Abstract
N-acetylneuraminate lyase (NAL) and dihydrodipicolinate synthase (DHDPS) belong to the NAL subfamily of (β/α)8-barrels. They share a common catalytic step but catalyze reactions in different biological pathways. By rational design, we have introduced various mutations into the NAL scaffold from Escherichia coli to switch the activity toward DHDPS. These mutants were tested with respect to their catalytic properties in vivo and in vitro as well as their stability. One point mutation (L142R) was sufficient to create an enzyme that could complement a bacterial auxotroph lacking the gene for DHDPS as efficiently as DHDPS itself. In vitro, this mutant had an increased DHDPS activity of up to 19-fold as defined by the specificity constant kcat/KM for the new substrate l-aspartate-β-semialdehyde when compared with the residual activity of NAL wild-type, mainly because of an increased turnover rate. At the same time, mutant L142R maintained much of its original NAL activity. We have solved the crystal structure of mutant L142R at 1.8 Å resolution in complex with the inhibitor β-hydroxypyruvate. This structure reveals that the conformations of neighboring active site residues are left virtually unchanged by the mutation. The high flexibility of R142 may favor its role in assisting in catalysis. Perhaps, nature has exploited the catalytic promiscuity of many enzymes to evolve novel enzymes or biological pathways during the course of evolution.
Keywords: protein|engineering|aldolase
Enzymes cover a broad range of activities. They usually exhibit high regio- and stereoselectivity, and the reactions proceed under benign reaction conditions. These properties make enzymes an attractive target for the synthesis of organic compounds in the pharmaceutical industry (1). However, the variety of catalysts that nature offers is not enough to satisfy the needs for the production of new compounds. Although there are numerous examples where biocatalysts have been improved or modified by protein engineering and directed evolution techniques (2–4), we have to understand more fully the relationships between structure, stability, and function to create tailor-made enzymes on a wider level. The postgenomic era, with its abundance of sequence data and the ever increasing number of available protein structures, has put us in a more favorable position to tackle the question of how nature has managed to graft new activities into an existing scaffold.
The (β/α)8-barrel fold is the most commonly occurring motif in enzyme structures. Its structural framework consists of eight repeating β/α-modules. The β-strands are located in the interior of the protein, where they form a barrel-shaped parallel β-sheet, whereas the α-helices are packed at the exterior of the barrel. It has been suggested that all contemporary (β/α)8-barrels have evolved from a common ancestor (5, 6). These studies have also shown that recruitment of enzymes plays a significant role in the formation of biological pathways rather than retrograde evolution. The (β/α)8-barrel fold itself may have evolved from an ancestral half-barrel (7, 8).
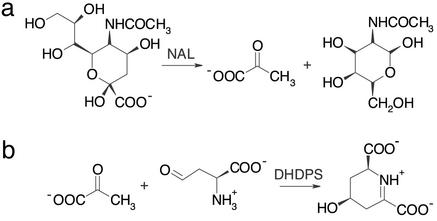
The N-acetylneuraminate lyase (NAL) subfamily of (β/α)8-barrel enzymes comprises several pyruvate-dependent class I aldolases. They all share a common catalytic step but catalyze different reactions in different biological pathways (9). The best-characterized members of this subfamily are NAL and dihydrodipicolinate synthase (DHDPS); the overall reactions for both enzymes are in Fig. 1. In bacteria, NAL is involved in the regulation of intracellular sialic acid, where it catalyzes the cleavage of N-acetylneuraminate (Neu5Ac) to produce pyruvate and N-acetyl-d-mannosamine. It has a widespread application as biocatalyst for the synthesis of sialic acid and its derivatives (10). DHDPS, on the other hand, catalyzes the aldol condensation of pyruvate and l-aspartate-β-semialdehyde (ASA), the first step in the biosynthesis of lysine via the diaminopimelate pathway.
Figure 1.
Reactions catalyzed by NAL (a) and DHDPS (b), two members of the NAL subfamily of (β/α)8-barrel enzymes. The common catalytic step is a Schiff-base formation between a conserved lysine and the keto group of the respective substrate followed by the aldol condensation/cleavage reaction. In DHDPS, subsequent transimination occurs to yield the cyclic product. On the basis of NMR experiments, Blickling et al. (14) proposed that (4S)-4-hydroxy-2,3,4,5-tetrahydro-(2S)-dipicolinic acid is the immediate product of the reaction catalyzed by DHDPS. The elimination of a water molecule to yield dihydrodipicolinate is supposed to occur spontaneously after the initial product is released into the solvent.
The catalytic action of both enzymes has been thoroughly studied and crystal structures have been solved for NAL from Escherichia coli (11, 12), NAL from Haemophilus influenzae (9), DHDPS from E. coli (13, 14), and DHDPS from Nicotiana sylvestris (15). The fold of all four structurally characterized enzymes is a (β/α)8-barrel with a C-terminal extension of three α-helices. The quaternary structure is a tetramer, which can be best described as a dimer of dimers. In DHDPS, different orientations of the dimers within the biologically active tetramer have been accounted for the different regulatory properties in the respective organisms (15). The high structural homology and the fact that there is a structural partition between residues involved in binding the common substrate pyruvate and those binding the specific acceptor aldehyde make this subfamily a very good model system for protein engineering and directed evolution experiments. Here, we present studies on transplanting activities within members of this subfamily using NAL as starting scaffold to mimic the crucial mutational step in pathway recruitment during evolution.
