Abstract
FCP1 [transcription factor IIF (TFIIF)-associated carboxyl-terminal domain (CTD) phosphatase] is the only identified phosphatase specific for the phosphorylated CTD of RNA polymerase II (RNAP II). The phosphatase activity of FCP1 is enhanced in the presence of the large subunit of TFIIF (RAP74 in humans). It has been demonstrated that the CTD of RAP74 (cterRAP74; residues 436–517) directly interacts with the highly acidic CTD of FCP1 (cterFCP; residues 879–961 in human). In this manuscript, we have determined a high-resolution solution structure of a cterRAP74/cterFCP complex by NMR spectroscopy. Interestingly, the cterFCP protein is completely disordered in the unbound state, but forms an α-helix (H1′; E945–M961) in the complex. The cterRAP74/cterFCP binding interface relies extensively on van der Waals contacts between hydrophobic residues from the H2 and H3 helices of cterRAP74 and hydrophobic residues from the H1′ helix of cterFCP. The binding interface also contains two critical electrostatic interactions involving aspartic acid residues from H1′ of cterFCP and lysine residues from both H2 and H3 of cterRAP74. There are also three additional polar interactions involving highly conserved acidic residues from the H1′ helix. The cterRAP74/cterFCP complex is the first high-resolution structure between an acidic residue-rich domain from a holoenzyme-associated regulatory protein and a general transcription factor. The structure defines a clear role for both hydrophobic and acidic residues in protein/protein complexes involving acidic residue-rich domains in transcription regulatory proteins.
RNA polymerase II (RNAP II) is a multisubunit enzyme complex that enters the initiation complex with the carboxyl-terminal domain (CTD) of its largest subunit in an unphosphorylated form (RNAP IIA). The CTD contains a heptapeptide repeat (YSPTSPS) that becomes extensively phosphorylated (RNAP IIO) primarily at serine-2 and -5 during early stages of transcription (1–3). In the last several years, numerous protein kinases have been implicated in the phosphorylation of the CTD (4–8). This phosphorylation of the CTD enables RNAP II to progress from the initiation phase to a stable elongation complex, and the CTD remains extensively phosphorylated throughout the elongation phase of transcription (4–6). After completion of the transcription cycle, this same RNAP II must be in the unphosphorylated form (RNAP IIA) to be recruited back to the initiation complex (9). Therefore, dephosphorylation of the CTD by a phosphatase(s) is essential to generating a form of the polymerase (RNAP IIA) that is capable of reinitiating transcription.
FCP1 [transcription factor IIF (TFIIF)-associating component of the CTD phosphatase], the only known RNAP II CTD-specific phosphatase, was originally partially purified from HeLa cell (10) and yeast (11) extracts. From experiments with this partially purified CTD phosphatase, it was determined that both general transcription factors IIB (TFIIB) and IIF (TFIIF) play important roles in regulating its activity (12). It was found that the evolutionarily conserved CTD of the large subunit of TFIIF (RAP74 in humans) strongly stimulates the CTD phosphatase activity (12). In contrast, TFIIB was shown to regulate the CTD phosphatase activity through its ability to block the stimulatory effect produced by the RAP74 subunit (12). Recent studies have shown that FCP1 can also stimulate elongation, and that the roles of FCP1 in elongation are distinct from its function as a phosphatase and independent of RAP74 (13).
FCP1 was first cloned on the basis of its ability to interact with the CTD of RAP74 [cterRAP74; residues 436–517 (Fig. 1A)] in a yeast two-hybrid screen (14, 15). Based on sequence homology between human and yeast FCP1, three domains have been identified in FCP1, and they are responsible for distinct functions (14, 15). The amino-terminal domain of FCP1 corresponds to the catalytic unit of the phosphatase and has been referred to as the FCP1 homology domain (14–17). The remaining central and CTD of FCP1 appear to be required for protein/protein interactions that are crucial in regulating the activity of the catalytic domain (14, 15, 18). The central region of FCP1 contains a BRCT homology domain related to the breast cancer tumor-suppressor protein BRCA1 (19). The CTD of FCP1 bears resemblance to acidic activation domains found in numerous DNA-binding proteins and has been shown to function independently as a transactivation domain (18, 20). In vitro and in vivo binding experiments have demonstrated that this acidic CTD of FCP1 [cterFCP; residues 879–961 in human (Fig. 1A)] directly interacts with cterRAP74 (14, 15, 18), and yeast deletion studies indicate that failure of FCP1 to interact with RAP74 appears to lead to loss of viability under conditions of severe DNA damage or other stresses (16, 18).
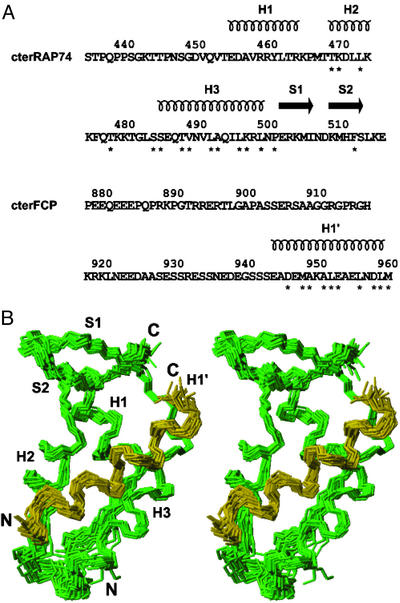
Figure 1.
