Abstract
We report here that a bacterial toxin, anthrax lethal toxin (LeTx), at very low concentrations represses glucocorticoid receptor (GR) transactivation in a transient transfection system and the activity of an endogenous GR-regulated gene in both a cellular system and an animal model. This repression is noncompetitive and does not affect ligand binding or DNA binding, suggesting that anthrax lethal toxin (LeTx) probably exerts its effects through a cofactor(s) involved in the interaction between GR and the basal transcription machinery. LeTx-nuclear receptor repression is selective, repressing GR, progesterone receptor B (PR-B), and estrogen receptor α (ERα), but not the mineralocorticoid receptor (MR) or ERβ. GR repression was also caused by selected p38 mitogen-activated protein (MAP) kinase inhibitors, suggesting that the LeTx action may result in part from its known inactivation of MAP kinases. Simultaneous loss of GR and other nuclear receptor activities could render an animal more susceptible to lethal or toxic effects of anthrax infection by removing the normally protective antiinflammatory effects of these hormones, similar to the increased mortality seen in animals exposed to both GR antagonists and infectious agents or bacterial products. These finding have implications for development of new treatments and prevention of the toxic effects of anthrax.
Death from anthrax toxin is reported to result from systemic shock (1) resembling lipopolysaccharide (LPS)-induced shock (2, 3) although the role of inflammatory cytokines in this process and the precise mechanism of this shock have not been determined (4). Anthrax toxin is composed of three proteins: protective antigen (PA), edema factor (EF), and lethal factor (LF) (for a recent review, see refs. 5 and 6). PA and EF comprise the edema toxin and PA and LF the lethal toxin (LeTx). It is this lethal toxin produced by Bacillus anthracis that causes death of the infected host (7). The mechanism of entry of this toxin into the cell is now well understood. PA binds to the anthrax toxin receptor (8), is cleaved (9), oligomerizes, and then binds LF and/or EF, facilitating internalization of these proteins into the cell (10, 11). Translocation of LF and EF to the cytosol is by a pH- and voltage-dependent mechanism (12–14).
The mechanism of action of LF inside the cell is less well understood. LF is a metalloprotease that cleaves the mitogen activation protein (MAP) kinase kinases (MAPKK/MEK), including MEK1, MEK2, MKK3, MKK4, MKK6, and MKK7 but not MEK5 (15–19). However, the fact that LeTx-resistant and -sensitive cells show similar internalization of LF (20) and similar MEK cleavage in response to LF (17, 18) suggests that these factors cannot alone account for differential susceptibility or resistance to the toxin. Other factors that have been proposed to play a role in toxicity of LeTx include the proteosome (21), intracellular calcium stores (22, 23), calmodulin (23), a calyculin A-sensitive protein phosphatase (24), protein synthesis (25), and reactive oxygen intermediates (26). It is not known which of these or other unknown factors contribute to the well-described differential cell line and rodent strain sensitivities to toxic effects of LeTx. Recently, the gene Kif1C has been determined to be different between resistant and sensitive strains although the implication of this finding is not understood (27).
Fischer (F344/N) rats have long been known to be particularly susceptible to the LeTx (28), with death occurring within 40 min after exposure to a lethal dose (29). F344/N rats are also known to be relatively inflammatory disease resistant, due in part to their hypothalamic-pituitary-adrenal (HPA) axis hyperresponsiveness and resultant hypersecretion of glucocorticoids from the adrenal glands in response to proinflammatory and other stimuli. Similar to F344/N rats, BALB/c mice have a hyperresponsive HPA axis (30) and are also susceptible to LeTx (31). Ordinarily this hyper-HPA axis responsiveness protects against inflammatory/autoimmune disease, including shock through the antiinflammatory and immunosuppressive effects of the glucocorticoids. However, F344/N rats and other inflammatory-resistant rodent strains become highly susceptible to inflammation and rapid death after simultaneous glucocorticoid receptor (GR) or HPA axis blockade and exposure to proinflammatory or infectious stimuli, including bacterial products such as streptococcal cell walls (SCW) or bacterial lipopolysaccharide (LPS) (32–37).
Here, we report that the LF and PA proteins comprising B. anthracis LeTx selectively and specifically repress GR and other nuclear hormone receptors. To our knowledge there have been no previous reports showing that a bacterial product interferes with nuclear hormone receptor function. This provides a previously uncharacterized explanation for how such agents might contribute to the pathogenesis of bacterial infections.
Materials and Methods
Materials.
The recombinant proteins LF and PA were produced as described (38, 39). All MEK inhibitors were purchased from Calbiochem except PD98059, which was purchased from Cell Signaling Technology (Beverly, MA).
Cell Culture.
Cos7 and HTC cells were grown at 37°C and 5% CO2 in DMEM containing 10% serum, 10 μg/ml penicillin-streptomycin, and 2 mM glutamine.
