Abstract
RNA polymerase II transcription complexes stalled shortly after initiation over a repetitive segment of the template can undergo efficient transcript slippage, during which the 3′ end of the RNA slides upstream and then re-pairs with the template, allowing transcription to continue. In the present study, we have used transcript slippage as an assay to identify possible structural transitions that occur as the polymerase passes from the initiation to the elongation phase of transcription. We reasoned that transcript slippage would not occur in fully processive complexes. We constructed a series of templates that allowed us to stall RNA polymerase II after the synthesis of a repetitive sequence (5′-CUCUCU-3′) at varying distances downstream of +1. We found that polymerase must synthesize at least a 23-nt RNA to attain resistance to transcript slippage. The ability to undergo slippage was lost in two discrete steps, suggestive of two distinct transitions. The first transition is the formation of the 8- to 9-bp mature RNA–DNA hybrid, when slippage abruptly dropped by 10-fold. However, easily detectable slippage continued until 14 more bonds were made. Thus, although the transcript becomes tightly constrained within the transcription complex once the hybrid reaches its final length, much more RNA synthesis is required before the RNA is no longer able to slip upstream along the template. This last point may reflect an important stabilizing role for the interaction of the polymerase with the transcript well upstream of the RNA–DNA hybrid.
Both RNA polymerase II (pol II) and bacterial RNA polymerase pass through a stage of abortive initiation immediately after the start of transcription, during which the polymerase may release the nascent RNA (1–4). After the synthesis of about a 10-nt RNA, the transcription complex (TC) begins its transition into the stable transcript elongation form (5–7). This transition is of major interest in understanding the mechanism of RNA synthesis in eukaryotic cells, because it is clear that regulation may be imposed at or very near this transition (reviewed in ref. 8). The pol II transcript elongation complex (TEC) must ultimately be able to function without dissociation over transcription units that may continue for a million base pairs. Thus, an understanding of the molecular mechanisms involved in the transition from initiation to transcript elongation is of central importance in the study of gene expression.
A major determinant of TC stability is the RNA–DNA hybrid (9). This duplex must be at least 8 bp long to achieve stability in complexes assembled from pure pol II and DNA (10). It has also been shown that the hybrid is 9 bp within a yeast pol II ternary complex containing a 14-nt RNA (9). Because pol II TC stability increases considerably after the synthesis of a 10-nt RNA (5, 6, 11), it might seem that the initiation–elongation transition is simply the process of extending the RNA–DNA hybrid to its mature length. However, results with both bacterial and eukaryotic RNA polymerase show that TC properties are also strongly dependent on RNA more than 10 nt upstream of the active site, indicating that the complete transition into the elongation complex must occur further downstream than +10 (12–15). For example, many pol II complexes halted from 18 to 32 nt downstream of transcription start have a strong tendency to translocate upstream, or backtrack; in some sequence contexts, this backtracking results in arrest (16, 17). Pol II does not acquire complete resistance to backtracking until about 50 nt have been added to the nascent RNA (16, 18). Bacterial RNA polymerases also arrest when paused at +27 on one particular template (12).
None of these observations reveal the mechanism by which RNA polymerase achieves the final, highly processive form of the TC. A major advance in this area was provided by the determination of the high-resolution crystal structure of a yeast pol II ternary complex containing 14 nt of RNA (9). In this complex, the RNA–DNA hybrid, particularly at the 3′ end, is tightly constrained by the so-called clamp domain, which gives a physical basis for processivity. However, because only a single ternary complex structure has been solved, both the ultimate form of the elongation complex and the pathway to this form from the initiating state are still unknown. The clamp has clearly moved toward the polymerase body in the 14-mer complex structure, relative to its location in free pol II (9, 19, 20). However, it is not known whether further movement of the clamp takes place as transcription continues, nor is it known at what point before the synthesis of a 14-mer the clamp begins to close.