(A) Sequences of cterRAP74 (residues 436–517 of RAP74) and cterFCP (residues 879–961 of human FCP1) used in the NMR structure determinations, with residue numbering and secondary structural elements represented above both protein sequences. Those amino acids that show intermolecular NOEs are indicated by an asterisk below the residue. (B) A stereoview of the 20 lowest-energy NMR structures showing the backbone trace (N, Cα, and C′) of residues 451–517 of cterRAP74 (green) and residues 945–961 of cterFCP (gold).
In earlier studies, we have shown from NMR chemical shift mapping that RAP74 and TFIIB share a common binding site on cterFCP (21). In this manuscript, we describe a high-resolution NMR solution structure of this cterRAP74/cterFCP complex and we compare the structure with other relevant structures of protein/protein complexes involved in transcriptional regulation.
Methods
Plasmids.
The GST fusion protein constructs for the FCP1-binding domain of RAP74 (cterRAP74) and the RAP74-binding domain of FCP (cterFCP) have been described (15, 21). The QuikChange site-directed mutagenesis kit (Stratagene) was used with the appropriate oligonucleotide to prepare the GST fusion protein expressing the E956A/L957A double-mutant cterFCP (cterFCPmut). The pGEX-5X vector containing the GST fusion cterFCP (15) was used as a template for the mutagenesis and the DNA sequence of the mutant resulting construct was verified.
NMR Samples.
The cterFCP and cterRAP74 proteins were expressed and purified by using published protocols (21). Uniform (>98%) 15N and 15N/13C labeling was obtained by growing the cells in a modified minimal medium containing 15NH4Cl and 13C6-glucose. NMR samples of the complex consisted of 1 mM cterRAP74/1 mM cterFCP in 20 mM sodium phosphate (pH 6.5) and 1 mM EDTA.
Binding Assays.
The GST-cterFCP fusion protein fragments were expressed from a GST-5X vector (Amersham Biosciences) in an Escherichia coli host strain, DH5α. The cells were grown at 37°C, and protein expression was induced for 3 h at 30°C with 0.7 mM isopropyl β-d-thiogalactoside (IPTG). The fusion protein was purified by affinity chromatography with glutathione-Sepharose resin (Amersham Biosciences). The purified GST-cterFCP fusion proteins were eluted with reduced glutathione (Sigma) and dialyzed into a storage buffer [50 mM Tris⋅Cl (pH 8.0)/100 mM NaCl/2 mM DTT/20% glycerol].
The purified GST-cterFCP fusion proteins were coupled to glutathione-Sepharose (10 μl) for the binding experiments. Each binding experiment was performed with 1 μM of the appropriate GST-cterFCP fusion protein (wild type/mutant) and varying concentrations (0.1–15 μM) of purified cterRAP74. The binding was performed in 500 μl of 40 mM Hepes (pH 7.9), 100 mM NaCl, 0.5% Nonidet P-40, and 10 mM DTT at 4°C for 1 h. After binding, the resin was collected by centrifugation and washed two times with 500 μl of binding buffer. The washed pellet was resuspended in 20 μl of 1× Mes gel loading buffer (Invitrogen) and proteins were resolved on a 12% NuPage gel (Invitrogen) in Mes running buffer (Invitrogen). The resolved proteins were then transferred by using the NuPage transfer buffer (Invitrogen) to an Immobilon-P membrane (Millipore). The transfer was carried out for 1 h at 25 V. The transferred proteins were detected with a primary anti-RAP74 antibody (C-18, Santa Cruz Biotechnology) and an anti-rabbit secondary antibody (Santa Cruz Biotechnology).
NMR Spectroscopy.
NMR spectra were collected at 27°C by using Varian Inova Unity 500 MHz, 600 MHz, and 800 MHz NMR spectrometers equipped with a z pulsed-field gradient unit and triple resonance probes. The backbone and aliphatic side chain signals (1H, 15N, and 13C) of cterRAP74 and cterFCP were assigned by using a combination of NMR experiments as described (21). Distance restraints were obtained from 2D and 3D NOESY experiments (22–25). Intermolecular nuclear Overhauser effects (NOEs) were obtained from a 2D 13C {F1}-filtered, {F2}-edited NOESY and a 3D 15N/13C {F1}-filtered, {F3}-edited NOESY (26). The NMR data were processed with NMRPIPE/NMRDRAW (27), and analyzed with pipp (28) and nmrview (29).
Structure Calculations.
