Abstract
Truncated hemoglobins (Hbs) are small hemoproteins, identified in microorganisms and in some plants, forming a separate cluster within the Hb superfamily. Two distantly related truncated Hbs, trHbN and trHbO, are expressed at different developmental stages in Mycobacterium tuberculosis. Sequence analysis shows that the two proteins share 18% amino acid identities and belong to different groups within the truncated Hb cluster. Although a specific defense role against nitrosative stress has been ascribed to trHbN (expressed during the Mycobacterium stationary phase), no clear functions have been recognized for trHbO, which is expressed throughout the Mycobacterium growth phase. The 2.1-Å crystal structure of M. tuberculosis cyano-met trHbO shows that the protein assembles in a compact dodecamer. Six of the dodecamer subunits are characterized by a double conformation for their CD regions and, most notably, by a covalent bond linking the phenolic O atom of TyrB10 to the aromatic ring of TyrCD1, in the heme distal cavity. All 12 subunits display a cyanide ion bound to the heme Fe atom, stabilized by a tight hydrogen-bonded network based on the (globin very rare) TyrCD1 and TrpG8 residues. The small apolar AlaE7 residue leaves room for ligand access to the heme distal site through the conventional “E7 path,” as proposed for myoglobin. Different from trHbN, where a 20-Å protein matrix tunnel is held to sustain ligand diffusion to an otherwise inaccessible heme distal site, the topologically related region in trHbO hosts two protein matrix cavities.
Truncated hemoglobins (trHbs) are a class of small oxygen-binding hemoproteins, dispersed in eubacteria, cyanobacteria, protozoa, and plants, recently recognized as a separate cluster within the hemoglobin (Hb) superfamily. On the basis of amino acid sequence analysis, three phylogenetic groups (groups I, II, and III) have been identified within the trHb family; some organisms contain genes from more than one group, suggesting different functions for trHbs belonging to the diverse groups (1). Crystal structures of three group I trHbs (2, 3) revealed that trHbs are clearly not just another variation on the motif of vertebrate myoglobin (Mb) and Hb. Neither are they similar to nonvertebrate Hbs, including the heme-containing domain of flavohemoglobins, nor to the plant symbiotic and nonsymbiotic Hbs (4–10). Major structural differences associated with known trHbs are an unprecedented 2-on-2 α-helical sandwich fold, resulting from striking editing of the classical 3-on-3 globin α-helical sandwich, and an extended hydrophobic tunnel/cavity network linking the solvent space and the distal heme pocket (1–3). Much smaller and topologically unrelated cavities, known by their ability to incorporate Xe atoms, have been found in Mb and interpreted as temporary ligand-docking sites (11, 12). In trHbs, the positioning and size of the hydrophobic tunnel suggest important roles in controlling ligand access to the heme, in ligand storage, and/or accumulation (1–3).
An additional major difference found between vertebrate Hbs/Mbs and trHbs is the presence, in the latter, of heme distal pocket hydrogen-bonding networks based on the TyrB10 residue, which may include residues E7(Gln/Thr/His/Ser), E11(Gln/Thr), CD1(Tyr), and G8(Trp). The different H-bond networks thus achievable may not only stabilize the bound O2 but also modulate the positioning and dynamics of distal pocket residues that participate in the control of ligand access to the iron, ligand diffusion within the distal heme pocket, and ligand diffusion into and out of the distal heme pocket (1–10).
In Mycobacterium tuberculosis and Mycobacterium bovis, two genes, glbN and glbO, encode distantly related trHbs from group I (trHbN) and from group II (trHbO), respectively. The extent of identity between their amino acid sequences is only 18%. Disruption of glbN in M. bovis bacillus Calmette–Guérin causes a dramatic reduction in the NO-consuming activity of stationary phase cells, resulting in marked NO-induced inhibition of aerobic respiration relative to wild-type cells, indicative of a key protective role played by trHbN against NO (13, 42). Recently, trHbN has also been reported to protect Escherichia coli cells from NO (14). In contrast, the physiological function of trHbO remains unknown. Studies performed with M. bovis bacillus Calmette–Guérin show that trHbO is expressed throughout the growth phase (15, 16), whereas trHbN expression is strikingly enhanced during the stationary phase (17). In a recent paper Pathania et al. (16) propose the involvement of trHbO in cell respiration, showing the enhancement of ATP levels in E. coli in which trHbO is overexpressed. On the contrary, the extremely low rates for O2 combination/dissociation seem to exclude the participation of trHbO in O2 delivery (15).