We reported earlier that the newly initiated human pol II TC can undergo transcript slippage (21). The initially transcribed region of the promoter in question consists of a series of dinucleotide repeats. We found that pol II pauses naturally at the end of this repeated element. In a substantial portion of these paused complexes, the 3′ end of the transcript can slip upstream by two bases, re-pair with the template strand, and allow transcription to continue. Because transcript slippage must involve freedom of movement of the transcript along the template (lateral mobility), slippage is an indication of the level of constraint imposed on transcript movement by the TC. Our original observations were made when pol II paused at +7. We supposed that one hallmark of the mature, highly processive TEC would be the absence of transcript slippage. This lack of slippage would ensure that the transcript is a faithful copy of the template. A study of the decline in transcript slippage as a function of transcript elongation might therefore reveal intermediates in the initiation-elongation transition, particularly those involved with clamp closure. We report here that the initially high levels of slippage drop abruptly at or very near the template location at which the hybrid reaches its final length (8- to 9-nt transcript; see refs. 9 and 10), consistent with the idea that most of the process of clamp closure is complete at this point. However, slippage was easily detectable for complexes with transcripts as long as 20 nt. TCs stalled after the synthesis of 23 or 30 nt RNA did not undergo detectable transcript slippage. These results suggest that the TEC does not reach its final state until well after hybrid formation has taken place. They also suggest that the nascent RNA well upstream of the RNA–DNA hybrid is important in stabilizing the ternary TC.
Experimental Procedures
Plasmid Constructions.
All plasmids used in these studies contained the adenovirus major late promoter and were made by replacing the segment between the BssHII and StuI sites on the pML20–40 plasmid with synthetic DNA fragments as described previously (17). All constructs were verified by DNA sequencing.
Template Preparation for the in Vitro Transcription Reaction.
Templates for transcription were made by PCR by using the same pair of primers, with the upstream primer biotinylated, as described (17). The PCR products ranged from 190 to 215 bp, depending on the template; on all templates, the transcription start site is 96 bp downstream from the biotinylated end.
In Vitro Transcription on Attached Templates.
Attachment of biotinylated templates on streptavidin-coated magnetic beads (Promega), and formation of preinitiation complexes using HeLa nuclear extract were performed as described (17). Briefly, preinitiation complexes were assembled at 30°C for 20 min. Unbound materials were removed by rinsing bead attached complexes twice with 100 μl of BC100 (20 mM Tris⋅HCl, pH 7.9/8 mM MgCl2/100 mM KCl/1 mM DTT/20% glycerol/0.5 mM EDTA). Rinsed complexes were resuspended in BC100 and stored on ice until used. Transcription reactions were carried out at 30°C with 1 mM dinucleotide (either ApC or ApU) or 50 μM ATP, 5 μM dATP (in all reactions except those initiated with ATP), 20 μM UTP/5Br-UTP and 0.25 μM [α-32P]CTP [Perkin–Elmer/New England Nuclear; 800 Ci/mmol (1 Ci = 37 GBq)] for 5 min followed by incubation with 20 μM CTP for 5 min. Chase reactions were carried out at 30°C for 5 min with 200 μM NTPs. Reactions were terminated and RNAs purified as described (17, 21). RNase T1 digestion was performed at 37°C for 15 min with 2 units of RNase T1 (MBI Fermentas, Hanover, MD)/10 μl of reaction with 0.25 μg of yeast tRNA as carrier. RNAs were suspended in 8 M urea before resolving in 25% acrylamide-3% bis-acrylamide gels with 7 M urea. Gels were visualized with a PhosphorImager (Molecular Dynamics), and bands were quantitated by imagequant software (Amersham Pharmacia/Molecular Dynamics). The lowest values of transcript slippage we report are in the range of 0.6–1% of total transcription (see Fig. 4). We estimate that we could have detected slippage at the 0.1% level.
Figure 4.
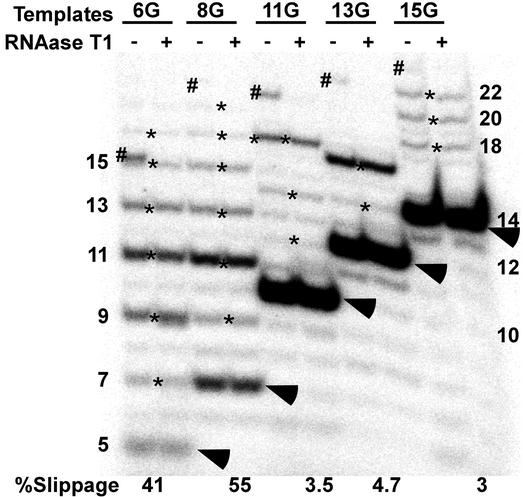
Slippage activity does not disappear until a 23-nt transcript is synthesized. Transcription was carried out on the indicated templates (see Fig. 1) with ApC primers in the absence of GTP as described in Experimental Procedures. RNAs in the indicated lanes were digested with RNase T1 or chased with 200 μM of all four NTPs. Arrowheads designate the expected G-stop (nonslipped) transcripts. Slippage and leakthrough transcripts are marked by asterisks and pound signs, respectively. The percentage slippage is the fraction of complexes that underwent at least one round of transcript slippage. Lengths of selected RNAs are given in the margin of the gel.
