Abstract
Bacterial transglycosylases are enzymes that couple the disaccharide subunits of peptidoglycan to form long carbohydrate chains. These enzymes are the target of the pentasaccharide antibiotic moenomycin as well as the proposed target of certain glycopeptides that overcome vancomycin resistance. Because bacterial transglycosylases are difficult enzymes to study, it has not previously been possible to evaluate how moenomycin inhibits them or to determine whether glycopeptide analogues directly target them. We have identified transglycosylase assay conditions that enable kinetic analysis of inhibitors and have examined the inhibition of Escherichia coli penicillin-binding protein 1b (PBP1b) by moenomycin as well as by various glycopeptides. We report that chlorobiphenyl vancomycin analogues that are incapable of binding substrates nevertheless inhibit E. coli PBP1b, which shows that these compounds interact directly with the enzyme. These findings support the hypothesis that chlorobiphenyl vancomycin derivatives overcome vanA resistance by targeting bacterial transglycosylases. We have also found that moenomycin is not competitive with respect to the lipid II substrate of PBP1b, as has long been believed. With the development of suitable methods to evaluate bacterial transglycosylases, it is now possible to probe the mechanism of action of some potentially very important antibiotics.
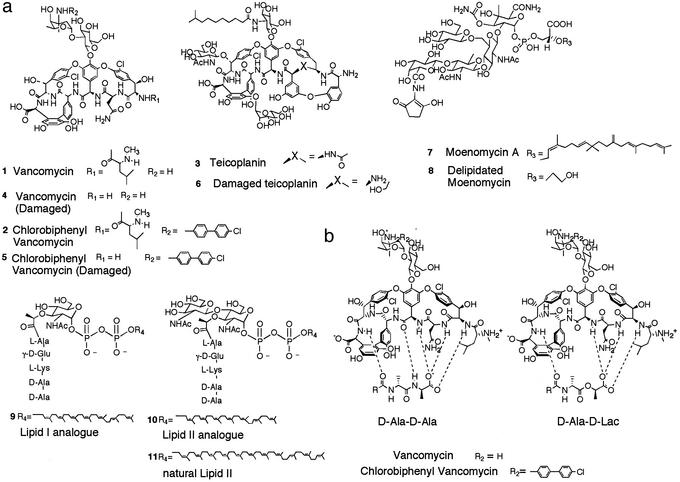
Vancomycin (1, Fig. 1a) is a glycopeptide antibiotic that inhibits the final steps of peptidoglycan biosynthesis by binding to the d-Ala-d-Ala dipeptide termini of peptidoglycan precursors so that they cannot be further processed (1, 2). The emergence of vancomycin resistance in the last decade has caused considerable concern because this drug is widely used to treat methicillin-resistant Gram-positive infections and there are few alternatives to it. The most common forms of resistance to vancomycin occur in enterococcal strains and involve the acquisition of a set of genes encoding proteins that direct peptidoglycan precursors to incorporate d-Ala-d-Lac (Lac = lactate) instead of d-Ala-d-Ala (3, 4). Because vancomycin does not bind d-Ala-d-Lac (Fig. 1b), bacterial cells expressing this modified peptide are not sensitive to the drug.
Figure 1.
(a) Structures of the compounds. (b) Structural basis for vancomycin resistance. Vancomycin binds d-Ala-d-Ala through a series of hydrogen bonds. Replacing d-Ala-d-Ala with d-Ala-d-Lac means that a critical hydrogen bond in the complex is replaced by a repulsive interaction.
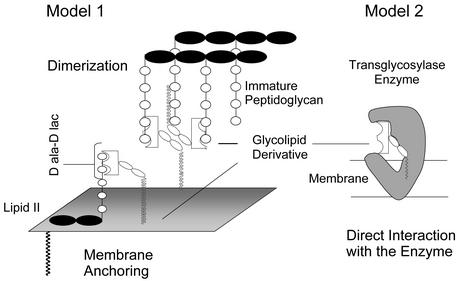
In 1989, Nagarajan et al. (5) reported that a vancomycin analogue (2) containing a chlorobiphenyl substituent attached to the vancosamino nitrogen on the disaccharide is active against vancomycin-resistant enterococcal strains. Because the disaccharide of vancomycin is not directly involved in binding to peptidoglycan precursors (Fig. 1b), it was surprising that changes to this portion of the molecule restored biological activity. Two fundamentally different theories have been proposed for how 2 overcomes vancomycin resistance. The first theory holds that the carbohydrate modification indirectly enhances binding to d-Ala-d-Lac by promoting dimerization and/or by facilitating membrane anchoring (Fig. 2, model 1; ref. 6). The second theory holds that 2 has a second mechanism of action that is independent of peptide binding (7).
Figure 2.