Transient Transfections.
Cos7 cells were plated in 24-well plates at a density of 5 × 105 cells per well in DMEM containing 10% charcoal-stripped serum, 10 μg/ml penicillin-streptomycin, and 2 mM glutamine one day before transfection. Cos7 cells were transfected overnight with 20 ng of receptor expression plasmid [SVGR, estrogen receptor (ER) α, ERβ, mineralocorticoid receptor (MR), or progesterone receptor B (PR-B)], 100 ng of reporter construct (GRE-TK luc, ERE-luc, phr-luc, or pGL3 control), 60 ng of pSG5 (Stratagene), and 20 ng of phRL TK (Promega, constitutive Renilla luciferase control) by using FuGENE6 (Roche Applied Science) according to the manufacturer's instructions. The medium was then replaced with DMEM containing 10% charcoal-stripped serum, the appropriate hormone, and LF and/or PA or inhibitor as required. After 24 h, the cells were lysed, and the firefly and Renilla luciferases were assayed by using the dual luciferase assay (Promega).
Assay of Tyrosine Aminotransferase (TAT) in HTC Cells.
HTC cells were plated in 6-cm plates at a density of 5 × 106 cells per plate in DMEM containing 10% FCS, 10 μg/ml penicillin-streptomycin, and 2 mM glutamine 1 day before treatment. The media was then replaced with DMEM containing increasing concentrations of dexamethasone (Dex) either alone or together with LF and PA. After 18 h, the cells were lysed by sonication, and TAT activity was assayed as described by Thompson et al. (40).
Animal Experiments.
Male and female BALB/cJ mice (10–12 wk old, The Jackson Laboratory) were injected i.p. with 50 μg of LF, 50 μg of PA, or the combination in 1 ml of sterile-filtered PBS 30 min before Dex treatment. Dex was injected i.p. in 0.25-ml volume (0.06 mg per mouse). Mice were killed by CO2 at various times postinjection, and livers were removed, homogenized in ice-cold lysis buffer (0.2 mM pyridoxal phosphate, 0.5 mM α-keto glutarate, 0.1 M potassium phosphate, pH 7.6) and then centrifuged at 100,000 × g at 4 C for 30 min. TAT activity of supernatants was assayed as described by Thompson et al. (40).
Western Blot Analysis.
Cos7 cells were plated in 6-cm plates at a density of 5 × 106 cells per plate 2 days before treatments. Cells were treated with MAP kinase inhibitors for 30 min. Cells were stimulated by addition of 10 μg/ml LPS or anisomycin for 30 min. Proteins were solubilized by using M-PER (Pierce), and 10 μg was separated by SDS/PAGE according to the method of Laemmli (41). Proteins were transferred to poly(vinylidene difluoride) and probed with antibodies against phospho-p38 MAP kinase (Thr-180/Tyr-182), phospho-p44/42 MAP kinase (Thr-202/Tyr-204), and phospho-c-Jun (Ser-63) (Cell Signaling Technology). Chemiluminescence was detected and analyzed by using the Chemidoc gel imaging system and volume analysis tool of the quantity one software (Bio-Rad).
Results
Cos7 cells, transiently transfected with the glucocorticoid receptor expression plasmid (SVGR) and a glucocorticoid response element (GRE)-luciferase reporter construct (GRE TK luc), were treated with 100 nM Dex and increasing concentrations of PA or LF in the presence or absence of saturating concentrations of the other LeTx component (Fig. 1). LF in the presence of 500 ng/ml PA (□), but not alone (▪), repressed GR (Fig. 1) at concentrations as low as 1 ng/ml. Also, PA in the presence of 50 ng/ml LF, but not alone, repressed GR activity at concentrations as low as 5 ng/ml (data not shown). Maximal repression of GR by LeTx was 50%. Further studies showed that, even in the presence of LeTx, the system can be additionally and fully repressed by coadministration of the GR/PR antagonist RU486 with LeTx (data not shown), indicating that repression by LeTx and RU486 are not competitive.
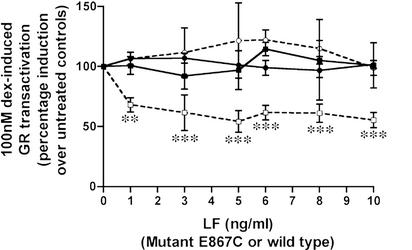
Figure 1.
LeTx repression of Dex-induced GR transactivation in Cos7 cells. GR-transfected Cos7 cells were treated with 100 nM Dex either with increasing concentrations of LF, alone (■) or in the presence of 500 ng/ml PA (□); or with increasing concentrations of E687C, either alone (●) or in the presence of 500 ng/ml PA (○). Means and standard deviations of eight (WT LF) or three (E687C LF) experiments are shown, and data are analyzed by a one-way ANOVA followed by a Dunnett post hoc test (*, P = 0.01–0.05; **, P = 0.001–0.01; ***, P ≤ 0.001).