Results
Transcript Slippage Is Strongly Reduced After the Synthesis of a 9-nt Transcript.
We showed (21) that pol II has a strong tendency to pause 7 bases downstream of transcription start at the adenovirus major late promoter, after the synthesis of the transcript ACUCUCU. A substantial proportion of the polymerases halted at this location undergo transcript slippage; that is, the 3′ segment of the transcript slips upstream by 2 nt and reforms the RNA–DNA hybrid, allowing transcription to continue (21). Multiple rounds of upstream slippage and reextension can take place, leading to transcripts that are longer than expected in multiples of 2 nt (21). We considered it surprising that a 7-nt transcript has the freedom of movement along the template, which is necessary to allow transcript slippage, given that the hybrid in the mature TEC is thought to be 8–9 bp (9, 22, 23). It seemed reasonable to suppose that, during normal transcript elongation, transcript slippage in repetitive regions of the template would not occur. This supposition led us to examine the ability of pol II TCs to undergo transcript slippage as a function of transcript length. Transcript slippage seems to require a pause after the synthesis of the repeated element, and it is most efficient if transcript elongation is temporarily halted at the appropriate point through the use of a subset of the NTPs (21, 24). We therefore constructed a set of 11 templates, based on the adenovirus major late promoter, which feature a G-less initially transcribed region ending in the sequence CTCTCTG on the non-template strand (Fig. 1). Transcription of these templates in the absence of GTP allowed us to stall pol II from 7 to 30 nt downstream of transcription start, with the same CUCUCU sequence at the 3′ end of the transcript and the same DNA sequence downstream of the stalling point.
Figure 1.
Sequences (nontemplate strand) of the initially transcribed regions of the templates used in these studies; note that these templates differ only between the initiating nucleotide (+1) and the first guanine residue (lowercase). The repeated element involved in transcript slippage is shown in italics.
The templates were assembled into preinitiation complexes by incubation with HeLa nuclear extract, followed by gentle rinsing with transcription buffer (without sarkosyl). Transcription was initiated with the dinucleotide primer ApC, radiolabeled CTP, and UTP for 10 min. These conditions should have resulted primarily in transcripts stalled before the G-stop, for example at +7 on the 8G template (Fig. 2; G-stop transcripts indicated by arrowheads). However, as expected from our earlier work (21), many transcripts on the 8G DNA were much longer than 7 nt (Fig. 2). Most of the 8G transcripts above the 7-nt band in Fig. 2 are longer than the G-stop RNA in increments of 2 nt, as would be expected from one or more rounds of upstream slippage and re-pairing of the 3′ end of the RNA (21). These transcripts (marked by asterisks) cannot have resulted from low levels of contaminating GTP causing readthrough of the G-stop, because they are resistant to digestion by RNase T1 (Fig. 2). Readthrough was very low in the 8G reaction shown in Fig. 2, but transcription of the other templates did yield detectable levels of T1-sensitive RNAs. These transcripts, marked by #, had lengths consistent with pausing before the second G residue downstream of +1 (see Fig. 1). If the transcripts marked by asterisks arose from misincorporation at the G-stop (25), instead of readthrough from contaminating GTP, then the resulting RNAs should have accumulated at the next downstream G-stop (i.e., at the # position) or at more distant positions, rather than at intermediate lengths. It should be noted that low levels of RNA were always observed at lengths that cannot be accounted for by transcript slippage; for example, in the 8G reaction in Fig. 2, bands were present between the transcripts marked by asterisks at 8, 10, 12, and 14 nt. We have not further characterized these transcripts, but we suspect that they originate from the slippage RNAs by transcript cleavage. The TCs used in these experiments were not detergent rinsed and thus should contain SII (see also ref. 18). Results from other groups indicate that SII should suppress misincorporation (25, 26).
Figure 2.