Amino acid sequence alignments (see supporting information on the PNAS web site, www.pnas.org) indicate that in trHbO the heme distal B10 and E7 sites host Tyr and Ala residues, respectively. Remarkably, residue CD1, which is invariably Phe in (non)vertebrate Hbs and Mbs, as well as in trHbs from groups I and III, is Tyr in trHbO and in other trHbs from group II. As observed for all currently known group II and III trHbs, trHbO displays a Trp residue at the G8 site, where smaller apolar residues (Val, Leu, or Ile) are constantly found in group I trHbs, as well as in (non)vertebrate Hbs/Mbs, flavohemoglobins, and plant Hbs (1, 4, 10, 18–20). Resonance Raman data indicate that in trHbO, a hydrogen-bonded network stabilizes the heme-bound O2 and CO ligands, as evidenced by frequencies of the Fe-O2 and Fe-CO stretching modes (13). These results have been taken as indicative of a trHbO rigid and polar distal pocket, significantly different from that of trHbN.
Considering that no 3D structures have been reported so far for groups II and III trHbs, we report here the crystallographic analysis of M. tuberculosis cyano-met trHbO at 2.1-Å resolution (Rgen/Rfree 18.6%/22.6%), as a first step in unraveling the structural properties and ligand recognition features of group II trHbs. Interestingly, trHbO is assembled as a tightly packed dodecamer, fully hosted in the crystallographic asymmetric unit.
Materials and Methods
trHbO was expressed and purified as reported (13). The cyano-met trHbO derivative was crystallized in hanging drops at 4°C at a protein concentration of 13.5 mg/ml, against reservoir solutions containing 2.15 M ammonium sulfate, 0.001 M KCN, 0.05 M phosphate buffer, pH 7.25. Well shaped bipyramidal red crystals grew in 2–3 weeks to a linear size of ≈200 μm. All x-ray diffraction data sets (Table 1) were collected at 100 K, adding 20% (vol/vol) glycerol, as cryoprotectant, to the crystal stabilizing medium (2.84 M ammonium sulfate/0.001 M KCN/0.05 M phosphate, pH 7.25). trHbO crystals belong to the tetragonal space group I4122, with unit cell constants a = b = 187.0 Å, c = 274.9 Å, 12 trHbO molecules (12 × 14,950 Da) per asymmetric unit (VM = 3.35 Å3/Da, 63% solvent content).
Table 1.
Data collection and refinement statistics
| EMBL BW7B | ESRF ID29
|
|||
|---|---|---|---|---|
| Peak | Inflection | Remote | ||
| Resolution, Å | 30.0–2.11 | 37.0–2.6 | 37.0–2.6 | 37.0–2.6 |
| Wavelength, Å | 0.8456 | 1.7387 | 1.7404 | 0.9801 |
| Rmerge, % | 8.1 (48.9)* | 9.0 (30.5)† | 8.7 (31.9)† | 6.3 (14.5)† |
| Completeness, % | 99.7 (97.5) | 100 (98.8) | 100 (100) | 99.9 (100) |
| Anomalous completeness, % | 100 (98.7) | 100 (100) | 99.9 (100) | |
| Mosaicity, ° | 0.285 | 0.45 | 0.45 | 0.47 |
| Total reflections | 1533455 | 523166 | 521145 | 539921 |
| Independent reflections | 138692 | 74954 | 74583 | 74728 |
| Redundancy | 11 (3) | 7.0 (6.8) | 7.0 (6.8) | 7.2 (7.3) |
| Average I/σ(I) | 16 (2.8) | 6.7 (2.5) | 7.1 (2.4) | 8.8 (5.0) |
| Space group | I4122 | I4122 | I4122 | I4122 |
| Unit cell, Å | a = b = 187.0 | a = b = 187.5 | a = b = 187.5 | a = b = 187.5 |
| c = 274.9 | c = 274.7 | c = 274.7 | c = 274.7 | |
| Refinement | ||||
| Rgen/Rfree, % | 18.6/22.6 | |||
| rms deviation | ||||
| Bonds, Å | 0.010 | |||
| Angles, ° | 1.250 | |||
| Ramachandran plot | ||||
| Most favored, % | 93.2 | |||
| Additionally allowed, % | 6.8 | |||
ESRF, European Synchrotron Radiation Facility; EMBL, European Molecular Biology Laboratory Hamburg Outstation.