Materials and Methods
Cloning and Mutagenesis.
DHDPS (dapA gene) and NAL (npl gene) were amplified from E. coli and cloned into the EcoRI and PstI restriction sites of vector pKK223–3 (Amersham Pharmacia). Point mutations were introduced using the QuikChange Site-Directed Mutagenesis kit (Stratagene). Dihydrodipicolinate reductase (DHDPR; dapB gene) and aspartate-β-semialdehyde dehydrogenase (asd gene) were amplified from E. coli with a C-terminal His-tag attached and cloned into the NcoI and PstI sites of vector pTrc99A (Amersham Pharmacia).
Genetic Complementation.
Functional complementation studies were performed by using E. coli strain AT997, an auxotroph lacking DHDPS activity (16). AT997 can be grown on medium complemented with diaminopimelate, a pathway intermediate, which can serve as a precursor for l-lysine synthesis. Medium for strain maintenance consisted of M9 minimal medium supplemented with 50 μg/ml dl-α,ɛ-diaminopimelate, 1 mM MgSO4, 1.7 mM thiamine hydrochloride, and 0.2% (wt/vol) glucose. Plasmids were transformed into AT997 by electroporation. For complementation studies, cells were recovered in M9 minimal medium with the same supplements as for strain maintenance except dl-α,ɛ-diaminopimelate. Transformed cells were plated on M9 minimal plates (Difco agar) with the same supplements as for cell recovery plus 100 μg/ml ampicillin for plasmid selection and incubated at 37°C. After elongated periods of >3 days, we occasionally observed background growth in controls with vector pKK223-3 without insert, indicating that results obtained after >3 days were no longer reliable.
Protein Purification.
All proteins were purified by using E. coli AT997 as host cells to avoid any contamination with DHDPS wild-type. The culture medium was, therefore, complemented with 50 μg/ml dl-α,ɛ-diaminopimelate. The purification protocol for DHDPS and NAL wild-type and mutants included a 2-min heat denaturation at 65–70°C in the presence of 5 mM pyruvate, 60–100 mM NaCl/KCl, and 20 mM Tris⋅HCl (pH 7.7–8.0), anion exchange chromatography on SOURCE 30Q (Amersham Pharmacia), and a final size exclusion chromatography on a Superdex 200 column (Amersham Pharmacia). Aspartate-β-semialdehyde dehydrogenase and DHDPR were purified as C-terminal His-tagged proteins with Ni-NTA agarose (Qiagen, Valencia, CA) followed by an anion exchange chromatography on a SOURCE 30Q column. Protein concentrations were measured photometrically by using calculated extinction coefficients (17).
Synthesis of ASA.
ASA was synthesized via ozonolysis of allylglycine and subsequent purification on a Dowex 50 column (18). ASA stock solutions were stored in 4 M HCl at −20°C and neutralized with ice-cold 1 M NaOH before use in enzymatic assays. Formation of ASA was monitored with aspartate-β-semialdehyde dehydrogenase (19). The concentrations of ASA stock solutions were determined enzymatically via end point titration using DHDPS and DHDPR (see below). All assays were performed by using the same stock solution of dl-ASA and l-ASA, respectively.
Activity Measurements.
Initial velocity data were collected at a constant temperature of 25°C on a Cary 500 spectrophotometer (Varian) after the disappearance of NADH at 340 nm. All assays were performed in a final volume of 1 ml. NAL activity was measured as described (20, 21) with lactate dehydrogenase as coupling enzyme. Typical assays contained 20 mM sodium phosphate (pH 7.2), 0.15 mM NADH, 30 μg of rabbit lactate dehydrogenase (Fluka), and varied concentrations of Neu5Ac (Sigma) ranging from 0.2 to 40 mM. DHDPS activity was monitored at 340 nm by coupling it to the reduction of the reaction product dihydrodipicolinate by NADH and DHDPR (22). Assays contained 100 mM Hepes (pH 7.0 or 8.0), 0.2 mM NADH, 25 μg of DHDPR, and varied concentrations of the substrates l-ASA and pyruvate. Inhibition studies were performed at pH 8.0 with fixed substrate concentrations of 2 mM dl-ASA, 5 mM pyruvate, and varied l-lysine concentrations ranging from 0.02 mM to 10 mM. Kinetic parameters were calculated from a fit of the initial velocity data at different substrate concentrations to the Michaelis–Menten equation by using the program kaleidagraph (Abelbeck Software, Reading, PA). The reaction of DHDPS follows a ping-pong mechanism with pyruvate binding first and l-ASA next. l-ASA acts as competitive inhibitor of pyruvate for DHDPS with a KI,ASA = 0.3 mM (22). This has to be considered when determining true Kpyruvate values. In our study, only apparent Kpyruvate values at a given concentration of 2.35 mM l-ASA were determined.