The NOE-derived restraints were subdivided into four classes, strong (1.8–2.7 Å), medium (1.8–3.3 Å), weak (1.8–5.0 Å), and very weak (1.8–6.0 Å), by comparison with NOEs involving protons separated by known distances, as described (21). An extra 0.2 Å was added to the upper distance limit for NOE restraints in the medium and strong distance range that involved NH protons, and 0.5 Å was added to the upper distance limit for restraints involving methyl protons. A total of 1,131 NOE-derived distance restraints was obtained for structure calculation. Backbone dihedral angle restraints (φ and ψ angles) were obtained from analysis of 1Hα, 1HN, 13Cα, 13Cβ, 13CO, and 15N chemical shifts by using the program talos (30). Structures were calculated by using the Torsion Angle Molecular Dynamics (TAD) protocol of cns (31), starting from two extended structures (one for cterFCP and one for cterRAP74) with standard geometry. The extended cterRAP74-(451–517) structure was placed horizontally ≈10 Å apart from the extended cterFCP-(941–961) before initiating the structure determination protocol. The quality of the structures was analyzed by using the PROCHECK_NMR program (32). All figures were generated by using molmol (33) and molscript (34).
Results and Discussion
Structure Determination of the Complex.
The 3D structures of the cterRAP74/cterFCP complex were calculated by using the TAD protocol of cns (ref. 31; Fig. 1). Because of the absence of medium-range, long-range, and intermolecular NOEs for amino acids 436–450 of cterRAP74 and amino acids 879–940 of cterFCP, these amino-terminal residues were not included in the calculations. From the 75 calculated structures, 50 were accepted because they did not display any distance restraint violation >0.2 Å and any dihedral angle restraint (ψ and ϕ) violation >5°. The 20 structures with the lowest energy were selected for further analysis (Table 1). A backbone superposition of these 20 structures (Fig. 1B) indicates that the overall topology of the cterRAP74/cterFCP complex is well defined by the NMR data.
Table 1.
Structural statistics
| Restraints used in the structure calculation* | |
| Number of distance restraints | |
| Intraresidue | 408 |
| Interresidue sequential (i, i + 1) | 252 |
| Interresidue medium range (1 < |i − j| < 5) | 228 |
| Long range [(|i − j|) ≥ 5] | 184 |
| Intermolecular | 58 |
| Total | 1,131 |
| Structural statistics | |
| rms deviations from distance restraints, Å | 0.0037 ± 0.0008 |
| Number of torsion angle restraints (φ, ψ)† | 136 |
| rms deviations from dihedral restraints, ° | 0.076 ± 0.021 |
| rms deviations from idealized geometry | |
| Bonds, Å | 0.00080 ± 0.00003 |
| Angle, ° | 0.30530 ± 0.0012 |
| Improper, ° | 0.1011 ± 0.0033 |
| Coordinate precision | |
| rms deviations from minimized average structure‡, Å | |
| Backbone atoms | 0.69 ± 0.08 |
| All heavy atoms | 1.25 ± 0.10 |
| Ramachandran analysis§ | |
| Most favored region, % | 84.5 |
| Additionally allowed region, % | 14.0 |
| Generously allowed region, % | 0.6 |
| Disallowed regions, % | 0.9 |
None of the 20 structures exhibited distance violations >0.2 Å or dihedral angles >5°.
Backbone dihedral angle restraints were generated with talos (30) based on analysis of 1Hα, 1HN, 13Cα, 13Cβ, 13C′, and 15N chemical shifts.
The coordinate precision is defined as average atomic rms deviation between the 20 structures with lowest energy and the minimized average structure for residues 452–476 and 486–514 of cterRAP74 and 945–961 of cterFCP. The backbone value refers to N, Cα, and C′.
procheck_nmr (32) was used to assess the quality of the structures.
Structure of cterRAP74 in the Complex.
Similarly to the cterRAP74-free form (21, 35), the cterRAP74 in the complex also forms a winged-helix domain consisting of three consecutive α-helices followed by an antiparallel β-sheet. The helices are of residues E456–R465 (H1), T470–K475 (H2), and S486–L500 (H3) and are linked by loops of 4 (L1) and 10 (L2) amino acids (Fig. 1A). The β-sheet is located at the carboxyl-terminal end of the domain and is formed by a first strand of residues E503–I507 (S1) and a second strand of residues K510–S514 (S2) (Fig. 1A). A superposition of the minimized average solution structures of cterRAP74 free and bound to cterFCP yields a backbone rmsd of 0.55 Å for residues 456–476 and 486–514. It is interesting to note that there is a reduction of ≈750 Å2 in the surface-accessible area of cterRAP74 in the presence of cterFCP.
Structure of cterFCP in the Complex.
The free cterFCP protein is devoid of any stable structural element but adopts a 17-residue α-helix (H1′) at its carboxyl terminus (E945-M961) on interaction with cterRAP74 (Fig. 1A). These residues of cterFCP that are involved in formation of the α-helix display significant changes in amide 1H and 15N chemical shifts between the free state and the complex. Chemical shift index (CSI) analysis of the Hα, Cα, Cβ, and C′ chemical shifts of cterFCP also supports the presence of the H1′ helix for residues E945–M961 in the complex, but not in the free form (36, 37). The importance of the H1′ helix for interaction with cterRAP74 is supported by numerous intermolecular NOEs between the H1′ α-helix of cterFCP and cterRAP74 (Fig. 2). It should be noted that an α-helix of residues 942–961 in cterFCP had been predicted based on primary sequence analysis in an earlier study (35), and the authors postulated that this helix may be important in binding to cterRAP74. Although there is no evidence for the α-helix in the free cterFCP, the predicted helix is fairly close to the 17-residue H1′ helix we observe in the cterRAP74/cterFCP complex.
