. G., Ridge K. D. Proc. Natl. Acad. Sci. USA. 1998;95:12854–12859. doi: 10.1073/pnas.95.22.12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oprian D. D. J. Bioenerg. Biomembr. 1992;24:211–217. doi: 10.1007/BF00762679. [DOI] [PubMed] [Google Scholar]
- 31.Farrens D. L., Altenbach C., Yang K., Hubbell W. L., Khorana H. G. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 32.Shi L., Liapakis G., Xu R., Guarnieri F., Ballesteros J. A., Javitch J. A. J. Biol. Chem. 2002;277:40989–40996. doi: 10.1074/jbc.M206801200. [DOI] [PubMed] [Google Scholar]
- 33.Ruprecht J. J., Mielke T., Vogel R., Villa C., Schertler G. F. EMBO J. 2004;23:3609–3620. doi: 10.1038/sj.emboj.7600374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okada T., Palczewski K. Curr. Opin. Struct. Biol. 2001;11:420–426. doi: 10.1016/s0959-440x(00)00227-x. [DOI] [PubMed] [Google Scholar]
- 35.Yan E. C. Y., Kazmi M. A., Ganim Z., Hou J. M., Pan D. H., Chang B. S. W., Sakmar T. P., Mathies R. A. Proc. Natl. Acad. Sci. USA. 2004;100:9262–9267. doi: 10.1073/pnas.1531970100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kenakin T. Trends Pharmacol. Sci. 2003;24:346–354. doi: 10.1016/S0165-6147(03)00167-6. [DOI] [PubMed] [Google Scholar]
- 37.Swaminath G., Xiang Y., Lee T. W., Steenhuis J., Parnot C., Kobilka B. K. J. Biol. Chem. 2004;279:686–691. doi: 10.1074/jbc.M310888200. [DOI] [PubMed] [Google Scholar]
- 38.Otwinowski Z., Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 39.Yeates T. O. Methods Enzymol. 1997;276:344–358. [PubMed] [Google Scholar]
- 40.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Acta. Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.