A catalytically inactive, single amino acid substitution mutant of LF, E687C, has been shown to be nontoxic in the LeTx-sensitive macrophage cell line, RAW264.7 (15). In these transient transfection assays, in contrast to the repression induced by wild-type LF in the presence of PA (□), the mutant LF (E687C) in the presence of 500 ng/ml PA (○) did not repress GR (Fig. 1). This result indicates that the proteolytic activity of LF may be required for GR repression.
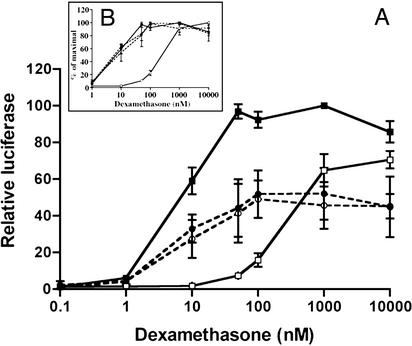
Full dose-response curves of the normalized luciferase activity to Dex are shown in Fig. 2. Fig. 2A shows the effect of 500 ng/ml PA and 10 ng/ml LF (●) or 5 ng/ml PA and 50 ng/ml LF (○) compared with Dex alone (▪) and with Dex plus the typical GR antagonist RU486 (□). It can be seen that both combinations of LF and PA caused an ≈50% repression of GR at all effective concentrations of Dex. This pattern is indicative of a noncompetitive repressor, in contrast to a competitive antagonist, such as RU486, the effect of which can be fully overcome by higher concentrations of Dex. Fig. 2 B Inset, with data presented as a percentage of the maximal activity in each case, shows that both combinations of LF and PA have no effect on the EC50, whereas the typical competitive antagonist RU486 causes a right shift in the curve and an increase in the EC50 value. Competitive ligand-binding studies showed that, in both whole cells and in cytosolic preparations, neither LF nor PA, nor a combination of both, were able to compete with a saturating concentration of [3H]Dex for binding to GR (data not shown). Gel shift analysis of GR-transfected Cos7 cell cytosol to a radiolabeled oligonucleotide showed that LF, PA, or a combination of both also had no effect on GR–DNA complex mobility, indicating that LeTx does not interfere with GR–DNA binding (data not shown), suggesting that it acts at a point downstream of GR-DNA binding either by interfering with a cofactor or acting itself as a corepressor.
Figure 2.
Comparison of the effects of RU486 and LeTx on the dose–response curve of Dex in GR-transfected Cos7 cells. GR-transfected Cos7 cells were treated with increasing concentrations of Dex, either alone (■) or in the presence of 0.2 μM RU486 (□), 500 ng/ml PA + 10 ngml LF (●) or 50 ng/ml LF + 5 ng/ml PA (□). (A) Means and standard deviations of three experiments are shown. (B) Data normalized as percentage of maximal for each treatment.
Steroid hormone receptor coactivator 1 (SRC1), transcriptional intermediary factor 2 (TIF2), and cAMP response element binding protein (CREB)-binding protein (CBP) are cofactors that are known to interact directly with nuclear hormone receptors such as GR. Cotransfection of SRC1, TIF2, or CBP expression vectors had no effect on LeTx repression of GR. An effect of TIF2 alone was observed, in which this cofactor significantly enhanced the GR transactivation. However, LeTx repressed GR transactivation 40–50% in the presence or absence of TIF2 (data not shown). This finding suggests that LeTx does not function directly through or prevent the action of these cofactors.
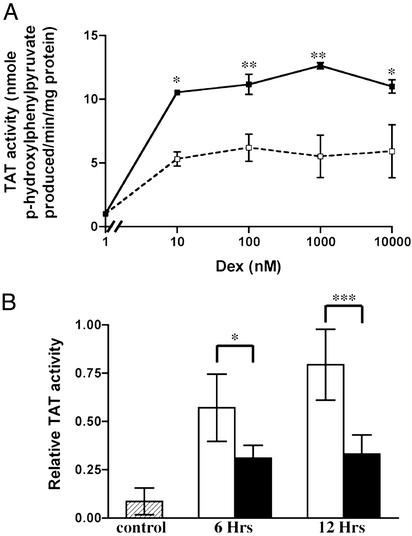
To determine whether the LeTx repression of GR gene activation observed in the transient transactivation system also occurs in a more natural system, the effects of LeTx on Dex induction of the GR-regulated enzyme TAT was investigated in a rat hepatoma cell line (HTC cells) and in mouse liver (Fig. 3). HTC cells were treated for 18 h with increasing concentrations of Dex either alone (▪) or together with 2 ng/ml LF in the presence of 500 ng/ml PA (□), and TAT enzyme activity was assayed (40). TAT activity was induced ≈10-fold by Dex concentrations as low as 10 nM. Cotreatment with LF and PA reduced Dex induction of TAT activity by 50%, in agreement with the transient transfection assays (Fig. 3A). BALB/cJ mice were injected with LeTx (50 μg each of PA and LF) 30 min before Dex treatment, and liver TAT activity was assayed at 6 and 12 h. Dex alone induced liver TAT activity (open bars). Pretreatment with LeTx significantly decreased Dex-induced TAT activity (filled bars) by 50% (Fig. 3B). Injection of LF or PA alone or PBS had no effect on either basal or Dex-induced TAT activity (data not shown).