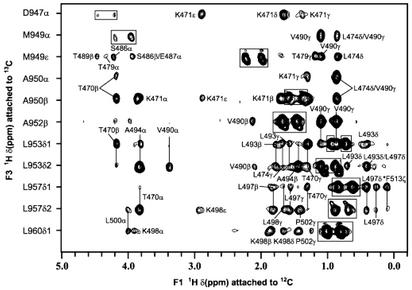
Figure 2.
Composite of 13C/1H strips taken from the 3D 13C {F1}-filtered, {F3}-edited NOESY-HSQC spectrum of the cterRAP74/cterFCP complex illustrating NOEs from protons of unlabeled cterRAP74 (attached to 12C along the F1 axis) to protons of 15N/13C-labeled cterFCP (attached to 13C). The 1H(F3)/13C(F2) strips were taken at chemical shifts (ppm) of D947α (4.35/57.74), M949α (4.11/59.25), M949ɛ (2.12/17.08), A950α (3.85/55.79), A950β (1.48/17.87), A952β (1.58/18.40), L953δ1 (0.84/26.34), L953δ2 (0.98/23.87), L957δ1 (0.73/26.31), L957δ2 (0.81/23.35), and L960δ1 (0.92/25.68).
Interface of the cterRAP74/cterFCP Complex.
In our previous studies, a cterFCP-binding site on cterRAP74 was identified on the basis of amide 1H, 15N, and 13C chemical shift changes between the free and bound states of cterRAP74 (21). The cterFCP-binding site was shown to consist of an exposed hydrophobic groove between α-helices H2 and H3 of cterRAP74. This binding site had also been previously postulated based on mutational data (18) mapped on the crystal structure of the zinc-bound cterRAP74 (35). In the NMR structure of the complex, this exposed hydrophobic groove formed by the side chain residues of H2 (L474), H3 (V490, L493, A494, L497, and L500), and S2 (F513) of cterRAP74 binds to the newly formed amphipathic helix H1′ in cterFCP (Fig. 3A). The hydrophobic groove in cterRAP74 provides a binding interface for the nonpolar face of the H1′ helix, which is composed of the side chains of M949, L953, L957, L960, and M961 (Fig. 3A). Specifically, the hydrophobic interface formed between cterRAP74 and cterFCP consists of van der Waals contacts between L474 and M949, V490 and M949, V490 and A950, V490 and A952, L493 and L953, A494 and L953, L497 and L957, L497 and F513, and L500 and L960. The formation of the cterRAP74/cterFCP interface is supported by the observation of numerous intermolecular NOEs between side chain methyl groups of cterRAP74 and cterFCP (Fig. 2). In addition, intermolecular NOEs are observed between the aromatic side chain of F513 (S2) in cterRAP74 and both δ methyl groups of L957 and L960 in cterFCP.
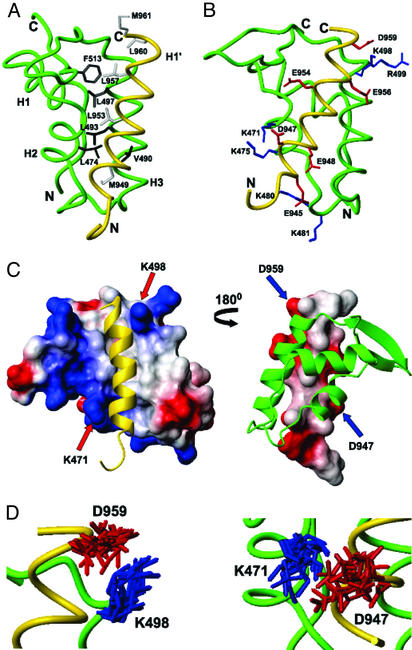
Figure 3.
Interactions between cterRAP74 and cterFCP. (A) Backbone trace of the cterRAP74 (green)/cterFCP (gold) complex highlighting the hydrophobic amino acids of cterRAP74 (dark gray) and cterFCP (pale gray) at the interface. (B) Backbone trace of the cterRAP74/cterFCP complex (green/gold) highlighting the basic amino acids of cterRAP74 (blue), and the acidic amino acids of cterFCP (red) at the interface. (C) Molecular surface representation of cterRAP74-(451–517) (Left) and cterFCP-(941–961) (Right) from the cterRAP74/cterFCP complex. In the representation of the cterRAP74 surface, cterFCP (yellow ribbon) fits within a hydrophobic groove on the surface of cterRAP74. Areas of positive (blue) and negative (red) charge are highlighted and the two lysines residues involved in formation of the well defined salt bridges are labeled (K471 and K498). In the representation of the cterFCP surface (Right), the hydrophobic side chains of cterFCP are directly inserted into the hydrophobic groove of cterRAP74 (green ribbon). Areas of negative charge (red) are highlighted and the two aspartic acid residues involved in formation of the salt bridges are labeled (D947 and D959). (D) Overlay of the 20 lowest-energy NMR structures, highlighting the lysine and aspartic acid side chains in the two salt bridges (K498/D959 and K471/D947).