CD Spectroscopy.
Temperature dependent far-UV CD spectroscopy was performed with a Jasco J-720 Spectropolarimeter in 50 mM Hepes (pH 7.5), 150 mM NaCl, and 1 mM Tris(2-carboxyethyl)phosphine hydrochloride. Protein concentrations were in the range of 0.7–1.3 mg/ml. Heating rates were 60°C h−1.
X-Ray Structure Determination.
Crystals of NAL mutant L142R in complex with β-hydroxypyruvate (BHP) were grown at 21°C using the hanging-drop vapor diffusion method. Three microliters of protein solution (8 mg/ml protein in 20 mM BHP and 20 mM Tris, pH 7.6) was mixed with 3 μl of reservoir buffer consisting of 100 mM sodium acetate (pH 4.6) and 8% (wt/vol) polyethylene glycol (PEG) 4000 above a reservoir solution of 700 μl. Crystals were cryo-protected by using crystallization buffer containing 25% (vol/vol) PEG 200. They belonged to the space group P21 with cell dimensions of a = 83.7 Å, b = 96.0 Å, c = 89.7 Å and β = 115.2°. Initial attempts to crystallize mutant L142R under conditions similar to those that had yielded crystals in the trigonal space group P3221 for wild-type NAL (11, 12) failed. A diffraction set to 1.8 Å resolution was collected from a single crystal at 100 K on beamline ID14–4 at the European Synchrotron Radiation Facility (Grenoble, France). Data processing was carried out by using CCP4 (23).
The structure was solved by molecular replacement using the program amore (24), using the tetrameric model of NAL E. coli in complex with BHP (12), without solvent and inhibitor, as search model. In addition, an alanine residue was introduced at position 142. There was one tetramer in the asymmetric unit. All subsequent structure refinement was carried out with cns (25). After simulated annealing, energy minimization and individual B factor refinement, BHP covalently linked to K165 (2-deoxy complex) and R142 were modeled into the structure. The observed electron density allowed the addition of residues at the N and C termini, which had not been detected in the original wild-type structure. Water molecules were added to the structure using the water pick option within cns. The atomic model was then further refined using repeated cycles of cns and rebuilding in o (26). procheck (27) was used to check the stereochemistry of the model. Figures were generated with molscript (28) and RASTER3D (29).
Results
Protein Design.
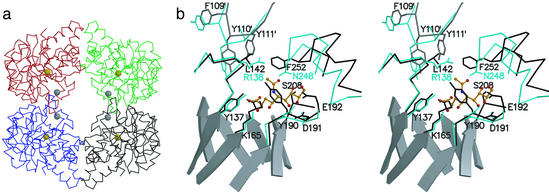
We chose NAL from E. coli as starting scaffold for our studies to transplant activities within members of the NAL subfamily, because in the reverse aldol condensation reaction NAL is highly specific for pyruvate but does not strongly discriminate between different aldehyde substrates (10). NAL tolerates a wide range of aldoses and, as such, this contemporary enzyme displays features typical of a multipotent ancestral enzyme. Despite the low sequence identity of <24% in E. coli, the three-dimensional structures of NAL and DHDPS are very similar. The first target residue in our attempt to incorporate DHDPS activity into the NAL scaffold was L142, which is located on a short helical turn within loop β5/α5 (Figs. 2 and 3). DHDPS has a conserved arginine at this position, which is involved in binding l-ASA and is furthermore supposed to assist in catalysis (14). When an arginine is modeled into the structure of NAL in a conformation as found in DHDPS, the guanidinium group lies right at the center of the hydrophobic network spun by the aromatic rings of F252, Y111 from a neighboring subunit, and the original leucine. To accommodate the arginine, additional mutations were designed: in loop β4/α4, we targeted the three-residue segment on the C-terminal side of Y110, a conserved residue among the whole subfamily, and replaced it with the most often occurring motif in DHDPS sequences from different species (module Y111N/P112K/F113P). Mutation F252S at the N terminus of helix α10 was added to increase the accessible volume of the active site in this region. A further target was loop β7/α7. In NAL, this loop contains residues involved in binding Neu5Ac, whereas it is supposed to coordinate the amino group of l-ASA in DHDPS (9). We, therefore, replaced this loop with the sequence of the DHDPS homolog in E. coli (module Y190D/E192A). The following mutants were created and tested with respect to their ability to complement the bacterial auxotroph AT997, which lacks DHDPS activity: L142R, L142R/F252S, L142R/Y190D/E192A, L142R/Y190D/E192A/F252S, Y111N/P112K/F113P/L142R, and a heptamutant combining all point mutations.
Figure 2.