Proposed mechanisms for how glycolipid derivatives overcome resistance. Model 1, dimerization facilitates polyvalent interactions with substrates and/or membrane anchoring enhances avidity for lipid II through proximity. Model 2, direct interaction with the transglycosylase enzyme. The cartoon is not intended to imply anything about the mode of inhibition.
We proposed the second theory after discovering that analogues of 2 in which the peptide-binding pocket is damaged (5) still have activity against resistant microorganisms (7). These analogues were shown to inhibit the transglycosylation step of peptidoglycan biosynthesis, and we thus suggested that 2 and 5 interact directly with a component of the transglycosylation complex. Sinha Roy et al. (8) provided support for this hypothesis recently when they showed that penicillin-binding protein 1b (PBP1b), the major transglycosylase in Escherichia coli, is retained on an affinity column derivatized with 2 or fragments thereof. We now report a study showing that 2 and 5 inhibit PBP1b by interacting directly with the enzyme (Fig. 2, model 2). The results confirm that vancomycin analogues containing biaryl substituents on the vancosamino sugar have a second mechanism of action, direct inhibition of bacterial transglycosylases, that may explain their ability to kill resistant microorganisms.
Materials and Methods
Synthesis of Substrate Analogues.
Lipid I analogue 9 was prepared as described in Men et al. (9) and Ye et al. (10) and enzymatically converted to lipid II analogue 10 by using E. coli MurG as described in Lo et al. (11).
Synthesis of Test Compounds.
Vancomycin analogues 4 and 5 were prepared as described in Booth et al. (12). Teicoplanin analogue 6 was prepared as described in Malabarba et al. (13). Moenomycin analogue 8 was prepared following the procedures described in Vogel et al. (14).
Transglycosylase Assays.
PBP1b preparations.
The gene encoding E. coli PBP1b was PCR amplified from MG1655 genomic DNA and cloned into the NdeI and XhoI sites of the pET21b vector (Novagen) for expression in BL21(DE3). The gene was sequenced and found to be identical to the sequence reported in Blattner et al. (15) except for a 3′ extension encoding the C-terminal histidine tag, insertion of a lysine codon after the methionine initiation codon, and a C-to-A transversion leading to a proline to glutamine mutation at amino acid 791. The alterations in the expressed protein lie outside the transglycosylase domain, which ends approximately at amino acid 423. Bacterial cultures were grown at 37°C to an OD of 0.6 at 595 nm and protein expression was induced by adding 1 mM isopropyl β-d-thiogalactoside. After another 3 h at 37°C, the cells were pelleted, lysed in a French press cell, and centrifuged to separate the soluble and insoluble fractions. The insoluble fraction was incubated at 60°C for 10 min to deactivate penicillin-binding proteins other than PBP1b (16). The insoluble fraction was then resuspended in a buffer containing 50 mM Tris (pH 8.0), 200 mM K2HPO4 (pH 8.0), 0.1 mg/ml DNase, 20 μl/ml protease inhibitor mixture (Sigma), 1 mM DTT, and 2% octyl glucoside, and was centrifuged at 20,000 × g for 5 min to remove insoluble material. The PBP1b concentration in the decanted supernatant was estimated to be 1.2 mg/ml based on the concentration of moenomycin required to fully inhibit the transglycosylation reaction (see Results). Protein was stored as a 50% glycerol stock at −70°C. A PBP1b knockout strain subjected to the same treatment had no detectable transglycosylase activity.
Enzymatic assays.
Assays were carried out by separately incubating various amounts of [14C]GlcNAc-labeled lipid II analogue 10 (specific activity = 273 cpm/pmol) and inhibitors (concentrations are indicated in the figure legends) in Eppendorf tubes containing 9 μl of buffer [50 mM Hepes at pH 7.5, 10 mM CaCl2, 1,000 units/ml penicillin G, 0.2 mM octaethylene glycol monodecyl ether (decyl-PEG; Anatrace, Maumee, OH), and 11% DMSO] and 1 μl of PBP1b (from a solution freshly prepared by diluting the 50% glycerol stock 20-fold into 5 mM Tris buffer (pH 8.0) containing 8 mM decyl-PEG) for 10 min. Reaction mixtures were started by adding 1 μl of the above PBP1b mixture to the substrate solution and were typically stopped after 15 min by adding 10 μl of ice-cold 10 mM Tris (pH 8.0) containing 0.2% Triton X-100. Reactions were left on ice until they were spotted on cellulose chromatography paper (3MM Whatman chromatography paper). Products and starting material were separated according to Anderson et al. (1) by using chromatography (isobutyric acid/1 M NH4OH, 5:3) and were quantitated by scintillation counting.