Figure 3.
LeTx repression of Dex-induced TAT in systems endogenously expressing GR. (A) HTC cells were treated with Dex either alone (■) or together with 2 ng/ml LF with 500 ng/ml PA (□) for 18 h, and TAT activity was assayed. Mean and standard deviations are shown, and a one-way ANOVA followed by a Bonferroni post hoc test was performed. (B) BALB/cJ mice were injected with LeTx, and 30 min later with Dex. After 6 and 12 h, liver TAT activity was assayed. Means and standard deviations of 6–10 animals are shown, and a two-way ANOVA followed by a Scheffé post hoc test was performed.
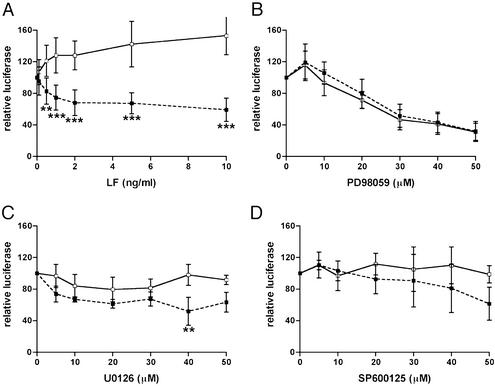
Known substrates for the proteolytic action of LF include some members of the MAP kinase family (MAPKKs). Cleavage of these proteins results in a blockade of the MAP kinase pathways. A panel of tumor cell lines exhibited similar patterns of sensitivity (i.e., growth inhibition) to LeTx and to the MEK1 inhibitor PD98059 (42, 43), a result that led to the discovery that LeTx inactivates MEK1. When the effect of the MEK1 inhibitors PD98059 and U0126 on GR transactivation was compared with the effects of LeTx, these inhibitors decreased luciferase activity of both the GRE-luciferase (▪) and the constitutive luciferase vector (pGL3 control) (□) to the same extent, with no significant difference between these two curves (Fig. 4 B and C), whereas LF in the presence of PA had no effect on the pGL3 control (Fig. 4A). Western blot analysis showed that 20 μM PD98059 and 10 μM U0126 completely inhibited MEK1 and prevented LPS induction of phospho-p44/42 MAP kinase (data not shown). Thus, at inhibitor concentrations at which MEK1 is inhibited there is no significant effect on the GR-induced gene induction compared with a constitutive control. This finding shows that PD98059 and U0126 have a nonspecific suppressive effect on luciferase, occurring through unknown mechanisms, in this transient transfection system. Furthermore, these data show that these inhibitors did not induce any GRE-specific changes in luciferase and therefore do not affect GR transactivation. Similarly, an inhibitor of the Jun-N-terminal kinase (JNK) pathway, SP600125, had no significant effect on either GRE-luciferase (▪) or pGL3 control luciferase (□) (Fig. 4D). Western blot analysis showed that 20 μM SP600125 inhibited anisomycin-induced phospho-c-Jun (data not shown), indicating that this inhibitor was able to repress the JNK pathway. These results indicate that the LF repression of GR probably does not occur through inhibition of the MEK1 or JNK pathways.
Figure 4.
Comparison of the effects of LeTx and inhibitors of MEK1 and JNK pathways on the response of a Dex-induced GRE luciferase and a constitutive luciferase. Cos7 cells were transfected with GR and GRE-TK luc (■) or with GR and the constitutive luciferase vector pGL3 control (Promega) (□) and treated with 100 nM Dex, and increasing concentrations of LF with 500 ng/ml PA (A), or increasing concentrations of the MEK1 inhibitors PD98059 (B) and U0126 (C) or the JNK inhibitor SP600126 (D). Means and standard deviations are shown, and data analyzed by using a two-way ANOVA followed by a Scheffé post hoc test.