Sequence alignment of NAL from E. coli, NAL from H. influenzae, DHDPS from E. coli, and DHDPS from N. sylvestris. Numbering for the latter is as in ref. 15. Secondary structural elements as found in NAL from E. coli are shown above the alignment. Residues identical in at least three of the four sequences are highlighted in dark gray, and similar residues are highlighted in light gray. Color coding: blue, residues forming l-lysine binding pocket in DHDPS (14, 15); red, residues involved in binding pyruvate or aldol condensation/cleavage reaction; magenta, residues specifically interacting with the aldol acceptor moiety of Neu5Ac (9, 37); green, residue binding the carboxylate group of l-ASA (14). Residues in NAL E. coli that were targeted for mutagenesis are marked with an asterisk. The alignment was done by using the program CLUSTALW (38) followed by manual editing.
Figure 3.
The structure of NAL. (a) The NAL tetramer. Cα-traces of individual subunits are shown in black (chain A), red (chain B), blue (chain C), and green (chain D). The golden spheres are centered on the Nζ-atoms of K165, which form a Schiff-base during catalysis; the gray spheres are centered on the Cα-atoms of L142. The subunit interface discussed in this work is between chains A/D and B/C respectively. (b) Stereoview of the active center of NAL from E. coli (black) in complex with BHP (12) superimposed on DHDPS (cyan) from E. coli (13). The superposition was calculated based on the coordinates for the main-chain atoms in β5, connective loop β5/α5, and β6. For clarity, active site loops are shown as Cα-traces with selected side chains. Residues contributed from an adjacent subunit are marked (′); e.g., Y111′. β-Strands forming the central barrel in NAL are shown as ribbon models. Also shown as ball-and-stick model (orange) is the NAL substrate analog sialic acid alditol in the conformation and position as found in the structure of NAL from H. influenzae (9).
Complementation Studies.
When the bacterial auxotroph AT997 was complemented with the gene of DHDPS wild-type in vector pKK223–3, small colonies reproducibly grew on minimal medium after 1.5 days. Surprisingly, complementation with NAL mutants L142R and L142R/Y190D/E192A was even better. Colonies appeared a few hours before those in controls with DHDPS wild-type and had grown to a bigger size after the same incubation period. If these mutations were combined with the F252S mutation, the ability to complement was significantly impaired. It took ≈1 day longer until colonies appeared. The constructs with changes in loop β4/α4 did not complement at all. No growth was observed within the first 3 days (see Materials and Methods). With NAL wild-type, no growth was observed within the first 3 days, but after elongated incubation periods we occasionally observed the tendency that more colonies grew than mere background. The expression levels of all tested proteins in AT997 were in the same range, as estimated by SDS gels. Accordingly, we purified mutants L142R, L142R/Y190D/E192A, and L142R/F252S as well as both NAL and DHDPS wild-type to investigate the in vitro properties of these enzymes.
NAL Activity.
We determined the kinetic parameters for the cleavage of Neu5Ac for the two mutants that gave the best results in complementation studies to see whether these mutant enzymes still maintain the original activity. Both mutants were still able to catalyze the original reaction, though less efficiently than NAL wild-type (Table 1). This was mainly caused by an increase in KNeu5Ac from 2.5 mM in wild-type to 13.6 mM in mutant L142R and >100 mM in mutant L142R/Y190D/E192A. kcat, on the other hand, was only slightly affected by the mutations. Remarkably, mutant L142R exhibited a 1.6-fold higher rate constant than wild-type NAL. The increase in KNeu5Ac is consistent with a stepwise perturbation of the Neu5Ac binding pocket. Mutation of E188 in NAL from Clostridium perfringens (the equivalent residue to E192 in E. coli) also results in a significant increase in KNeu5Ac, whereas kcat is virtually unchanged (30).
Table 1.
Kinetic constants for the cleavage of Neu5Ac
| kcat (per subunit), s−1 | KNeu5Ac, mM | kcat/KNeu5Ac, s−1⋅mM−1 | |
|---|---|---|---|
| NAL | 7.7 ± 0.5 | 2.5 ± 0.2 | 3.1 |
| L142R | 12.1 ± 0.9 | 13.6 ± 1.3 | 0.9 |
| L142R/Y190D/E192A | 6.8 ± 1.6 | 125 ± 36 | 0.05 |
DHDPS Activity.
The results of in vitro DHDPS assays at pH 7.0 and 8.0 are shown in Table 2. Interestingly, NAL wild-type, which was used as starting scaffold, had a very weak DHDPS activity. This reflects the somewhat indiscriminate nature of the active site of NAL in general. However, both the turnover rate and the affinity for l-ASA were much lower, and the specificity constant kcat/KASA was several orders of magnitude lower than that of DHDPS. The apparent binding constants for pyruvate were in the same range, consistent with the pyruvate binding pocket being conserved in both enzymes.
Table 2.