Results
To determine whether the chlorobiphenyl vancomycin derivatives inhibit bacterial transglycosylases, we first needed to develop an assay to monitor the activity of one of these enzymes. We chose to focus initially on the major transglycosylase in E. coli, PBP1b, because it has been previously purified in small amounts and shown to convert natural lipid II (11, Fig. 1a) to a polymeric product (17). It is the single best characterized transglycosylase (18); nevertheless, only one detailed kinetic analysis has been reported (19). Bacterial transglycosylases have generally been difficult to study because lipid II, their substrate, cannot readily be obtained in useful quantities from natural sources. Furthermore, lipid II contains a 55-carbon polyprenyl chain that renders it insoluble in water and difficult to handle in enzyme assays. To enable the study of bacterial transglycosylases, we and others (10, 20, 21) have established synthetic routes to natural lipid II and analogues containing truncated polyprenyl chains (10). We previously reported that compound 10, which contains a 35-carbon lipid chain, is a better substrate than natural lipid II to use for monitoring PBP1b activity in vitro (10). Compound 10 contains the features essential for recognition by PBP1b, but because the lipid chain is shorter, the compound does not aggregate as extensively as natural lipid II containing a 55-carbon chain. Because we have found that 10 reacts reliably under a wider range of conditions than does natural lipid II, we have used it for the studies reported below.
Initial efforts to use compound 10 in transglycosylase inhibition assays were hampered because the enzyme was found to display biphasic kinetics, with a lag phase lasting for several minutes after the start of the reaction. In the recent kinetic studies of PBP1b, a lag phase was also observed when synthetic, full-length lipid II was used as the substrate (19). Schwartz et al. (19), however, reported that no lag phase was observed when additional aliquots of lipid II were added to a reaction initiated earlier. Bacterial transglycosylases are believed to be processive enzymes that form polymeric products from lipid II. Schwartz et al. (19) thus attributed the lag phase to the slow coupling of lipid II molecules and the more rapid phase to the faster coupling of lipid II to a partially polymerized “primer.” An alternative explanation, which would also be consistent with the reported experiment, is that the enzyme undergoes a slow transition to an active conformation after being added to the reaction buffer. In fact, we have found that the lag phase can be eliminated by storing the enzyme in 50% glycerol and incubating it in decyl-PEG-containing buffer for several minutes before the addition of substrate. Decyl-PEG is not required for activity, but it does increase the reaction rate. The reaction rate is linear for at least 30 min at 2 μM lipid II and 100 min at 20 μM lipid II. Under the optimized reaction conditions, the enzyme obeys Michaelis–Menten kinetics, and the apparent Km for lipid II is ≈2 μM, in good agreement with the value determined previously (18, 19). The kcat, based on an estimated PBP1b concentration of 30 nM (see below), is calculated to be 0.07 s−1, which is also consistent with values determined previously (18, 19). With the development of conditions that produce a linear initial rate, we were finally able to evaluate inhibition of the enzyme by various glycopeptides and other compounds.
The Compounds.
The compounds 1–8 in Fig. 1 were examined. Compounds 1 and 2 are vancomycin and chlorobiphenyl vancomycin, respectively. Chlorobiphenyl vancomycin is active against both major forms of vancomycin-resistant enterococcal strains, vanA and vanB strains (22). Compound 3 is teicoplanin, a glycopeptide antibiotic currently in clinical use in Japan, which contains, like compound 2, a hydrophobic substituent on the A4 glycoside. Teicoplanin (3) is active against vanB resistant enterococcal strains but not vanA resistant enterococcal strains (23). It was included in the present studies because it contains a hydrophobic substituent and thus serves as a control for inhibitory effects caused by nonspecific interactions involving hydrophobic moieties. Compounds 4, 5, and 6 are derivatives of 1, 2, and 3 in which the peptide-binding pockets have been damaged so that they no longer bind d-Ala-d-Ala. Finally, we also examined moenomycin 7 and the analogue 8, in which the lipid chain has been removed by oxidative cleavage. Moenomycin is a commercially used transglycosylase inhibitor. It was proposed many years ago that moenomycin mimics the substrates in the transglycosylation reaction and is a competitive inhibitor of the transglycosylase enzymes (24, 25). However, its mode of inhibition has not been evaluated previously.
IC50 Measurements.
Compounds 1–8 were evaluated for their ability to inhibit PBP1b under a standard set of assay conditions. As expected, vancomycin (1), chlorobiphenyl vancomycin (2), and teicoplanin (3) were effective inhibitors of transglycosylase activity, with IC50 values in the low micromolar range (Table 1). However, the damaged versions of vancomycin (4) and teicoplanin (6) were both poor inhibitors, with IC50 values being two orders of magnitude higher than the parent compounds. Their inability to inhibit transglycosylase effectively is consistent with their inability to bind d-Ala-d-Ala. Compound 5, damaged chlorobiphenyl vancomycin, which similarly does not bind d-Ala-d-Ala, was nevertheless found to have an IC50 value comparable to compounds 1–3. Therefore, the chlorobiphenyl substituent attached to the disaccharide somehow confers transglycosylase inhibitory activity onto the glycopeptide analogue. We do not believe that the effects of the chlorobiphenyl group are related to nonspecific hydrophobic interactions because the damaged teicoplanin derivative (6), which also contains a lipid-substituted carbohydrate moiety, does not have transglycosylase inhibitory activity.