Transcript slippage by pol II is drastically reduced when the polymerase is halted 10 or more bases downstream of +1. Transcription was carried out on the indicated templates (see Fig. 1) with dinucleotide primers in the absence of GTP as described in Experimental Procedures. RNAs in the indicated lanes were digested with RNase T1. Arrowheads designate the expected G-stop (nonslipped) transcripts. Slippage and leakthrough transcripts are marked by asterisks and pound signs, respectively. The percentage slippage is the fraction of complexes that underwent at least one round of transcript slippage. Lengths of selected RNAs are given in the margins of the gel.
For the reaction shown in Fig. 2, slightly more than half of the TCs on the 8G template went through at least one round of transcript slippage. This result is consistent with the extent of transcript slippage seen in our earlier work with complexes that are stalled for many minutes at the end of a repetitive segment in the template (21). The most prominent of the 8G slippage products was 11 nt in length. We cannot determine whether this RNA resulted from two rounds of slippage by 2 nt or a single slippage event of 4 nt (see Fig. 1). However, on the 6G template, on which slippage can only take place by 2 nt, 9-nt RNAs were more abundant than 7-mers (Fig. 2).
In contrast to the high levels of transcript slippage seen with the 6G and 8G templates, the 11G, 13G, and 15G templates yielded much lower levels of T1-resistant RNAs longer than the G-stop transcript (Fig. 2). The fraction of 11G–15G complexes that went through at least one round of slippage is an order of magnitude less than the proportion of slipped complexes on with 6G and 8G templates. Multiple rounds of slippage were still possible on 11G–15G. Successive cycles of slippage resulted, in the case of the 15G template, in the generation of RNAs through slippage that were >20 nt in length. Because our original observations on slippage relied on a natural pause by pol II at +7, one might ask whether the lower levels of slippage seen with the 11G–15G complexes actually reflect slippage from pausing at +7 and not at +10, +12, or +14. All of our templates with G stops at +10 or further downstream (that is, 11G through 31G) were designed to eliminate the repetitive sequence element that allows upstream slippage for polymerases paused at +7 (see Fig. 1). Thus, we are confident that transcript slippage in these complexes resulted from polymerases stalled at the G stop and not further upstream.
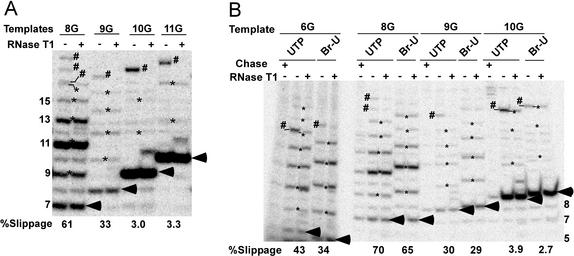
To determine the exact point at which the potential for transcript slippage falls off, reactions identical to those in Fig. 2 were performed on the 9G and 10G templates, which should synthesize 8 nt and 9 nt transcripts, respectively, in the absence of GTP (see Fig. 1). This assay (Fig. 3A) showed that slippage activity drops by half from the 8G to the 9G template, and then falls 10-fold on the 10G template to roughly the same level seen with the 11G–15G complexes. Thus, the formation of a 9-nt transcript significantly reduces the ability of the pol II complex to support slippage. This finding strongly suggests that the TEC undergoes a major structural transition after the synthesis of a 9-nt transcript, a point to which we will return in Discussion.
Figure 3.
(A) TCs initiating with a dinucleotide acquire most of their resistance to transcript slippage on synthesis of a 9-nt transcript. Transcription was carried out on the indicated templates (see Fig. 1) with dinucleotide primers in the absence of GTP as described in Experimental Procedures. RNAs in the indicated lanes were digested with RNase T1. It should be noted that, among the templates we tested, determining the extent of slippage was most problematic with 10G and 11G. All of the T1-resistant bands were scored as slippage products for these templates, although some of these bands do not seem to be the expected length for slippage products. We believe that these anomalous-length RNAs resulted from slippage followed by transcript cleavage from low levels of SII in the reactions, although we have not explored this point further. Pol II transcription complexes may vary considerably in their sensitivity to SII-mediated cleavage (see ref. 18). (B) Incorporation of 5Br-UTP instead of UTP does not significantly alter the transcript slippage activity of halted elongation complexes. Transcription of the indicated templates was performed as in Figs. 2 and 3A except that 5Br-UTP was substituted for UTP in the indicated reactions. An equal aliquot of each reaction mixture was chased with 200 μM of all four NTPs at 30°C for 5 min. Run-off transcripts are not shown. For both panels, arrowheads designate the expected G-stop (nonslipped) transcripts. Slippage and leakthrough transcripts are marked by asterisks and pound signs, respectively. The percentage slippage is the fraction of complexes that underwent at least one round of transcript slippage. Lengths of selected RNAs are given in the margin of the gels.