In previous structural studies mapping the cterFCP-binding site on cterRAP74, it was noted that numerous positively charged amino acid residues surrounded the exposed hydrophobic groove in cterRAP74 (21, 35). Because cterFCP is highly acidic, it was hypothesized that the binding of the cterRAP74/cterFCP complex would most likely involve specific electrostatic interactions. The basic residues surrounding the hydrophobic groove are located in H2 (K471 and K475), H3 (K498 and R499), and the connecting loop L2 (K480 and K481) of cterRAP74 (Fig. 3B). Interestingly, in the structure of the complex, several acidic residues from the H1′ helix of cterFCP (Glu-945, Asp-947, Glu-948, Glu-954, Glu-956, and Asp-959; Fig. 3B) form negatively charged patches adjacent to the hydrophobic residues (Fig. 3 B and C). In our energy-minimized structure, we clearly detect two salt bridges between cterRAP74 and cterFCP in the complex on the basis of close proximity (Fig. 3 C and D). The first salt bridge is formed by K471 of cterRAP74 and D947 of cterFCP, and this links the amino-terminal end of the H1′ helix of cterFCP to the H2 helix of cterRAP74 (Fig. 3 C and D). The formation of the salt bridge is supported by intermolecular NOEs observed between the side chains of K471 and D947 (Fig. 2). The second salt bridge is formed by K498 of cterRAP74 and D959 of cterFCP and bridges the carboxyl-terminal of the H1′ helix from cterFCP to the H3 helix of cterRAP74. The formation of the second salt bridge is supported by intermolecular NOEs between the side chains of K498 and L960 (Fig. 2). The two salt bridges are located on opposite ends of the H1′ helix, and they appear to clamp the H1′ helix in the hydrophobic groove between the H2 and H3 helices of cterRAP74.
Careful analysis of the ensemble of structures indicates that these are the only two ion pairs that are close enough in space to be clearly defined by the NMR data, despite the large number of residues of opposite charges in the complex. However, the cterRAP74/cterFCP complex interface does contain three potential polar interactions involving additional acidic residues from the H1′ helix of cterFCP based on proximity in the energy-minimized structure (see Table 2, which is published as supporting information on the PNAS web site, www.pnas.org). The polar interactions are between the side chains of T470 and E954, between the side chains of S486 and E945, and between the backbone of Q495 and the side chain of E956 (Fig. 3B). As was the case with the salt bridges, each of these interactions involves a strictly conserved acidic residue from the H1′ helix of cterFCP (Fig. 4A).
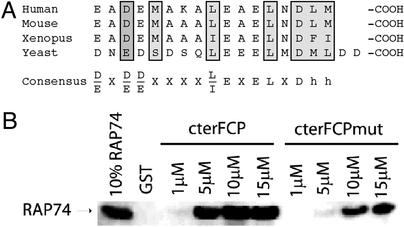
Figure 4.
The acidic hydrophobic motif in cterFCP. (A) Alignment of the amino acids in the H1′ α-helix of human FCP1-(E945–M961) with corresponding sequence in mouse FCP1, Xenopus laevis FCP1, and yeast FCP1. Residues that are important for interactions with RAP74 are boxed in gray. A consensus sequence is shown below. The mouse and Xenopus sequence are from databases and the mouse sequence is partial. (B) Varying concentrations (0.1–15 μM) of cterRAP74-(436–517) was incubated either with 1 μM GST-cterFCP (ELNDLM) or 1 μM GST-cterFCPmut (AANDLM) immobilized on GSH resin as described in Methods. In the GST lane, 15 μM of purified cterRAP74 was incubated with 1 μM of immobilized GST as a control for nonspecific binding. The input lane is 10% input of purified cterRAP74.
Genetic and biochemical studies with a RAP74 K471E/K475E double lysine mutant support a functional interaction between cterFCP and cterRAP74 and the importance of the K471/D947 salt bridge in the complex. Yeast strains carrying a mutant fcp1 allele are more temperature sensitive in a RAP74 double lysine mutant background than in the wild-type RAP74 background, except in the case of a mutant fcp1 allele, which lacks the FCP1 carboxyl-terminal RAP74-binding site (18). These results are consistent with the hypothesis that RAP74 stimulation of FCP1 occurs in vivo and becomes more important when FCP1 phosphatase activity is lowered by mutations outside of the carboxyl-terminal RAP74-binding site (16, 18). In addition, the ability of cterFCP to activate transcription when artificially tethered to a promoter is reduced by the RAP74 K471E/K475E mutant (18). These results with the double lysine mutant explain the importance of the salt bridge between K471 and D947 in the complex.
Other Protein/Protein Complexes Containing Acidic Activation Domains.
The formation of an α-helix in cterFCP after binding to cterRAP74 is very similar to what has been observed with complexes involving the acidic activation domains from the tumor suppressor protein p53 and the herpes simplex virus activation protein VP16 (38, 39). A high-resolution structure is available only for the p53/MDM2 complex (38). In the p53/MDM2 complex (38), a nine-residue amphipathic α-helix (residues 18–26) is formed in p53 on binding to MDM2. Amino acids F19, W23, and L26 in this newly formed helix insert deeply into a hydrophobic binding cleft on MDM2 and make contact with the α2 helix, the α2′ helix, and the middle β-sheet of MDM2. Like the p53/MDM2 binding interface, the cterRAP74/cterFCP binding interface relies on extensive hydrophobic interactions (Fig. 3A). However, there are very few polar interactions at the p53/MDM2 binding interface; in fact, there are only two intermolecular hydrogen bonds and there are no intermolecular electrostatic interactions involving residues of opposite charges. Finally, the secondary structure of the p53-binding domain of MDM2 does not appear to be structurally homologous to the cterFCP-binding domain of cterRAP74.