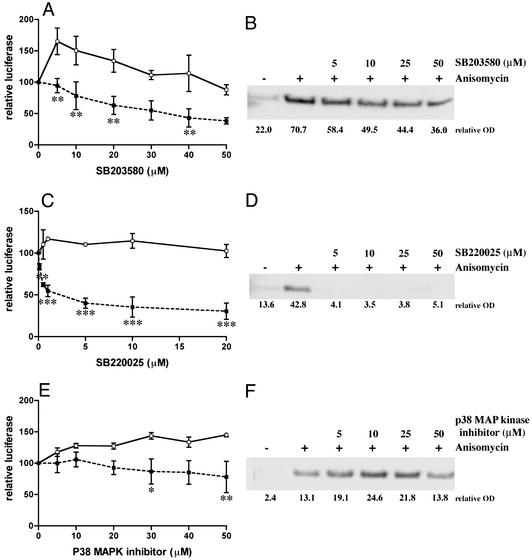
LeTx has also been shown to cleave MKK3, thereby inhibiting the p38 pathway (17, 44). In transient transfection experiments, the p38 MAP kinase inhibitors SB203580 and SB220025 significantly repressed the GRE-luciferase (▪) compared with the pGL3 control luciferase (□) (Fig. 5 A and C). Western blot analysis showed that anisomycin-induced phosphorylation of p38 was also inhibited (Fig. 5 B and D). However, a third inhibitor, “p38 MAP kinase inhibitor,” slightly repressed GRE luciferase (Fig. 5E) and did not repress anisomycin-induced phospho-p38 (Fig. 5F), indicating a correlation between extent of GR repression and efficiency of p38 inhibition.
Figure 5.
Effect of p38 MAP kinase inhibitors on the response of a Dex-induced GRE luciferase and a constitutive luciferase and on inhibition of p38. Cos7 cells were transfected with GR and GRE-TK luc (■) or with GR and the constitutive luciferase vector pGL3 control (□) and treated with 100 nM Dex, and increasing concentrations of the p38 MAP kinase inhibitors SB203580 (A), SB220025 (C), and p38 MAP kinase inhibitor (E). Means and standard deviations are shown, and data analyzed by using a two-way ANOVA followed by a Scheffé post hoc test. Cos7 cells were pretreated for 30 min with various concentrations of SB203580 (B), SB220025 (D), or p38 MAP kinase inhibitor (F) and then further incubated with 10 μg/ml anisomycin for 30 min. Proteins were then subjected to SDS/PAGE and Western blotting using an anti-phospho-p38 antibody.
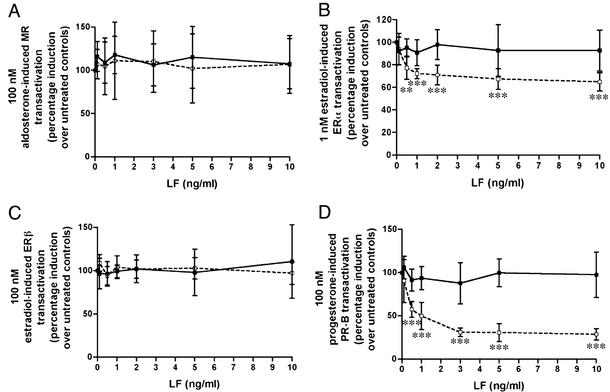
To determine whether the GR repression by LeTx is specific for GR or affects other nuclear hormone receptors, transient transfection experiments were performed using ERα, ERβ, MR, and PR-B, and their respective reporter plasmids. In contrast to its 50% repression of GR, LeTx had no effect on MR (Fig. 6A). LeTx repressed ERα by ≈40% (Fig. 6B) but had no effect of ERβ (Fig. 6C). Finally, LeTx repressed PR-B by 70% (Fig. 6D). Similar to their lack of effect on GR transactivation, cotransfection of SRC1, TIF2, or CBP had no effect on LeTx repression of PR-B. TIF2 similarly enhanced the progesterone-induced PR-B transactivation in the absence of LeTx, and in the presence of LeTx the 70–80% repression was maintained, even with addition of 100 ng of TIF2 (data not shown). This result suggests that these cofactors are not directly involved in LeTx repression of PR-B.
Figure 6.
Effect of the LeTx on hormone-induced activity of other nuclear hormone receptors in Cos7 cells. Cos7 cells were transfected with the expression plasmids for MR (A), ERα (B), ERβ (C), or PR-B (D) and their respective reporters and treated with 100 nM aldosterone (A), 1 nM 17β-estradiol (B), 100 nM 17β-estradiol (C), or 100 nM progesterone (D), together with increasing concentrations of LF either alone (■) or in the presence of 500 ng/ml PA (□). Means and standard deviations are shown, and data analyzed by using a one-way ANOVA followed by a Dunnett post hoc test.