Because transcript slippage requires the transcript to transiently release from the DNA to slip upstream, slippage could be influenced by the strength of the RNA–DNA hybrid. To address this point without changing the sequence of the transcript, we compared levels of slippage on the 6G, 8G, 9G, and 10G templates in reactions that contained either UTP or 5Br-UTP. We found (Fig. 3B) that increasing the RNA–DNA hybrid strength with 5Br-UTP resulted in either a slight reduction or no change in slippage. Thus, the transcript slippage reaction is primarily influenced by the length of the RNA at the point of slippage and not by the strength of the hybrid.
Elongation Complexes Containing a 23-nt Transcript Do Not Undergo Detectable Transcript Slippage.
Transcript slippage remains at low but detectable levels for TCs stalled between +9 and +14. To explore the possibility that slippage disappears as elongation continues, we assayed for slippage in complexes stalled after the synthesis of 16-, 18-, 20-, 23-, and 30-nt RNAs (Fig. 4). These complexes all contained the same CUCUCU sequence at the 3′ ends of their transcripts when stalled at the G-stop. Low but detectable levels of transcript slippage (≈1% of total complexes) were observed on the 17G, 19G, and 21G templates, but no slippage products were seen with 24G and 31G. Failure of complexes stalled on the 24G and 31G templates to undergo slippage was not the result of arrest at +23 or +30, because nearly all of the stalled complexes could be chased by the addition of GTP to the reactions (Fig. 4). Substituting 5Br-UTP for UTP in reactions like those in Fig. 4 did not change the results (data not shown). Note that failure of the 23-mer complexes to slip was not caused by insufficient residence time at the pause site. Slippage reactions were carried out for 10 min, and 23-nt RNAs had accumulated after 1 min (data not shown).
Transcription Complexes Initiated with ATP Show Much Less Transcript Slippage on Synthesis of an 8-nt Transcript Than ApC-Initiated Complexes with 8-nt RNAs.
In all of the above experiments, transcription was primed with the dinucleotide ApC. In our initial study on transcript slippage, we noted lower levels of slippage in some tests when comparing initiation with ATP to ApC priming (21). However, those were cases in which slippage depended on the natural pause at +7, and the use of ATP instead of ApC reduced the duration of the +7 pause (21). In the present study, the presence of a G-stop at the end of the repetitive segment allowed us to remove pause time as a variable. When we assayed for the extent of transcript slippage for ATP-initiated complexes stalled at the G-stop on the 6G, 8G, 9G, and 10 G templates (Fig. 5), we found a significant difference from the results seen on the same templates with ApC-initiated transcription. ATP-initiated complexes showed relatively high slippage when stalled at +5 (54%), less slippage at +7 (30%), and much less slippage at +8 (3%). Thus, ATP-initiated complexes behaved in the transcript slippage reaction like ApC-primed complexes with transcripts 1 nt longer. ATP-initiated complexes behaved indistinguishably from ApC-initiated complexes in slippage assays on the 11G–31G templates (data not shown).
Figure 5.
ATP-initiated TCs acquire most of their resistance to transcript slippage on synthesis of an 8-nt RNA. Transcription of the indicated templates was carried as in Fig. 2 except that 50 μM ATP was used instead of a dinucleotide to support initiation. (A) Transcript slippage on templates 6G and 8G. (B) Transcript slippage on templates 8G through 10G. Slippage and leakthrough transcripts are marked by asterisks and pound signs, respectively. The percentage slippage is the fraction of complexes that underwent at least one round of transcript slippage. Lengths of selected RNAs are given in the margins of the gel.
Discussion
We have used transcript slippage by human pol II during the initial phases of transcription as a means of studying the process by which the stable TEC is established. For transcript slippage to occur, the 3′ end of the nascent RNA must separate from the template, move upstream, and then re-form base pairs with the DNA. We reported earlier that the slippage reaction is robust in complexes with RNAs as long as 7 nt (21). One would expect the transcript to be unable to slip upstream in the mature, fully processive TC. It was important to establish that this supposition is correct, and if it is true, to determine at what point during transcript elongation the TC is no longer capable of transcript slippage. We found that the ability of the TC to undergo slippage decreases in a stepwise fashion as transcript elongation proceeds, suggesting at least two discrete transitions. The first transition occurs concomitant with the formation of the full-length RNA–DNA hybrid, at 8–9 nt downstream of transcription start. The essentially complete loss of slippage occurs between 21 and 23 nt downstream of +1.