Given these structural differences between the cterFCP/cterRAP74 and p53/MDM2 complexes, one would predict that cterRAP74 is not an ideal target for all proteins containing acidic activation domains. In support of this, the VP16 activation domain does not directly interact with TFIIF (40). However, it has been shown, in vitro, that RAP74 is the target of activation domains from the androgen receptor (41), the serum response factor (42), and Fos-Jun (43). In addition, mutations of lysine residues in the H2 helix of cterRAP74 significantly reduce the ability of cterFCP to activate transcription (18). Thus, it has been suggested that cterRAP74 may be the target of a distinct subset of activators (18). In support of this hypothesis, we have used NMR to map the binding site of three additional acidic residue-rich domains on cterRAP74, and preliminary analysis indicates that all three bind in a similar manner as cterFCP (unpublished data). These experiments were conducted by using high concentrations of protein and therefore the interactions might not be physiologically relevant.
cterFCP Contains an Acidic Hydrophobic Motif.
Structural analysis of the cterRAP74/cterFCP complex indicates the presence of crucial hydrophobic and acidic residues in the H1′ helix of cterFCP that are important for interaction with cterRAP74. To determine whether these residues are conserved in other species, we aligned primary sequences of FCP1 from the mouse FCP1, the X. laevis FCP1, and the Saccharomyces cerevisiae FCP1 with the 17 amino acids that make up the H1′ helix of cterFCP (Fig. 4A). Interestingly, the six acidic residues in the H1′ are completely conserved and every acidic residue with the exception of E948 forms an important interaction with cterRAP74. Based on this alignment, we have derived a consensus sequence for the H1′ helix of cterFCP (Fig. 4A). The alignment clearly demonstrates that the last nine amino acids with the consensus L/IEXELXDhh are highly conserved in these eukaryotes, particularly three acidic residues and four hydrophobic residues (Fig. 4A).
The L/IEXELXDhh motif found in the H1′ helix of cterFCP resembles the LXXLL motif found in nuclear hormone receptor (NR) coactivators and the LXXI/HIXXXL/I motif found in NR corepressor proteins (44, 45). To investigate whether the motif located in the H1′ helix of cterFCP is related to the motifs found in the NR coactivator and corepressor proteins, we made a mutant of cterFCP in which we changed the final nine amino acids from LEAELNDLM to LEAAANDLM (cterFCPmut). In vitro binding studies indicate that the cterFCPmut protein binds significantly more weakly to cterRAP74 than does wild-type cterFCP (Fig. 4B). A similar double alanine mutations has been made in the LXXLL motif-containing α-helix of the GRIP1 coactivator protein, and it was shown to significantly decrease the ability of a GRIP1 peptide to bind to ligand-binding domain (LBD) of the thyroid receptor (46).
Comparison to Complexes Containing LXXLL and LXXI/HIXXXL/I Motifs.
Several years ago, it was proposed that interactions of an amphipathic α-helix containing a hydrophobic motif with a complementary hydrophobic surface represent a very common mode of recognition in transcriptional regulatory protein/protein complexes (38, 39, 46, 47). Like the cterFCP/cterRAP74 complex, the α-helices from the NR coactivators and corepressors bind in a hydrophobic groove formed by several α-helices in the LBD of the NR (46, 48–53). In addition, NR coactivators, NR corepressors, and cterFCP appear to rely predominantly on nonaromatic side chains. The only exception is the presence of a single phenylalanine in the second to last hydrophobic position in Xenopus FCP1 (Fig. 4A). This predominant use of nonaromatic side chains distinguishes these complexes from the p53/MDM2 complex and the VP16/TAF∥31 complexes, where it has been shown that multiple aromatic side chains are crucial for complex formation (38, 39). As mentioned above, the VP16 activation domain does not directly interact with TFIIF, and this may be in part due to the presence of multiple aromatic amino acids in VP16 (40). Finally, the complexes involving NR coactivator binding and NR corepressor binding to NR LBDs contain an important charge clamp (46, 48–51). These interactions occur at both the carboxyl-terminal and amino-terminal ends of the NR coactivators LXXLL-containing helix, and they are similar to the two salt bridges observed at both the carboxyl- and amino-terminal ends of the H1′ helix of cterFCP in the cterRAP74/cterFCP complex. In both cases, these interactions seem to orient the binding of the helix in the hydrophobic groove. Surprisingly, despite a number of acidic residues in p53, these residues do not appear to play a role in binding to MDM2 and no charge clamps were observed in the p53/MDM2 complex (38).