Table 1.
IC50 values of the compounds
| Compound | IC50, μM |
|---|---|
| Vancomycin (1) | 0.38 |
| Damaged vancomycin (4) | 144 |
| Chlorobiphenyl vancomycin (2) | 1.50 |
| Damaged chlorobiphenyl vancomycin (5) | 3.88 |
| Teicoplanin (3) | 2.04 |
| Damaged teicoplanin (6) | 698 |
| Moenomycin (7) | 0.006 |
| Delipidated moenomycin (8) | 2.33 |
IC50 values were measured at [10] = 4 μM. Data were fit by using prism software to the equation: Y = Ymin + (Ymax − Ymin)/(1 + 10(X−log IC50)h), where X is the logarithm of the inhibitor concentration, Y is the reaction rate, and h is the Hill slope.
Moenomycin (7), the proposed active site binder, proved to be the best inhibitor, with an IC50 value in the nanomolar range. At the enzyme concentration used in these assays, 30 nM moenomycin shuts down enzymatic activity completely. Assuming that moenomycin inhibits PBP1b by binding to the enzyme in a 1:1 ratio, this concentration provides an estimate for the upper limit of the enzyme concentration.
The presence of a long lipid chain on moenomycin has been shown to be essential for good antimicrobial activity (26, 27). To assess the role of the lipid chain in enzyme inhibition, we tested the delipidated moenomycin derivative 8. It was found to be more than two orders of magnitude less potent as an inhibitor, implying that the lipid chain enhances binding to the enzyme. Nevertheless, the potency of the delipidated derivative is comparable to that of the glycopeptides.
Inhibition Kinetics.
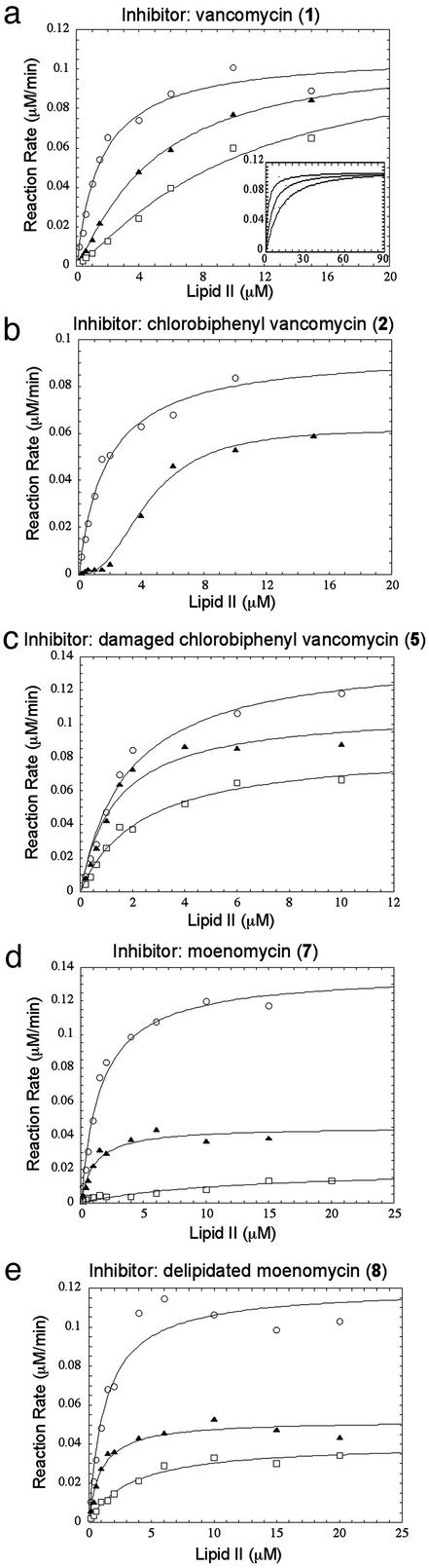
Compounds 1, 2, 5, 7, and 8 were subjected to more detailed kinetic analysis to probe their modes of inhibition. The velocity versus [substrate] curve for vancomycin (1) indicates that inhibition is competitive with respect to lipid II. Closer inspection of the curves reveals a sigmoidal shape in the presence of vancomycin, with lower-than-expected activity at low substrate concentrations (Fig. 3a). This sigmoidal shape is characteristic of inhibitors that bind to substrates (28). The inhibition curves observed with chlorobiphenyl vancomycin (2) are also sigmoidal, suggesting that at low substrate concentrations 2 similarly inhibits PBP1b by binding to the lipid II substrate (Fig. 3b). However, at higher substrate concentrations 2 differs from 1 in that the inhibition cannot be competed away. This result suggests that compound 2, in addition to binding to lipid II, has another mode of inhibition. Compound 5, the damaged version of 2, behaves similarly to 2 in that the inhibition cannot be competed away at high concentrations of lipid II; however, the velocity versus substrate curves for 5 are not sigmoidal (Fig. 3c), which is consistent with the inability of 5 to bind the d-Ala-d-Ala-containing substrate. The kinetic results reveal a marked difference in the manner in which chlorobiphenyl derivatives of vancomycin and vancomycin itself inhibit bacterial transglycosylase.