Outer shell statistics (2.15–2.11 Å) within parentheses.
Outer shell statistics (2.73–2.60 Å) within parentheses.
A multiwavelength anomalous dispersion (MAD) experiment on the heme Fe absorption edge (data sets collected at 0.980-, 1.738-, and 1.740-Å wavelengths), performed at European Synchrotron Radiation Facility ID29 beam line (Grenoble, France), provided initial phases to build the protein model. In particular, the data set collected at the absorption peak (using data to 3.5-Å resolution) allowed location of 12 Fe atoms in the crystallographic asymmetric unit, by using the program shelxd (21). The anomalous scatterer positions were refined with solve (22) by using the three MAD datasets. Improvement of the starting MAD phases was achieved by solvent flattening at 2.6-Å resolution (resolve, ref. 22) (the figure of merit varied from 0.20 to 0.54), resulting in automatic model building of 43% of the entire dodecamer. Phase extension to 2.2 Å (data collected at ELETTRA, Trieste, Italy) was based on the arp/warp program suite (23) and used to build manually a full model of the 12 trHbO molecules. A subsequent dataset (2.1 Å; Table 1) was used for model correction and refinement (o, ref. 24; refmac5, ref. 25; and cns, ref. 26). In consideration of the good quality of the experimental electron density, noncrystallographic symmetry restraints were never applied. In the last refinement steps, stereochemically sound water molecules, located at 3σ level in difference Fourier maps, were added. The final model contains 12 trHbO molecules, corresponding to 12,856 protein atoms (average B factor, 25 Å2), 12 hemes, 12 CN− ions, 18 sulfate anions, and 1,618 water molecules (average B factor, 34 Å2; Table 1). Atomic coordinates and structure factors have been deposited with the Protein Data Bank (27).
To confirm the observation of a covalent bond between residues TyrB10-23 and TyrCD1-36, the trHbO structure was further subjected to simulated annealing omit refinement (T = 2,500 K). Moreover, trHbO tryptic peptides were examined with a LCQ DecaXP ion trap mass spectrometer (ThermoFinnigan, San Jose, CA). The presence of the peptide VYPEDDLAGAEER linked to the peptide FYAQVAEDEVLR, expected as a consequence of the TyrB10-23—TyrCD1-36 covalent bond, was unambiguously detected in the acquired mass spectra (for details, see supporting information).
Results
A trHb Dodecamer.
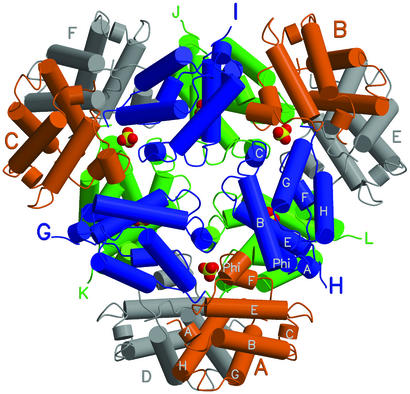
The trHbO crystallographic asymmetric unit hosts a full protein dodecameric assembly displaying ≈32 point group symmetry (individual subunits are labeled A, B, .., L; see Fig. 1). The compact dodecamer can be visualized as a stack of two equilateral triangles (with sides of ≈100 Å and thickness of ≈45 Å each), which are defined by the vertex-subunits A, B, C and D, E, F (orange and gray in Fig. 1) in the upper and lower layers, respectively. Each triangle includes one additional subunit at the center of its three sides (side-subunits G, H, I and J, K, L; blue and green in the upper and lower layers, respectively). Local 2-fold axes, hosted between the two triangular protein layers, relate pairs of trHbO subunits (A/D, B/E, C/F, and G/K, H/L, I/J pairs).