A plausible mechanism for transcript slippage is diagrammed in the left hand column of Fig. 6. To shift upstream, the 3′ end of the transcript must transiently release from the template, as shown in step 2. Because the complexes we studied were paused after the synthesis of a transcript ending in ..CUCUCU3′, breathing of the 3′ end of the RNA-DNA hybrid can result in re-pairing of the RNA with the template 2 nt upstream of the original position, as shown in step 3. If the body of the polymerase translocates upstream along with the RNA 3′ end (or, if the active site reassociates with the re-paired 3′ end), then transcription can continue (step 4). On the templates we used, the 2-nt upstream sliding and re-pairing of the transcript would leave the 4 nt of RNA at the 3′ end in continuous hybrid with the DNA, but some or all of the next four upstream RNA bases would not be paired with the template. We cannot determine how this unpaired RNA is accommodated within the TC. It may be looped out within the polymerase, as suggested by the diagram; alternatively, some or all of this RNA may be extruded down the RNA exit channel.
Figure 6.
A diagrammatic comparison of transcript slippage and backtracking. The rounded rectangle represents pol II, the paired lines are the two strands of DNA, and the gray boxes are the RNA exit channel and the funnel, respectively. The asterisk is the active site, and the transcript is shown by either the designated dashed (RNA in RNA–DNA hybrid) or solid (RNA not in hybrid) lines. The transcript in this case has a repetitive sequence at the 3′ end, which would allow transcript slippage (steps in left column); an alternative pathway involving backtracking is shown in the right column. Transcript cleavage occurs at step 3 in the backtracking pathway. Resumption of RNA synthesis is shown in step 4 in both cases.
Because the polymerase active center must move upstream relative to the template strand during transcript slippage, it is useful to contrast slippage with the backtracking that accompanies transcriptional pausing and arrest. As diagramed in the right column of Fig. 6, backtracking involves the upstream translocation of the body of the polymerase, the transcription bubble, and the RNA–DNA hybrid as a unit, which removes the transcript 3′ end from the active site. The displaced RNA is thought to be extruded into a channel, referred to as the funnel, which opens beneath the active center (27). Backtracking separates the transcript 3′ end from both the template and the active site. Thus, backtracking should not be an intermediate in the transcript slippage reaction. Note that backtracking requires the withdrawal of some RNA from the RNA exit channel (reverse threading; step 2, right column, Fig. 6). Recovery from backtracking involves either a reversal of polymerase movement or cleavage of the transcript in the backtracked complex (step 3, Fig. 6) to realign the active site with the RNA 3′ end.
The significance of our results is most apparent in the context of the high resolution crystal structure of the yeast pol II TC containing a 14-nt RNA (9). A key feature of that structure is the striking inward movement, relative to free pol II (19), of a clamp-like domain that embraces the RNA–DNA hybrid. This constraint of the hybrid, which is tightest over the last 3–4 bases of the transcript, is thought to be a central feature in maintaining the processive nature of the TEC. The interpretation of the yeast ternary complex structure is complicated by the fact that it was assembled without general transcription factors directly from polymerase and DNA by using 3′-extended (“tailed”) templates. It has also been suggested (20) that the 14-mer complex represents a paused form of the TC, rather than a form that is directly poised to add the next nucleotide to the nascent RNA. It is not known when clamp closure takes place during the synthesis of a 14-mer transcript, nor is it known how (or if) the structure of the ternary complex changes as transcription proceeds past +14.
Our results on transcript slippage as a function of transcript length, obtained with promoter-initiated pol II, may shed light on both of these points. The 2-fold reduction in slippage that is seen as the nascent RNA is extended from 7 to 8 nt (Fig. 3) suggests that closure of the processivity clamp begins at this point. The correlation of clamp closure with the completion of the 9-bp RNA–DNA hybrid agrees with the prediction (9, 19, 20) that interactions of the hybrid with previously disordered switch regions at the base of the clamp drive the formation of helices in the switch regions and thus force inward movement of the clamp.