Discussion
Taken together, these experiments show that, at extremely low concentrations, LeTx selectively represses transactivation of transiently transfected nuclear hormone receptors (GR, PR-B, and ERα, but not MR or ERβ; Figs. 1 and 6) and also suppresses the enzyme activity of an endogenous GR-regulated gene in both a cellular system and an animal model (Fig. 3). This repression is noncompetitive and does not affect ligand or DNA binding, suggesting that LeTx may exerts its effects through a cofactor(s) involved in the interaction between GR and the basal transcription machinery. The specificity of repression of some but not all members of the nuclear hormone receptor family tested also supports the notion that LeTx is working through a cofactor rather than through a direct interaction with the GR receptor. Cotransfection of the coactivators SRC1, TIF2, or CBP did not overcome LeTx repression of GR or PR-B. Because there are many proteins involved in the interaction of nuclear hormone receptors with the basal transcription machinery, the fact that these three did not overcome the LeTx repression does not, however, rule out the possibility that any one or more of the other cofactors are the target of LeTx.
The MEK1 and JNK pathways appear not involved in LeTx repression of GR and PR-B as the MEK1 inhibitors PD98059 and U0126 and the JNK inhibitor SP600125 did not induce a GRE-specific effect in the transient transfection assay (Fig. 4). However, the p38 MAP kinase inhibitors SB203580 and SB220025 specifically inhibited GR-induced gene activation with a correlated inhibition of p38 phosphorylation (Fig. 5). These findings suggest that LeTx cleavage/inhibition of p38 may be involved in GR repression. Previous findings have shown that stimuli-induced activation, not basal repression/inhibition, of the MEK pathways can repress GR (45–52). However, one study has shown that SB203580 prevents Dex-induced Hsp27 production in osteoclasts (53), suggesting that the relationship between LeTx repression of GR and p38 requires further investigation.
If the LeTx repression of nuclear hormone receptors reported here also occurs in vivo in the course of anthrax infection, this mechanism could potentially contribute to some of the toxic effects of anthrax. Because the glucocorticoid receptor is essential for survival and also for modulation of immune responses to infectious agents, loss of glucocorticoid receptor activity during infection could render the host more susceptible to the lethal or toxic effects of anthrax bacteria. Simultaneous loss of activity of other nuclear receptors, particularly PR, could potentially amplify these effects. Indeed, this scenario is consistent with the well-described increased mortality in rodents that have been adrenalectomized or treated with the GR/PR receptor antagonist RU486, and simultaneously exposed to infectious agents or proinflammatory bacterial products (32, 34–37). Identification of nuclear receptor cofactor interactions as a potential mechanism of toxicity of anthrax lethal factor could thus have important implications for development of new avenues for treatment and prevention of the toxic effects of anthrax and possibly for toxicities of related bacterial products.
Acknowledgments
We thank Jan-Åke Gustafsson, Jan Carlstedt-Duke, and Samuel Listwak for helpful discussions. Also, we thank Gordon Hager, Jan Carlstedt-Duke, Jan-Åke Gustafsson, Samuel Listwak, Henrich Gronemeyer, Bert O'Malley, and Richard Goodman for various plasmids.
Abbreviations
- Dex
dexamethasone
- ER
estrogen receptor
- GR
glucocorticoid receptor
- GRE
glucocorticoid response element
- LeTx
lethal toxin
- LF
lethal factor
- MAP kinase
mitogen-activated protein kinase
- MR
mineralocorticoid receptor
- PA
protective antigen
- PR-B
progesterone receptor B
- TAT
tyrosine aminotransferase
- LPS
lipopolysaccharide
- MEK
MAP kinase kinase
- SRC1
steroid hormone receptor coactivator-1
- TIF2
transcriptional intermediary factor-2
- CBP
cAMP response element binding protein-binding protein
- JNK