Figure 3.
Inhibition kinetics of vancomycin derivatives. (a) [Vancomycin (1)] = 0 μM (○), 0.5 μM (▴), or 1.0 μM (□). The data were fit to the Michaelis–Menten equation: v = (Vmax × S)/(Km + S), where Vmax is the maximum reaction rate, Km is the Michaelis–Menten constant, and S represents the concentration of free substrate available to the enzyme. In the presence of vancomycin, S was calculated for any given total inhibitor and total substrate concentration by using 1 μM as the dissociation constant of the vancomycin–lipid II complex (42), based on a simple substrate-depletion mechanism (28). (Inset) Data over a greater concentration range. (b) [Chlorobiphenyl vancomycin (2)] = 0 μM (○) or 3 μM (▴). (c) [Damaged chlorobiphenyl vancomycin (5)] = 0 μM (○), 2 μM (▴), or 5 μM (□). (d) [Moenomycin (7)] = 0 nM (○), 4 nM (▴), or 8 nM (□). (e) [Delipidated moenomycin (8)] = 0 μM (○), 1 μM (▴), or 4 μM (□).
Chlorobiphenyl derivatives of vancomycin are reported to form dimers, and possibly, other aggregates in solution (22). Therefore, we considered the possibility that the transglycosylase inhibition observed for both 2 and 5 at higher substrate concentrations might be due not to the compounds themselves, but to aggregates of the compounds. McGovern et al. (29) have recently described the kinetic behavior of inhibitory aggregates and have suggested a set of criteria that enable one to distinguish noncompetitive inhibition caused by a compound aggregate from that caused by a compound itself. In general, the IC50 values of compound aggregates are highly dependent on reaction conditions such as ionic strength, pH, and the presence of detergents or other proteins such as BSA. The IC50 values of the tested compounds were unaffected by the salt concentration (from 10 to 250 mM) or the presence of decyl-PEG (up to 2 mM). BSA affected the activity of the enzyme in the absence of inhibitors, making it difficult to evaluate its effect on the inhibitors. Nevertheless, BSA had a significantly smaller effect on the IC50 of these compounds than on the aggregates reported by McGovern et al. (29). Therefore, the simplest interpretation of the inhibition results for the chlorobiphenyl derivatives is that 5 interacts directly with PBP1b, and that the noncompetitive component of the inhibition caused by the parent compound 2 is similarly related to its ability to interact with the enzyme.
Moenomycin (7) was also tested and, like compounds 2 and 5, found to inhibit PBP1b in a manner that cannot be overcome by adding more substrate (Fig. 3d). To determine whether the lipid chain, which increases the affinity of moenomycin for the enzyme, also influences the mode of inhibition, we examined the inhibition patterns for delipidated derivative 8. It, too, is not competitive with respect to lipid II (Fig. 3e). Thus, we have been able to establish that the inability of lipid II to compete away the inhibition is not an artifact due to the presence of the hydrophobic chain, but rather reflects the manner in which the pentasaccharide portion of the molecule interacts with the enzyme.
Discussion
Bacterial transglycosylases are targets for a number of natural products, including the pentasaccharide moenomycin and the glycopeptides vancomycin and teicoplanin. The transglycosylation reaction was originally described in 1965 by Strominger (1), but it was not feasible to test how various compounds inhibit bacterial transglycosylases until now. A major impediment to studying these enzymes has been that adequate supplies of lipid II cannot be obtained from natural sources. The development of synthetic approaches to make lipid II and analogues has solved the substrate problem (10, 20, 21), making it possible to assay inhibitors.