Although we may assign the initial drop in the extent of slippage to clamp closure coincident with completion of the RNA–DNA hybrid, it is less clear why the TC can continue to undergo slippage with transcripts as long as 20 nt but lose this ability by +23. This transition may be related to the filling of the RNA binding domain upstream of the hybrid. One could imagine that occupancy of the RNA exit channel triggers a conformational change in the polymerase, which results in a tighter grip of the clamp domain on the RNA–DNA hybrid and perhaps on the upstream RNA as well. This idea is consistent with the fact that, in bacterial RNA polymerase, interruption of the interaction between RNA and the flap domain (analogous to the distal segment of the pol II exit channel) can cause pausing by the polymerase (28, 29). Both the pol II ternary complex structure (9) and the results of RNase treatment of pol II complexes (13, 30) indicate that the RNA binding domain in pol II should be filled in complexes bearing 17-nt RNAs. However, we found that complexes with 18- or 20-nt transcripts can undergo transcript slippage (Fig. 4). This finding suggests that the polymerase and the nascent RNA undergo additional stabilizing interactions upstream of the point at which the transcript emerges from the polymerase (14, 28, 29, 31). Our current study cannot shed any light on what those interactions might be, but we note that such interactions have also been suggested by earlier work (18, 32).
It is important to emphasize that, whereas it is plausible to attribute the two reductions in transcript slippage to two stages of closure of the processivity clamp, we have not made direct observations of clamp position or any other aspect of pol II structure. Thus, the actual physical bases for the changes in slippage competence may be different from those we have proposed. In particular, because the slippage reaction requires some upstream translocation of the active site, one might think that the loss of slippage competence between positions +20 and +23 is simply the result of a blocking of backtracking, through folding of the 5′ end of the RNA into a secondary structure or the interaction of the 5′ end with the polymerase itself. For several reasons, this explanation seems unlikely to us. First, we cannot identify any potential secondary structures in the RNA upstream of the RNA–DNA hybrid for TCs paused at the G-stop on either the 24G or 31G templates (see Fig. 1). Second, as shown in Fig. 4, essentially all of the 15G, 17G, 19G, and 21G complexes chased on addition of all four NTPs. Thus, we have no evidence for a tendency toward stable upstream translocation and arrest in these complexes, which are nevertheless able to undergo transcript slippage. The most important point is the difference in displacement of the main body of the polymerase and the transcript during backtracking and transcript slippage. As noted in Fig. 6, backtracking requires that RNA withdraw up the RNA exit channel in compensation for the extrusion of the 3′ end into the funnel. Structures or interactions that prevent reverse threading would also prevent backtracking. However, whereas slippage must require some upstream translocation by the active site, it should not require reverse threading of the transcript (Fig. 6). Thus, even if the 5′ segment of the RNA in, for example, complexes stalled at +23 can form secondary structures or interact with the polymerase, this result should not prevent transcript slippage.
In summary, we have used transcript slippage as an assay for the transition of pol II from the initiating to the elongating state. This measure of the freedom of the transcript to move laterally within the TC falls off in two discrete steps. A minimum of 23 nt of RNA must be synthesized before pol II loses the ability to undergo slippage. Because the structural core of the multisubunit RNA polymerases is highly conserved (20, 33, 34), our results should illuminate a general mechanism through which processivity is achieved.
Acknowledgments
This work was supported by Grant GM-29487 from the National Institutes of Health.