Kinetic constants for the aldol condensation of l-ASA and pyruvate
|
kcat (per subunit), s−1
|
Kl-ASA, mM
|
Kapp, pyruvate, mM*
|
Relative kcat/Kl-ASA†
|
|||||
|---|---|---|---|---|---|---|---|---|
| pH 7.0 | pH 8.0 | pH 7.0 | pH 8.0 | pH 7.0 | pH 8.0 | pH 7.0 | pH 8.0 | |
| DHDPS‡ | 8.5 ± 0.5 | 51 ± 3 | 0.075 ± 0.006 | 0.172 ± 0.012 | 0.23 ± 0.01 | 0.51 ± 0.03 | 6,000 | 14,000 |
| NAL | 0.47 ± 0.05 | 0.27 ± 0.02 | 24 ± 3 | 12.4 ± 1.2 | 0.67 ± 0.05 | 0.46 ± 0.04 | 1 | 1 |
| L142R | 3.9 ± 0.3 | 3.4 ± 0.3 | 15.6 ± 1.5 | 8.3 ± 0.9 | 1.0 ± 0.1 | 0.9 ± 0.1 | 13 | 19 |
| L142R/Y190D/E192A | 1.3 ± 0.1 | 0.98 ± 0.07 | 16.0 ± 1.6 | 7.6 ± 0.7 | 1.1 ± 0.1 | 1.0 ± 0.1 | 4.2 | 6 |
| L142R/F252S | 0.65 ± 0.1 | 0.44 ± 0.05 | 12 ± 2 | 4.5 ± 0.5 | 1.4 ± 0.1 | 1.5 ± 0.1 | 2.7 | 4 |
Measured at a fixed l-ASA concentration of 2.35 mM.
Compared with NAL wild type.
Karsten (22) reported a similar KASA value (0.124 mM) at pH 8.0. Our kcat value is significantly lower than 188 s−1 as reported by Karsten. We measured the same activity with a sample stored in liquid nitrogen over a period of several months. However, we noticed an ≈50% activity increase when a diluted sample of the protein in the buffer (100 mM Hepes) used for our kinetic measurements was left at 4°C for several weeks and assayed again. This was not observed for NAL and its mutant.
Mutants L142R and L142R/Y190D/E192A, which gave the best results in complementation studies with AT997, had increased DHDPS activity in vitro. Compared with NAL wild-type, this was mainly caused by an increase in kcat, whereas the affinity for l-ASA was only slightly improved. At pH 8.0, kcat for mutant L142R was increased from 0.27 to 3.4 s−1 and KASA was slightly lowered from 12.4 to 8.3 mM. Mutant L142R/Y190D/E192A had an almost identical KASA of 7.6 mM to L142R but kcat was only 1 s−1. It seems that the mutations in loop β7/α7 counteract the effects of R142 on catalysis. For mutant L142R/F252S, which was less efficient in complementation studies, kcat was only 0.44 s−1 but KASA was lower than in the other mutants. Compared with NAL wild-type, the KASA of 4.5 mM in mutant L142R/F252S indicates an appreciable increase in affinity. The apparent Kpyruvate values for the mutants were somewhat higher than in wild-type, especially in mutant L142R/F252S, but were still in a comparable range.
The overall improvement can be best compared when looking at the specificity constant kcat/KASA. The increase was 19-fold in L142R, 6-fold in L142R/Y190D/E192D, and 4-fold in L142R/F252S. Similar results were observed at pH 7.0, though the differences between the mutants and NAL wild-type were less pronounced. In addition, the comparison of the data at different pH values suggests different pH profiles for activity as well as substrate binding in both scaffolds.
Feedback Inhibition Studies.
In plants and some bacteria, DHDPS is feedback-regulated by l-lysine, the end product of the pathway (31). For the E. coli and N. sylvestris enzyme, the allosteric binding site has been localized by x-ray crystallography (14, 15). The inhibitor binds at the subunit interface of the dimer forming monomers around a local 2-fold axis. In E. coli, lysine binding connects via Y107 with Y133 and R138 in the active site. It is suggested that lysine inhibition is at least partly accomplished through reduction of the flexibility of this arginine (14).
No feedback inhibition by l-lysine was expected for the in vitro-evolved DHDPS activity because the specific l-lysine binding residues are not conserved in the starting scaffold (Figs. 2 and 3). Instead of Y107 in DHDPS, NAL has a phenylalanine at the structurally equivalent position. For DHDPS E. coli we observed 50% inhibition at 0.2 mM l-lysine under the conditions used, and 7% activity was retained at 10 mM l-lysine (data not shown). This is consistent with data reported by Karsten (22). For NAL mutants L142R and L142R/Y190D/E192A, no inhibition by l-lysine was observed up to 10 mM l-lysine. This lack of feedback inhibition may explain the fact that these proteins complement the DHDPS-deficient E. coli strain AT997 as well as or even better than DHDPS wild-type despite the much lower activity in the in vitro assays. Previous studies on lysine-sensitive DHDPS enzymes from plants have shown that the ability to complement AT997 not only depends on the expression of a functional DHDPS enzyme but also on the regulatory properties of the enzyme (32).
Stability Measurements.