Having an efficient route to lipid II and assay conditions to monitor PBP1b activity has allowed us to analyze the inhibition kinetics of moenomycin, vancomycin, and various derivatives. Our results confirm hypotheses for how some of these compounds function, but they raise questions about others. For example, the sigmoidal velocity versus substrate curves generated in the presence of vancomycin confirm that the molecule inhibits PBP1b by binding to the lipid II substrate. As expected for a simple substrate binder (28, 30), the inhibition can be overcome by competition at high concentrations of lipid II (Fig. 3a Inset). In contrast to vancomycin, the experiments on moenomycin do not support the proposed mechanism for inhibition. Although this compound was predicted to be a competitive inhibitor with respect to lipid II (24, 31, 32), our results show that it is not. Lipid II cannot overcome the inhibition. We considered the possibility that lipid II might not compete effectively with moenomycin because the moenomycin lipid chain prevents dissociation. However, the delipidated derivative shows a very similar inhibition pattern, albeit at higher concentrations. The inability of lipid II to displace the water-soluble, delipidated moenomycin implies that this inhibitor does not bind in the immediate vicinity of lipid II. Because PBP1b makes a polymer, it is presumed to have an extended saccharide-binding site for the growing chain and it remains possible that moenomycin occupies at least part of this site, thereby blocking elongation. It should be possible to glean more information about the mode of binding of moenomycin and its derivatives once additional substrate probes are developed. In the meantime, it is worth noting that because our results suggest that the lipid chain on moenomycin plays a nonspecific role in activity, it should be possible to replace it with groups having more desirable physical properties (33).
The results on moenomycin are intriguing, but the initial motivation for doing these studies was to probe the mechanism of action of lipid-linked derivatives of vancomycin. We have previously made many glycopeptide derivatives containing lipid substituents at different positions, either on the aglycone or on the carbohydrates (7, 33–36). We have found that the potency of most lipid-linked glycopeptides decreases dramatically when their peptide-binding pockets are damaged, implying that peptide binding is important for activity. However, a small subset of lipid-linked glycopeptides retain good activity against both vanA and vanB strains when their binding pockets are damaged. Activity against vanA strains requires specific positioning of a hydrophobic substituent attached to the vancomycin disaccharide (7, 34, 36).
Overcoming vanA resistance is challenging because it requires that glycopeptide analogues be able to kill bacterial cells that express d-Ala-d-Lac. Several models have been proposed to explain the activity of glycolipid analogues of vancomycin against vanA strains. According to one model, the vancomycin analogues have enhanced avidity for d-Ala-d-Lac because membrane anchoring localizes them near the altered lipid II precursors that are produced. Many lipid-linked glycopeptides, including teicoplanin, can anchor to bacterial membranes, but only those with a particular glycolipid structure have activity against vanA strains. Therefore, we do not think that vanA activity is explained by the membrane-anchoring hypothesis, which involves a nonspecific mechanism.
Williams and coworkers (37–39) have shown that some glycopeptides form dimers in solution and that attaching lipid substituents to the carbohydrate portion lowers the concentration at which they dimerize. They have proposed that these derivatives have enhanced avidity for d-Ala-d-Lac moieties presented on bacterial cell surfaces because the interactions are effectively divalent (40). There is considerable biophysical data to support the hypothesis that lipid-substituted glycopeptides have enhanced avidity for both d-Ala-d-Ala- and d-Ala-d-Lac-modified surfaces (41), and it has also been shown that covalent dimers and trimers of vancomycin bind covalent multimers of d-Ala-d-Ala many orders of magnitude better than the monomers bind (42). Nevertheless, there is no evidence that chlorobiphenyl vancomycin derivatives kill vanA strains by dimerizing near cell surfaces and binding d-Ala-d-Lac (43, 44). Furthermore, it is virtually impossible to design biophysical experiments that report on the extent to which enhanced d-Ala-d-Lac binding contributes to killing resistant microorganisms.
One requirement of the Williams model (37–39) is that glycopeptides must bind peptidoglycan precursors to kill cells. This requirement can be tested by damaging the peptide-binding pockets of lipid-substituted glycopeptides and evaluating their activities against relevant strains. Because glycopeptide analogues containing hydrophobic substituents on the vancosamino nitrogen retain activity against vanA strains when the binding pocket is damaged (7, 34–36), the Williams model cannot explain the activity of these compounds. Therefore, we have proposed that certain glycopeptide analogues have a second mechanism of action that does not involve peptide binding (7, 45). In this paper, we have presented kinetic evidence that chlorobiphenyl-vancosamino derivatives of vancomycin can inhibit PBP1b, the major transglycosylase in E. coli, even in the absence of substrate binding. This result is consistent with previous work (7) showing that damaged chlorobiphenyl vancomycin blocks the transglycosylation step of peptidoglycan synthesis. PBP1b shares homology with Gram-positive transglycosylases and so it is possible that the chlorobiphenyl derivatives similarly inhibit the Gram-positive transglycosylases. If so, this finding would explain the activity of chlorobiphenyl vancomycin and related compounds against vanA strains. The ability of glycopeptide derivatives to inhibit Gram-positive transglycosylases remains to be tested.
Acknowledgments
We thank Michael Gelb for the careful reading of this manuscript. This work was supported by National Institutes of Health Grants R01GM66174 (to D.K.) and R01AI50855 (to S.W.).