Jun-N-terminal kinase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Smith H, Keppie J, Stanley J L, Harris-Smith P W. Br J Exp Pathol. 1955;36:323–335. [PMC free article] [PubMed] [Google Scholar]
- 2.Hanna P. J Appl Microbiol. 1999;87:285–287. doi: 10.1046/j.1365-2672.1999.00891.x. [DOI] [PubMed] [Google Scholar]
- 3.Hanna P C, Ireland J A. Trends Microbiol. 1999;7:180–182. doi: 10.1016/s0966-842x(99)01507-3. [DOI] [PubMed] [Google Scholar]
- 4.Erwin J L, DaSilva L M, Bavari S, Little S F, Friedlander A M, Chanh T C. Infect Immun. 2001;69:1175–1177. doi: 10.1128/IAI.69.2.1175-1177.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhatnagar R, Batra S. Crit Rev Microbiol. 2001;27:167–200. doi: 10.1080/20014091096738. [DOI] [PubMed] [Google Scholar]
- 6.Leppla S H. In: Bacterial Protein Toxins. Aktories K, Just I, editors. Berlin: Springer; 2000. pp. 445–472. [Google Scholar]
- 7.Pezard C, Berche P, Mock M. Infect Immun. 1991;59:3472–3477. doi: 10.1128/iai.59.10.3472-3477.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradley K A, Mogridge J, Mourez M, Collier R J, Young J A. Nature. 2001;414:225–229. doi: 10.1038/n35101999. [DOI] [PubMed] [Google Scholar]
- 9.Klimpel K R, Molloy S S, Thomas G, Leppla S H. Proc Natl Acad Sci USA. 1992;89:10277–10281. doi: 10.1073/pnas.89.21.10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh Y, Klimpel K R, Goel S, Swain P K, Leppla S H. Infect Immun. 1999;67:1853–1859. doi: 10.1128/iai.67.4.1853-1859.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedlander A M. J Biol Chem. 1986;261:7123–7126. [PubMed] [Google Scholar]
- 12.Zhao J, Milne J C, Collier R J. J Biol Chem. 1995;270:18626–18630. doi: 10.1074/jbc.270.31.18626. [DOI] [PubMed] [Google Scholar]
- 13.Wesche J, Elliott J L, Falnes P O, Olsnes S, Collier R J. Biochemistry. 1998;37:15737–15746. doi: 10.1021/bi981436i. [DOI] [PubMed] [Google Scholar]
- 14.Blaustein R O, Koehler T M, Collier R J, Finkelstein A. Proc Natl Acad Sci USA. 1989;86:2209–2213. doi: 10.1073/pnas.86.7.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klimpel K R, Arora N, Leppla S H. Mol Microbiol. 1994;13:1093–1100. doi: 10.1111/j.1365-2958.1994.tb00500.x. [DOI] [PubMed] [Google Scholar]
- 16.Duesbery N S, Webb C P, Leppla S H, Gordon V M, Klimpel K R, Copeland T D, Ahn N G, Oskarsson M K, Fukasawa K, Paull K D, Vande Woude G F. Science. 1998;280:734–737. doi: 10.1126/science.280.5364.734. [DOI] [PubMed] [Google Scholar]
- 17.Pellizzari R, Guidi-Rontani C, Vitale G, Mock M, Montecucco C. FEBS Lett. 1999;462:199–204. doi: 10.1016/s0014-5793(99)01502-1. [DOI] [PubMed] [Google Scholar]
- 18.Pellizzari R, Guidi-Rontani C, Vitale G, Mock M, Montecucco C. Int J Med Microbiol. 2000;290:421–427. doi: 10.1016/S1438-4221(00)80056-9. [DOI] [PubMed] [Google Scholar]
- 19.Vitale G, Bernardi L, Napolitani G, Mock M, Montecucco C. Biochem J. 2000;352:739–745. [PMC free article] [PubMed] [Google Scholar]
- 20.Singh Y, Leppla S H, Bhatnagar R, Friedlander A M. J Biol Chem. 1989;264:11099–11102. [PubMed] [Google Scholar]
- 21.Tang G, Leppla S H. Infect Immun. 1999;67:3055–3060. doi: 10.1128/iai.67.6.3055-3060.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin S, Hur G H, Kim Y B, Park K J, Park Y M, Lee W S. Cell Biol Toxicol. 2000;16:137–144. doi: 10.1023/a:1007646227674. [DOI] [PubMed] [Google Scholar]
- 23.Bhatnagar R, Singh Y, Leppla S H, Friedlander A M. Infect Immun. 1989;57:2107–2114. doi: 10.1128/iai.57.7.2107-2114.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kau J H, Lin C G, Huang H H, Hsu H L, Chen K C, Wu Y P, Lin H C. Curr Microbiol. 2002;44:106–111. doi: 10.1007/s00284-001-0059-8. [DOI] [PubMed] [Google Scholar]
- 25.Bhatnagar R, Friedlander A M. Infect Immun. 1994;62:2958–2962. doi: 10.1128/iai.62.7.2958-2962.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanna P C, Kruskal B A, Ezekowitz R A, Bloom B R, Collier R J. Mol Med. 1994;1:7–18. [PMC free article] [PubMed] [Google Scholar]
- 27.Watters J W, Dewar K, Lehoczky J, Boyartchuk V, Dietrich W F. Curr Biol. 2001;11:1503–1511. doi: 10.1016/s0960-9822(01)00476-6. [DOI] [PubMed] [Google Scholar]