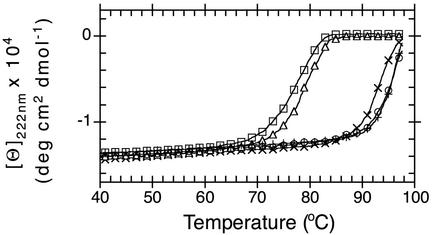
Temperature dependent CD-spectroscopy was used to determine the effects of the introduced mutations on the stability of the protein. Because the thermal unfolding process of NAL and DHDPS was not reversible over the whole temperature range, only differences in apparent Tm values are discussed. Mutants L142R and L142R/Y190D/E192A exhibited a similarly high thermostability as the wild-type enzyme (Fig. 4). In contrast, the apparent Tm for double mutant L142R/F252S was decreased by ≈20°C compared with the single mutant L142R and the wild-type. DHDPS wild-type was less stable than NAL under the used conditions. The thermostability was comparable to NAL mutant L142R/F252S.
Figure 4.
Thermal denaturation of DHDPS E. coli (□), NAL E. coli wild-type (+) and mutants L142R (×), L142R/F252S (▵), and L142R/Y190D/E192A (○) monitored by far-UV CD spectroscopy. Data were measured in 50 mM Hepes (pH 7.5)/150 mM NaCl/1 mM Tris(2-carboxyethyl)phosphine hydrochloride with protein concentrations in the range of 0.7–1.3 mg/ml and normalized to amino acid residue concentrations.
It has been suggested that the reason for the high thermostability of the NAL scaffold is the tetrameric structure (30). The side chain of F252, in wild-type, is part of a cluster of aromatic residues that contribute to the dimer interface. Our data suggest that the alteration of this interface results in a weakened dimer interface in mutant L142R/F252S and consequently in a significantly lower thermostability of this mutant enzyme.
The Crystal Structure of Mutant L142R.
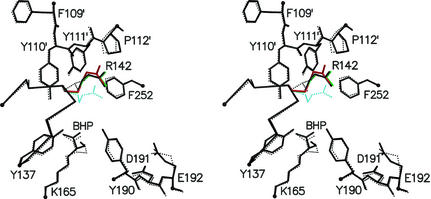
We have solved the structure of mutant L142R in complex with the inhibitor BHP in a new crystal form at 1.8 Å resolution (Table 3). The crystallographic R-factor for the final model was 18.3% (Rfree = 21.0%). The structure reveals no significant changes compared with the wild-type BHP complex at 2.45 Å resolution. The introduction of an arginine at position 142 has only minor effects on the conformations of neighboring active site residues (Fig. 5), leaving the overall active site geometry intact. R142 adopts the same conformation in the three monomers (A, B, and D) where there was well defined electron density for this side chain. Relative to the conformation in DHDPS, the arginine folds back toward its own backbone and the phenolic hydroxyl of Y190. The only specific interaction stabilizing this conformation of the arginine is a hydrogen bond between the Nɛ-atom and the carbonyl oxygen of Y110 from an adjacent subunit (Fig. 5). In monomer C, the electron density beyond the Cδ-atom of R142 suggested alternative conformations for the guanidinium group besides the one found in the other three monomers. The relatively high B factors of the arginine in monomers A, B, and D and the absence of a unique conformation for the guanidinium group in monomer C indicate that R142 is highly flexible.
Table 3.
Data collection and refinement statistics
| Data collection and processing | |
| Resolution range, Å | 37.4–1.8 |
| Total measurements | 412,908 |
| Unique reflections | 117,329 |
| Completeness, %* | 98.8 (97.7) |
| Rsym, %*† | 7.1 (30) |
| 〈I/σ〉* | 5.2 (2.4) |
| Refinement statistics | |
| Resolution range, Å | 37.4–1.8 |
| Rcryst/Rfree, %‡ | 18.3/21.0 |
| rms deviation bond lengths, Å | 0.009 |
| rms deviation angles, ° | 1.5 |
| Number of atoms (protein/water) | 9,113/832 |
| Average B factors (protein§/water), Å2 | 26.1/36.7 |
The numbers in parentheses refer to the 1.9- to 1.8-Å resolution shell.
Rsym = Σ(Ih,i − 〈Ih〉)/ΣIh,i
Rcryst and Rfree = Σ|Fobs − Fcalc|/Σ|Fobs|, where Rfree was calculated over 1,164 amplitudes chosen at random and not used in the refinement.
For individual subunits, the average B factors were 25.6 Å2 (A), 27.5 Å2 (B), 26.8 Å2 (C), and 24.7 Å2 (D).
Figure 5.
Stereoview of the active center of NAL mutant L142R monomer A (black solid) superimposed on the wild-type structure (black dotted). The conformations of R142 in monomers B and D are shown in red and green, respectively. The conformation of the equivalent R138 in the structure of substrate-free DHDPS from E. coli (13) is shown for comparison (cyan dotted).
The α-keto acid moiety of the inhibitor BHP was well defined in the same orientation as in wild type, covalently linked via its C2-atom to Nζ of K165. The observed electron density beyond C2 was much weaker and less conclusive. The drop in density for C3 suggests that BHP is not only bound in a R-configured keto form as described for wild-type (12) and shown for mutant L142R in Fig. 5. Instead, there might be a dynamic equilibrium of different orientations and tautomeric forms caused by the lower pH value in our crystallization conditions.