Abbreviations
- Lac
lactate
- PBP1b
penicillin-binding protein 1b
- decyl-PEG
octaethylene glycol monodecyl ether
References
- 1.Anderson J S, Matsuhashi M, Haskin M A, Strominger J L. Biochemistry. 1965;53:881–889. doi: 10.1073/pnas.53.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perkins H R. Biochem J. 1969;111:196–205. doi: 10.1042/bj1110195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bugg T D, Dutka-Malen S, Arthur M, Courvalin P, Walsh C T. Biochemistry. 1991;30:2017–2021. doi: 10.1021/bi00222a002. [DOI] [PubMed] [Google Scholar]
- 4.Walsh C T, Fisher S L, Park I S, Prahalad M, Wu Z. Chem Biol. 1996;3:21–28. doi: 10.1016/s1074-5521(96)90079-4. [DOI] [PubMed] [Google Scholar]
- 5.Nagarajan R, Schabel A A, Occolowitz J L, Counter F T, Ott J L, Felty-Duckworth A M. J Antibiot. 1989;42:63–72. doi: 10.7164/antibiotics.42.63. [DOI] [PubMed] [Google Scholar]
- 6.Sharman G J, Try A C, Dancer R J, Younghoon R C, Staroske T, Bardsley B, Maguire A J, Cooper M A, O'Brien D P, Williams D H. J Am Chem Soc. 1997;119:12041–12047. [Google Scholar]
- 7.Ge M, Chen Z, Onishi H R, Kohler J, Silver L L, Kerns R, Fukuzawa S, Thompson C, Kahne D. Science. 1999;284:507–511. doi: 10.1126/science.284.5413.507. [DOI] [PubMed] [Google Scholar]
- 8.Sinha Roy R, Yang P, Kodali S, Xiong Y, Kim R M, Griffin P R, Onishi H R, Kohler J, Silver L L, Chapman K. Chem Biol. 2001;8:1095–1106. doi: 10.1016/s1074-5521(01)00075-8. [DOI] [PubMed] [Google Scholar]
- 9.Men H, Park P, Ge M, Walker S. J Am Chem Soc. 1998;120:2484–2485. [Google Scholar]
- 10.Ye X Y, Lo M-C, Brunner L, Walker D, Kahne D, Walker S. J Am Chem Soc. 2001;123:3155–3156. doi: 10.1021/ja010028q. [DOI] [PubMed] [Google Scholar]
- 11.Lo M-C, Men H, Branstrom A, Helm J, Yao N, Goldman R, Walker S. J Am Chem Soc. 2000;122:3540–3541. [Google Scholar]
- 12.Booth P M, Stone D J M, Williams D H. J Chem Soc Chem Commun. 1987;22:1694–1695. [Google Scholar]
- 13.Malabarba A, Ciabatti R, Kettenring J, Ferrari P, Vekey K, Bellasio E, Denaro M. J Org Chem. 1996;61:2137–2150. [Google Scholar]
- 14.Vogel S, Stembera K, Hennig L, Findeisen M, Giesa S, Welzel P, Tillier C, Bonhomme C, Lampilas M. Tetrahedron. 2001;57:4147–4160. [Google Scholar]
- 15.Blattner F R, Plunkett G, 3rd, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 16.Nakagawa J, Matsuzawa H, Matsuhashi M. J Bacteriol. 1979;138:1029–1032. doi: 10.1128/jb.138.3.1029-1032.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakagawa J, Tamaki S, Tomioka S, Matsuhashi M. J Biol Chem. 1984;34:13937–13946. [PubMed] [Google Scholar]
- 18.Terrak M, Ghosh T K, van Heijenoort J, Van Beeumen J, Lampilas M, Aszodi J, Ayala J A, Ghuysen J-M, Nguyen-Disteche M. Mol Microbiol. 1999;34:350–364. doi: 10.1046/j.1365-2958.1999.01612.x. [DOI] [PubMed] [Google Scholar]
- 19.Schwartz B, Markwalder J A, Seitz S P, Wang Y, Stein R L. Biochemistry. 2002;41:12552–12561. doi: 10.1021/bi026205x. [DOI] [PubMed] [Google Scholar]
- 20.VanNieuwenhze M S, Mauldin S C, Zia-Ebrahimi M, Winger B E, Hornback W J, Saha S L, Aikins J A, Blaszczak L C. J Am Chem Soc. 2002;124:3656–3660. doi: 10.1021/ja017386d. [DOI] [PubMed] [Google Scholar]
- 21.Schwartz B, Markwalder J A, Wang Y. J Am Chem Soc. 2001;123:11638–11643. doi: 10.1021/ja0166848. [DOI] [PubMed] [Google Scholar]
- 22.Nicas T I, Mullen D L, Flokowitsch J E, Preston D A, Snyder N J, Zweifel M J, Wilkie S C, Rodriguez M J, Thompson R C, Cooper R D G. Antimicrob Agents Chemother. 1996;40:2194–2199. doi: 10.1128/aac.40.9.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baptista M, Depardieu F, Courvalin P, Arthur M. Antimicrob Agents Chemother. 1996;40:2291–2295. doi: 10.1128/aac.40.10.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welzel P, Kunisch F, Kruggel F, Stein H, Scherkenbeck J, Hiltmann A, Duddeck H, Müller D, Maggio J E, Fehlhaver H-W, et al. Tetrahedron. 1987;43:585–598. [Google Scholar]