Conclusion
In this manuscript, we have determined the solution structure of the cterRAP74/cterFCP complex. The cterFCP protein is completely disordered in the unbound state but forms an amphipathic α-helix (H1′) in the complex. The hydrophobic face of the H1′ α-helix is inserted within a complementary hydrophobic groove on RAP74, and this interface is composed of predominantly nonaromatic hydrophobic amino acids. In addition, the cterRAP74/cterFCP complex contains two intermolecular salt bridges and three other intermolecular polar interactions involving five highly conserved acidic side chains from cterFCP. We found some interesting similarities between the recognition of the H1′ helix of cterFCP and that of the hydrophobic binding motifs found in NR coactivator and corepressors. However, we have found significant differences between the cterRAP74/cterFCP complex and that of the p53/MDM2 complex, which also contains an acidic activation domain. This cterRAP74/cterFCP complex structure clearly points to a role for acidic chains in protein/protein complexes involving acidic residue-rich activation domains that had not been previously demonstrated.
Supplementary Material
Acknowledgments
We thank Frank Delaglio, Daniel Garrett, and Bruce Johnson for NMR data processing/analysis programs and Lewis Kay for NMR pulse sequences. This work was supported by National Institutes of Health Grant RO1 GM60298-01 (to J.G.O. and P.L.) and by a grant from the Canadian Institutes of Health Research (to J.G.).
Abbreviations
- CTD
carboxyl-terminal domain
- TFIIF
transcription factor IIF
- FCP1
TFIIF-associating CTD phosphatase
- cterRAP74
CTD of RAP74
- cterFCP
CTD of FCP1
- NOE
nuclear Overhauser effect
- NR
nuclear hormone receptor
- RNAP
RNA polymerase
Note Added in Proof.
An x-ray crystal structure of a complex containing similar domains of RAP74 and FCP1 was recently reported (54).
Footnotes
Data deposition: The atomic coordinates for the cterFCP/cterRAP74 complex have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1ONV).
References
- 1.Cadena D L, Dahmus M E. J Biol Chem. 1987;262:12468–12474. [PubMed] [Google Scholar]
- 2.Lu H, Flores O, Weinmann R, Reinberg D. Proc Natl Acad Sci USA. 1991;88:10004–10008. doi: 10.1073/pnas.88.22.10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Brien T S, Hardin S, Greenleaf A, Lis J T. Nature. 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- 4.Feaver W J, Gileadi O, Li Y, Kornberg R D. Cell. 1991;67:1223–1230. doi: 10.1016/0092-8674(91)90298-d. [DOI] [PubMed] [Google Scholar]
- 5.Lu H, Zawel L, Fisher L, Egly J M, Reinberg D. Nature. 1992;358:641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- 6.Serizawa H R, Conaway R C, Conaway J W. Proc Natl Acad Sci USA. 1992;89:7476–7480. doi: 10.1073/pnas.89.16.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marshall N F, Peng J, Xie Z, Price D H. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 8.Cujec T P, Okamoto H, Fujinaga K, Meyer J, Chamberlin H, Morgan D O, Peterlin B M. Genes Dev. 1997;11:2645–2657. doi: 10.1101/gad.11.20.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dahmus M E. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 10.Chambers R S, Dahmus M E. J Biol Chem. 1994;269:26243–26248. [PubMed] [Google Scholar]
- 11.Chambers R S, Dahmus M E. J Biol Chem. 1996;271:24498–24504. doi: 10.1074/jbc.271.40.24498. [DOI] [PubMed] [Google Scholar]
- 12.Chambers R S, Wang B Q, Burton Z F, Dahmus M E. J Biol Chem. 1995;270:14962–14969. doi: 10.1074/jbc.270.25.14962. [DOI] [PubMed] [Google Scholar]
- 13.Mandal S S, Cho H, Kim S, Cabane K, Reinberg D. Mol Cell Biol. 2002;22:7543–7552. doi: 10.1128/MCB.22.21.7543-7552.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Archambault J, Chambers R S, Kobor M S, Ho Y, Cartier M, Bolotin D, Andrews B, Kane C M, Greenblatt J. Proc Natl Acad Sci USA. 1997;94:14300–14305. doi: 10.1073/pnas.94.26.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Archambault J, Pan G, Dahmus G K, Cartier M, Marshall N, Zhang S, Dahmus M E, Greenblatt J. J Biol Chem. 1998;273:27593–27601. doi: 10.1074/jbc.273.42.27593. [DOI] [PubMed] [Google Scholar]
- 16.Kobor M S, Archambault J, Lester W, Holstege F C P, Gileadi O, Jansman D B, Jennings E G, Kouyoumdjian F, Davidson A R, Young R A, Greenblatt J. Mol Cell. 1999;4:55–62. doi: 10.1016/s1097-2765(00)80187-2. [DOI] [PubMed] [Google Scholar]
- 17.Cho H, Kim T-K, Mancebo H, Lane W S, Flores O, Reinberg D. Genes Dev. 1999;13:1540–1552. doi: 10.1101/gad.13.12.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobor M S, Simon L S, Omichinski J G, Zhong G, Archambault J, Greenblatt J. Mol Cell Biol. 2000;20:7438–7449. doi: 10.1128/mcb.20.20.7438-7449.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bork P, Hofmann K, Bucher P, Neuwald A F, Altschul S, Koonin E V. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- 20.Licciardo P, Ruggiero L, Lania L, Majello B. Nucleic Acids Res. 2001;29:3539–3545. doi: 10.1093/nar/29.17.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen B D, Chen H-T, Kobor M S, Greenblatt J, Legault P, Omichinski J G. Biochemistry. 2003;42:1460–1469. doi: 10.1021/bi0265473. [DOI] [PubMed] [Google Scholar]