Discussion
We have shown that transplanting a conserved arginine from DHDPS into the NAL-scaffold significantly increases the potential to catalyze the condensation of pyruvate and l-ASA mainly by improving the turnover rate. By rational design we could enable NAL from E. coli to replace an enzyme from another pathway in vivo while still maintaining much of its original activity. A similar scenario has been described for two structurally similar (β/α)8-enzymes from the biosynthetic pathways of histidine and tryptophan, where random mutagenesis on one of these two homologs yielded a mutant exhibiting both activities (33).
Our findings also offer a structural rationalization for the observed activities. The structure of mutant L142R reveals that introduction of R142 does not significantly affect the pyruvate binding pocket. This finding is consistent with the apparent Kpyruvate value being in the same range for both enzymes. The mutation also leaves the binding pocket for Neu5Ac mainly intact but changes the environment at the binding position for the methyl group of the acetyl moiety from mainly hydrophobic to positively charged (see Fig. 3), hence the increased KNeu5Ac. The kinetic constants for DHDPS activity reveal that the arginine can fulfill only one of its functions observed in DHDPS. In DHDPS, specific binding of the carboxylate group of l-ASA is achieved in conjunction with residues from loop α9/α10. In mutant L142R the arginine cannot provide the same binding platform, and consequently the affinity for l-ASA is only marginally improved compared with the starting scaffold. What the arginine does, however, is facilitate catalysis, as reflected by the increased kcat value. This could be achieved by following the substrate along the reaction pathway or simply by providing a more favorable electrostatic environment for Schiff-base formation and transimination reaction.
Our results give further insights into general principles of protein evolution. There is wide consensus that most contemporary enzymes have evolved by divergent evolution. Recruitment of enzymes across different pathways can occur by duplicating genes and conserving the catalytic mechanism of the respective protein but evolving new substrate specificity, as first shown by Jensen (34). It has been suggested that the promiscuous nature of many active sites has played and probably still does play an essential role in the creation of novel enzymes by divergent evolution (35). Recent statistically significant analyses of sequences and protein structures show that nature seems to have applied exactly this strategy when evolving novel enzymes or complete pathways (6, 36). We have recreated such a recruitment event in our experiments. More precisely, we have mimicked the crucial event transforming a dormant precursor with residual activity into an enzyme that is competent enough to efficiently play its role as part of another biological pathway and thus conferring a selective advantage. This threshold activity is particular to the specific reaction in a network of overlapping pathways. Starting with a suitable scaffold, which has the required or very similar chemistry and a similar or simply indiscriminate enough binding site to allow for more or less unspecific initial binding, this can be achieved with relatively few mutations. In our particular case, a single mutation was sufficient. In evolutionary terms, this avoids extended periods of random drift, which increases the likelihood that the duplicated gene is anchored into the genome. Once the gene product confers this initial selective advantage, the catalytic properties can be optimized and additional regulatory properties can be evolved. This, however, seems to be a much longer process requiring many more mutations. Although nature undoubtedly explores alternative strategies as well, given the random nature of evolution, the presented strategy seems to have proven the most successful as mirrored in the distribution of contemporary enzymes across different pathways.
Acknowledgments
We thank Dr. Martin Smith for assistance with the ozonolysis of allylglycine and Dr. Yu Wai Chen for helpful advice in structure solution and refinement procedures. We thank the Escherichia coli Genetic Stock Center at Yale University (New Haven, CT) for providing E. coli strain AT997. A.C.J. is supported by Marie Curie Fellowship MCFI-2000-02118.
Abbreviations
- ASA
aspartate-β-semialdehyde
- BHP
β-hydroxypyruvate
- DHDPR
dihydrodipicolinate reductase
- DHDPS
dihydrodipicolinate synthase
- NAL
N-acetylneuraminate lyase
- Neu5Ac
N-acetylneuraminate
Footnotes
Data deposition: The atomic coordinates of mutant L142R have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1HL2).