- 25.van Heijenoort Y, Leduc M, Singer H, van Heijenoort J. J Gen Microbiol. 1987;133:667–674. doi: 10.1099/00221287-133-3-667. [DOI] [PubMed] [Google Scholar]
- 26.Marzian S, Happel M, Wagner U, Müller D, Welzel P, Fehlhaber H-W, Stark A, Schutz H-J, Markus A, Limbert M, et al. Tetrahedron. 1994;50:5299–5308. [Google Scholar]
- 27.Kempin U, Hennig L, Welzel P, Marzian S, Müller D, Fehlhaber H-W, van Heijenoort Y, van Heijenoort J. Tetrahedron. 1995;51:8471–8482. [Google Scholar]
- 28.Segel I H. Enzyme Kinetics. New York: Wiley; 1975. [Google Scholar]
- 29.McGovern S L, Caselli E, Grigorieff N, Shoichet B K. J Med Chem. 2002;45:1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- 30.Barna J C, Williams D H. Annu Rev Microbiol. 1984;38:339–357. doi: 10.1146/annurev.mi.38.100184.002011. [DOI] [PubMed] [Google Scholar]
- 31.Stembera K, Buchynskyy A, Vogel S, Knoll D, Osman A A, Ayala J A, Welzel P. ChemBioChem. 2002;3:332–340. doi: 10.1002/1439-7633(20020402)3:4<332::AID-CBIC332>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 32.Stembera K, Vogel S, Buchynskyy A, Ayala J A, Welzel P. ChemBioChem. 2002;3:559–565. doi: 10.1002/1439-7633(20020603)3:6<559::AID-CBIC559>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 33.Sun B, Chen Z, Eggert U S, Shaw S J, LaTour J V, Kahne D. J Am Chem Soc. 2001;123:12722–12723. doi: 10.1021/ja0166693. [DOI] [PubMed] [Google Scholar]
- 34.Kerns R, Dong S D, Fukuzawa S, Carbeck J, Kohler J, Silver L, Kahne D. J Am Chem Soc. 2000;122:12608–12609. [Google Scholar]
- 35.Dong S D, Oberthur M, Losey H C, Anderson J W, Eggert U S, Peczuh M W, Walsh C T, Kahne D. J Am Chem Soc. 2002;124:9064–9065. doi: 10.1021/ja026342h. [DOI] [PubMed] [Google Scholar]
- 36.Chen Z, Eggert U S, Dong S D, Shaw S J, Sun B, LaTour J V, Kahne D. Tetrahedron. 2002;58:6585–6594. [Google Scholar]
- 37.Williams D H, Searle M S, Mackay J P, Gerhard U, Maplestone R A. Proc Natl Acad Sci USA. 1993;90:1172–1178. doi: 10.1073/pnas.90.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beauregard D A, Williams D H, Gwynn M N, Knowles D J. Antimicrob Agents Chemother. 1995;39:781–785. doi: 10.1128/AAC.39.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beauregard D A, Maguire A J, Williams D H, Reynolds P E. Antimicrob Agents Chemother. 1997;41:2418–2423. doi: 10.1128/aac.41.11.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams D H, Maguire A J, Tsuzuki W, Westwell M S. Science. 1998;280:711–714. doi: 10.1126/science.280.5364.711. [DOI] [PubMed] [Google Scholar]
- 41.Rao J, Yan L, Lahiri J, Whitesides G M, Weis R M, Warren H S. Chem Biol. 1999;6:353–359. doi: 10.1016/S1074-5521(99)80047-7. [DOI] [PubMed] [Google Scholar]
- 42.Rao J, Lahiri J, Isaacs L, Weis R M, Whitesides G M. Science. 1998;280:708–711. doi: 10.1126/science.280.5364.708. [DOI] [PubMed] [Google Scholar]
- 43.Kim S J, Cegelski L, Studelska D R, O'Connor R D, Mehta A K, Schaefer J. Biochemistry. 2001;41:6967–6977. doi: 10.1021/bi0121407. [DOI] [PubMed] [Google Scholar]
- 44.Cegelski L, Kim S K, Hing A W, Studelska D R, O'Connor R D, Mehta A K, Schaefer J. Biochemistry. 2002;41:13053–13058. doi: 10.1021/bi0202326. [DOI] [PubMed] [Google Scholar]
- 45.Eggert U S, Ruiz N, Falcone B V, Branstrom A A, Goldman R C, Silhavy T J, Kahne D. Science. 2001;294:361–364. doi: 10.1126/science.1063611. [DOI] [PubMed] [Google Scholar]