- 22.Bax A, Sparks S W, Torchia D A. Methods Enzymol. 1989;176:134–150. doi: 10.1016/0076-6879(89)76009-2. [DOI] [PubMed] [Google Scholar]
- 23.Marion D, Kay L E, Sparks S W, Torchia D A, Bax A. J Am Chem Soc. 1989;111:1515–1517. [Google Scholar]
- 24.Pascal S M, Muhandiram D R, Yamazaki T, Forman-Kay J D, Kay L E. J Magn Reson. 1994;103:197–201. [Google Scholar]
- 25.Zuiderweg E R P, McIntosh L P, Dahlquist F W, Fesik S W. J Magn Reson. 1990;86:210–216. [Google Scholar]
- 26.Zwahlen C, Legault P, Vincent S J F, Greenblatt J, Konrat R, Kay L E. J Am Chem Soc. 1997;119:6711–6721. [Google Scholar]
- 27.Delaglio F, Grzesiek S, Vuister G W, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 28.Garrett D S, Powers R, Gronenborn A M, Clore G M. J Magn Reson. 1991;95:214–220. doi: 10.1016/j.jmr.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 29.Johnson B A, Blevins R A. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 30.Cornilescu G, Delaglio F, Bax A. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 31.Brunger A T, Adams P D, Clore G M, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges M, Pannu N S, Read R J, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 32.Laskowski R A, Antoon J, Rullmann C, Macarthur M W, Kaptein R, Thornton J M. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 33.Koradi R, Billeter M, Wüthrich K. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 34.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 35.Kamada K, Angelis J D, Roeder R G, Burley S K. Proc Natl Acad Sci USA. 2001;98:3115–3120. doi: 10.1073/pnas.051631098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wishart D S, Sykes B D, Richards F M. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 37.Wishart D S, Sykes B D. J Biomol NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 38.Kussie P H, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine A J, Pavletich N P. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 39.Uesugi M, Nyanguile O, Lu H, Levine A J, Verdine G L. Science. 1997;277:1310–1313. doi: 10.1126/science.277.5330.1310. [DOI] [PubMed] [Google Scholar]
- 40.Xiao H, Pearson A, Coulombe B, Truant R, Zhang S, Regier J L, Trienzenberg S J, Reinberg D, Flores O, Ingles C J, Greenblatt J. Mol Cell Biol. 1994;14:7013–7024. doi: 10.1128/mcb.14.10.7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reid J, Murray I, Watt K, Betney R, McEwan I J. J Biol Chem. 2002;277:41247–41253. doi: 10.1074/jbc.M205220200. [DOI] [PubMed] [Google Scholar]
- 42.Jollot V, Demma M, Prywes R. Nature. 1995;373:632–635. doi: 10.1038/373632a0. [DOI] [PubMed] [Google Scholar]
- 43.Mitchell M L, Lieberman P M, Curran T. Mol Cell Biol. 1996;16:2110–2118. doi: 10.1128/mcb.16.5.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heery D M, Kalkhoven E, Hoare S, Parker M G. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 45.Hu X, Lazar M A. Nature. 1999;402:93–96. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- 46.Darimont B D, Wagner R L, Apriletti J W, Stallcup M R, Kushner P J, Baxter J D, Fletterick R J, Yamamoto K R. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Radhakrishnan I, Perez-Alvarado G C, Parker D, Dyson H J, Montminy M R, Wright P E. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 48.Shiau A K, Barstad D, Loria P M, Cheng L, Kushner P J, Agard D A, Greene G L. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 49.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Wilson T M, Glass C K, Milburn M V. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 50.Xu H E, Stanley T B, Montana V G, Lambert M H, Shearer B G, Cobb J E, McKee D D, Galardi C M, Plunket K D, Nolte R T, et al. Nature. 2002;415:813–817. doi: 10.1038/415813a. [DOI] [PubMed] [Google Scholar]
- 51.Bledsoe R K, Montana V G, Stanley T B, Delves C J, Apolito C J, McKee D D, Consler T G, Parks D J, Stewart E L, Wilson T M, et al. Cell. 2002;110:93–105. doi: 10.1016/s0092-8674(02)00817-6. [DOI] [PubMed] [Google Scholar]
- 52.Freedman S J, Sun Z-Y J, Poy F, Kung A L, Livingston D M, Wagner G, Eck M J. Proc Natl Acad Sci USA. 2002;99:5367–5372. doi: 10.1073/pnas.082117899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Demarest S J, Martinez-Yamout M, Chung J, Chen H, Xu W, Dyson H J, Evans R M, Wright P E. Nature. 2002;415:549–553. doi: 10.1038/415549a. [DOI] [PubMed] [Google Scholar]
- 54.Kamada K, Roeder R G, Burley S K. Proc Natl Acad Sci USA. 2003;100:2296–2299. doi: 10.1073/pnas.262798199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.