Abbreviations
- pol II
RNA polymerase II
- TC
transcription complex
- TEC
transcript elongation complex
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.McClure W R. Proc Natl Acad Sci USA. 1980;77:5634–5638. doi: 10.1073/pnas.77.10.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luse D S, Jacob G A. J Biol Chem. 1987;262:14990–14997. [PubMed] [Google Scholar]
- 3.Uptain S M, Kane C M, Chamberlin M J. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 4.Kugel J F, Goodrich J A. Mol Cell Biol. 2002;22:762–773. doi: 10.1128/MCB.22.3.762-773.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holstege F C P, Fiedler U, Timmers H T M. EMBO J. 1997;16:7468–7480. doi: 10.1093/emboj/16.24.7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dvir A, Conaway J W, Conaway R C. Curr Opin Genet Dev. 2001;11:209–214. doi: 10.1016/s0959-437x(00)00181-7. [DOI] [PubMed] [Google Scholar]
- 7.Dvir A, Tan S Y, Conaway J W, Conaway R C. J Biol Chem. 1997;272:28175–28178. doi: 10.1074/jbc.272.45.28175. [DOI] [PubMed] [Google Scholar]
- 8.Lis J. Cold Spring Harbor Symp Quant Biol. 1998;63:347–356. doi: 10.1101/sqb.1998.63.347. [DOI] [PubMed] [Google Scholar]
- 9.Gnatt A L, Cramer P, Fu J, Bushnell D A, Kornberg R D. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 10.Kireeva M L, Komissarova N, Waugh D S, Kashlev M. J Biol Chem. 2000;275:6530–6536. doi: 10.1074/jbc.275.9.6530. [DOI] [PubMed] [Google Scholar]
- 11.Coppola J A, Luse D S. J Mol Biol. 1984;178:415–437. doi: 10.1016/0022-2836(84)90151-7. [DOI] [PubMed] [Google Scholar]
- 12.Nudler E, Goldfarb A, Kashlev M. Science. 1994;265:793–796. doi: 10.1126/science.8047884. [DOI] [PubMed] [Google Scholar]
- 13.Komissarova N, Kashlev M. Proc Natl Acad Sci USA. 1998;95:14699–14704. doi: 10.1073/pnas.95.25.14699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson K S, Conant C R, von Hippel P H. J Mol Biol. 1999;289:1179–1194. doi: 10.1006/jmbi.1999.2814. [DOI] [PubMed] [Google Scholar]
- 15.Darst S A. Curr Opin Struct Biol. 2001;11:155–162. doi: 10.1016/s0959-440x(00)00185-8. [DOI] [PubMed] [Google Scholar]
- 16.Samkurashvili I, Luse D S. Mol Cell Biol. 1998;18:5343–5354. doi: 10.1128/mcb.18.9.5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pal M, McKean D, Luse D S. Mol Cell Biol. 2001;21:5815–5825. doi: 10.1128/MCB.21.17.5815-5825.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Újvári A, Pal M, Luse D S. J Biol Chem. 2002;277:32527–32537. doi: 10.1074/jbc.M201145200. [DOI] [PubMed] [Google Scholar]
- 19.Cramer P, Bushnell D A, Kornberg R D. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 20.Landick R. Cell. 2001;105:567–570. doi: 10.1016/s0092-8674(01)00381-6. [DOI] [PubMed] [Google Scholar]
- 21.Pal M, Luse D S. Mol Cell Biol. 2002;22:30–40. doi: 10.1128/MCB.22.1.30-40.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 23.Korzheva N, Mustaev A, Kozlov M, Malhotra A, Nikiforov V, Goldfarb A, Darst S A. Science. 2000;289:619–625. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- 24.Severinov K, Goldfarb A. J Biol Chem. 1994;269:31701–31705. [PubMed] [Google Scholar]
- 25.Sijbrandi R, Fiedler U, Timmers H T. Nucleic Acids Res. 2002;30:2290–2298. doi: 10.1093/nar/30.11.2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas M J, Platas A A, Hawley D K. Cell. 1998;93:627–637. doi: 10.1016/s0092-8674(00)81191-5. [DOI] [PubMed] [Google Scholar]
- 27.Cramer P, Bushnell D A, Fu J H, Gnatt A L, Maier-Davis B, Thompson N E, Burgess R R, Edwards A M, David P R, Kornberg R D. Science. 2000;288:640–649. doi: 10.1126/science.288.5466.640. [DOI] [PubMed] [Google Scholar]
- 28.Artsimovitch I, Landick R. Genes Dev. 1998;12:3110–3122. doi: 10.1101/gad.12.19.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Artsimovitch I, Landick R. Proc Natl Acad Sci USA. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gu W G, Reines D. J Biol Chem. 1995;270:30441–30447. doi: 10.1074/jbc.270.51.30441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yarnell W S, Roberts J W. Science. 1999;284:611–615. doi: 10.1126/science.284.5414.611. [DOI] [PubMed] [Google Scholar]
- 32.Rice G A, Chamberlin M J, Kane C M. Nucleic Acids Res. 1993;21:113–118. doi: 10.1093/nar/21.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klug A. Science. 2001;292:1844–1846. doi: 10.1126/science.1062384. [DOI] [PubMed] [Google Scholar]
- 34.Ebright R H. J Mol Biol. 2000;304:687–698. doi: 10.1006/jmbi.2000.4309. [DOI] [PubMed] [Google Scholar]