- 28.Klein F, Haines B W, Mahlandt B G, DeArmon I A, Jr, Lincoln R E. J Bacteriol. 1963;85:1032–1038. doi: 10.1128/jb.85.5.1032-1038.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ezzell J W, Ivins B E, Leppla S H. Infect Immun. 1984;45:761–767. doi: 10.1128/iai.45.3.761-767.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shanks N, Griffiths J, Zalcman S, Zacharko R M, Anisman H. Pharmacol Biochem Behav. 1990;36:515–519. doi: 10.1016/0091-3057(90)90249-h. [DOI] [PubMed] [Google Scholar]
- 31.Welkos S L, Keener T J, Gibbs P H. Infect Immun. 1986;51:795–800. doi: 10.1128/iai.51.3.795-800.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edwards C K I, Yunger L M, Lorence R M, Dantzer R, Kelley K W. Proc Natl Acad Sci USA. 1991;88:2274–2277. doi: 10.1073/pnas.88.6.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuckerman S H, Qureshi N. Infect Immun. 1992;60:2581–2587. doi: 10.1128/iai.60.7.2581-2587.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sternberg E M, Hill J M, Chrousos G P, Kamilaris T, Listwak S J, Gold P W, Wilder R L. Proc Natl Acad Sci USA. 1989;86:2374–2378. doi: 10.1073/pnas.86.7.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruzek M C, Pearce B D, Miller A H, Biron C A. J Immunol. 1999;162:3527–3533. [PubMed] [Google Scholar]
- 36.MacPhee I A M, Antoni F A, Mason D W. J Exp Med. 1989;169:431–445. doi: 10.1084/jem.169.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gomez S A, Fernandez G C, Vanzulli S, Dran G, Rubel C, Berki T, Isturiz M A, Palermo M S. Clin Exp Immunol. 2003;131:217–224. doi: 10.1046/j.1365-2249.2003.02057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park S, Leppla S H. Protein Expression Purif. 2000;18:293–302. doi: 10.1006/prep.2000.1208. [DOI] [PubMed] [Google Scholar]
- 39.Ramirez D M, Leppla S H, Schneerson R, Shiloach J. J Ind Microbiol Biotechnol. 2002;28:232–238. doi: 10.1038/sj/jim/7000239. [DOI] [PubMed] [Google Scholar]
- 40.Thompson E B, Tomkins G M, Curran J F. Proc Natl Acad Sci USA. 1966;56:296–303. doi: 10.1073/pnas.56.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laemmli U K. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 42.Koo H M, VanBrocklin M, McWilliams M J, Leppla S H, Duesbery N S, Vande Woude G F. Proc Natl Acad Sci USA. 2002;99:3052–3057. doi: 10.1073/pnas.052707699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duesbery N S, Vande Woude G F. J Appl Microbiol. 1999;87:289–293. doi: 10.1046/j.1365-2672.1999.00892.x. [DOI] [PubMed] [Google Scholar]
- 44.Park J M, Greten F R, Li Z W, Karin M. Science. 2002;297:2048–2051. doi: 10.1126/science.1073163. [DOI] [PubMed] [Google Scholar]
- 45.Krstic M D, Rogatsky I, Yamamoto K R, Garabedian M J. Mol Biol Cell. 1997;17:3947–3954. doi: 10.1128/mcb.17.7.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lopez G N, Turck C W, Schaufele F, Stallcup M R, Kushner P J. J Biol Chem. 2001;276:22177–22182. doi: 10.1074/jbc.M010718200. [DOI] [PubMed] [Google Scholar]
- 47.Rogatsky I, Logan S K, Garabedian M J. Proc Natl Acad Sci USA. 1998;95:2050–2055. doi: 10.1073/pnas.95.5.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lucibello F C, Slater E P, Jooss K U, Beato M, Muller R. EMBO J. 1990;9:2827–2834. doi: 10.1002/j.1460-2075.1990.tb07471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schule R, Rangarajan P, Kliewer S, Ransone L J, Bolado J, Yang N, Verma I M, Evans R M. Cell. 1990;62:1217–1226. doi: 10.1016/0092-8674(90)90397-w. [DOI] [PubMed] [Google Scholar]
- 50.Herrlich P. Oncogene. 2001;20:2465–2475. doi: 10.1038/sj.onc.1204388. [DOI] [PubMed] [Google Scholar]
- 51.Karin M, Chang L. J Endocrinol. 2001;169:447–451. doi: 10.1677/joe.0.1690447. [DOI] [PubMed] [Google Scholar]
- 52.Irusen E, Matthews J G, Takahashi A, Barnes P J, Chung K F, Adcock I M. J Allergy Clin Immunol. 2002;109:649–657. doi: 10.1067/mai.2002.122465. [DOI] [PubMed] [Google Scholar]
- 53.Kozawa O, Niwa M, Hatakeyama D, Tokuda H, Oiso Y, Matsuno H, Kato K, Uematsu T. J Cell Biochem. 2002;86:357–364. doi: 10.1002/jcb.10221. [DOI] [PubMed] [Google Scholar]