References
- 1.Schmid A, Dordick J S, Hauer B, Kiener A, Wubbolts M, Witholt B. Nature. 2001;409:258–268. doi: 10.1038/35051736. [DOI] [PubMed] [Google Scholar]
- 2.Arnold F H. Nature. 2001;409:253–257. doi: 10.1038/35051731. [DOI] [PubMed] [Google Scholar]
- 3.Walsh C. Nature. 2001;409:226–231. doi: 10.1038/35051697. [DOI] [PubMed] [Google Scholar]
- 4.Zhao H, Chockalingam K, Chen Z. Curr Opin Biotechnol. 2002;13:104–110. doi: 10.1016/s0958-1669(02)00291-4. [DOI] [PubMed] [Google Scholar]
- 5.Reardon D, Farber G K. FASEB J. 1995;9:497–503. doi: 10.1096/fasebj.9.7.7737457. [DOI] [PubMed] [Google Scholar]
- 6.Copley R R, Bork P. J Mol Biol. 2000;303:627–641. doi: 10.1006/jmbi.2000.4152. [DOI] [PubMed] [Google Scholar]
- 7.Lang D, Thoma R, Henn-Sax M, Sterner R, Wilmanns M. Science. 2000;289:1546–1550. doi: 10.1126/science.289.5484.1546. [DOI] [PubMed] [Google Scholar]
- 8.Höcker B, Beismann-Driemeyer S, Hettwer S, Lustig A, Sterner R. Nat Struct Biol. 2001;8:32–36. doi: 10.1038/83021. [DOI] [PubMed] [Google Scholar]
- 9.Barbosa J A, Smith B J, DeGori R, Ooi H C, Marcuccio S M, Campi E M, Jackson W R, Brossmer R, Sommer M, Lawrence M C. J Mol Biol. 2000;303:405–421. doi: 10.1006/jmbi.2000.4138. [DOI] [PubMed] [Google Scholar]
- 10.Wong C-H, Halcomb R L, Ichikawa Y, Kajimoto T. Angew Chem Int Ed Engl. 1995;34:412–432. [Google Scholar]
- 11.Izard T, Lawrence M C, Malby R L, Lilley G G, Colman P M. Structure (London) 1994;2:361–369. doi: 10.1016/s0969-2126(00)00038-1. [DOI] [PubMed] [Google Scholar]
- 12.Lawrence M C, Barbosa J A, Smith B J, Hall N E, Pilling P A, Ooi H C, Marcuccio S M. J Mol Biol. 1997;266:381–399. doi: 10.1006/jmbi.1996.0769. [DOI] [PubMed] [Google Scholar]
- 13.Mirwaldt C, Korndorfer I, Huber R. J Mol Biol. 1995;246:227–239. doi: 10.1006/jmbi.1994.0078. [DOI] [PubMed] [Google Scholar]
- 14.Blickling S, Renner C, Laber B, Pohlenz H D, Holak T A, Huber R. Biochemistry. 1997;36:24–33. doi: 10.1021/bi962272d. [DOI] [PubMed] [Google Scholar]
- 15.Blickling S, Beisel H G, Bozic D, Knäblein J, Laber B, Huber R. J Mol Biol. 1997;274:608–621. doi: 10.1006/jmbi.1997.1393. [DOI] [PubMed] [Google Scholar]
- 16.Yeh P, Sicard A M, Sinskey A J. Mol Gen Genet. 1988;212:105–11. doi: 10.1007/BF00322451. [DOI] [PubMed] [Google Scholar]
- 17.Gill S C, von Hippel P H. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 18.Black S, Wright N G. J Biol Chem. 1955;213:39–50. [PubMed] [Google Scholar]
- 19.Karsten W E, Viola R E. Biochim Biophys Acta. 1991;1077:209–219. doi: 10.1016/0167-4838(91)90060-d. [DOI] [PubMed] [Google Scholar]
- 20.Comb D G, Roseman S. Methods Enzymol. 1962;5:391–394. [Google Scholar]
- 21.Lilley G G, von Itzstein M, Ivancic N. Protein Expression Purif. 1992;3:434–440. doi: 10.1016/s1046-5928(05)80047-6. [DOI] [PubMed] [Google Scholar]
- 22.Karsten W E. Biochemistry. 1997;36:1730–1739. doi: 10.1021/bi962264x. [DOI] [PubMed] [Google Scholar]
- 23.Collaborative Computational Project No. 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 24.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 25.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 26.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 27.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 28.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 29.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 30.Krüger D, Schauer R, Traving C. Eur J Biochem. 2001;268:3831–3839. doi: 10.1046/j.1432-1327.2001.02297.x. [DOI] [PubMed] [Google Scholar]
- 31.Blickling S, Knäblein J. Biol Chem. 1997;378:207–210. doi: 10.1515/bchm.1997.378.3-4.207. [DOI] [PubMed] [Google Scholar]
- 32.Vauterin M, Frankard V, Jacobs M. Plant J. 2000;21:239–248. doi: 10.1046/j.1365-313x.2000.00668.x. [DOI] [PubMed] [Google Scholar]
- 33.Jürgens C, Strom A, Wegener D, Hettwer S, Wilmanns M, Sterner R. Proc Natl Acad Sci USA. 2000;97:9925–9930. doi: 10.1073/pnas.160255397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jensen R A. Annu Rev Microbiol. 1976;30:409–25. doi: 10.1146/annurev.mi.30.100176.002205. [DOI] [PubMed] [Google Scholar]
- 35.O'Brien P J, Herschlag D. Chem Biol. 1999;6:R91–R105. doi: 10.1016/S1074-5521(99)80033-7. [DOI] [PubMed] [Google Scholar]
- 36.Teichmann S A, Rison S C, Thornton J M, Riley M, Gough J, Chothia C. J Mol Biol. 2001;311:693–708. doi: 10.1006/jmbi.2001.4912. [DOI] [PubMed] [Google Scholar]
- 37.Smith B J, Lawrence M C, Barbosa J A. J Org Chem. 1999;64:945–949. doi: 10.1021/jo981960v. [DOI] [PubMed] [Google Scholar]
- 38.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]