Figure 1.
A view of the assembled trHbO dodecamer down the local 3-fold axis direction. α-Helices are represented by cylinders (white labels in two subunits); heme groups have been omitted for clarity. The upper triangle six subunits are A, B, C, G, H, and I (orange and blue, large color-coded labels). The lower triangular assembly (green and gray subunits) is labeled with smaller lettering. Six sulfate anions are shown as yellow/red space filling models (partly hidden). The dodecamer's local 2-fold axes stretch from the center to the periphery of the assembly, through the six sulfate anions shown. All figures were drawn with MOLSCRIPT and RASTER3D (36, 37).
Because of the tight dodecameric packing, several types of trHbO subunit dimers can, in principle, be identified. The two different kinds of dimers built around the dodecamer's local 2-fold axes (A/D, B/E, C/F and G/K, H/L, I/J) are characterized by small association interfaces (<300 Å2). On the other hand, among other possible dimer types (e.g., D/G, D/K, or D/L), the vertex/side subunit association (D/G, E/H, F/I, A/L, B/J, and C/K), based on a 450-Å2 interface and stabilized by two intermolecular salt bridges (Arg-79–SerH13-122′ and ArgF5-72–Asn-125′),‖ appears to be the best candidate to account for a dimeric species (≈20% of the whole population) observed in gel filtration experiments at 10 μM protein concentration (10 mM Tris⋅HCl, pH 7.5/50 μM EDTA/15 mM NaCl; refs. 15 and 16). Despite the tight packing, all of the hemes in the assembled dodecamer have their propionate groups accessible to solvent.
The charge distribution at the trHbO subunit surface highlights the presence of a positive patch next to the heme proximal site, related to the localization of five Arg and one His residues (ArgE10-47, Arg-66, His-68, Arg-70, ArgF5-72, and ArgF7-74); Arg is the most abundant amino acid in trHbO, accounting for 12% of the total residues. Interestingly, the positively charged patches locate at the center of the trHbO dodecamer, finding electrostatic compensation in six sulfate anions from the solvent (the positive patch residues are totally conserved in seven of the 21 group II trHbs discovered so far, being conservatively mutated or partly maintained in the remaining 14). Three of the stabilizing sulfate anions fall on the dodecamer's 2-fold axes, relating the green and blue side subunits, acting as electrostatic bridges between their Φ-F facing helical regions (see below). The remaining three sulfates fall on the local 2-fold axes relating the vertex subunit pairs (orange and gray in Fig. 1) and find electrostatic compensation in residues ArgF5-72 and Arg-79, from each subunit. On the contrary, the dodecamer outer surface is negatively charged.
The Group II trHb Fold.
The good-quality electron density permitted building of almost the whole trHbO polypeptide chain (residues 2–3, up to 128), for all of 12 subunits. Although trHbO displays low sequence homology relative to group I trHbs (<20% amino acids identity), the overall 2-on-2 α-helical fold observed for group I trHbs (2, 3) is conserved, but with some evident modifications (Fig. 2). A structural overlay of the different trHbO subunits on group I trHbN structure (based on 69 Cα atoms of the B, E, F, G, H helices) yields rms deviation (rmsd) values in the 1.8- to 1.9-Å range, the main differences being located in the regions CD (rmsd of ≈3 Å, over amino acids 35–40), EF, and GH. Within the trHbO dodecamer, the rmsd values for comparisons of trHbO vertex vs. side subunits vary in the 0.5- to 0.7-Å range (calculated over 122 Cα pairs). More specifically, in their respective CD regions (a short six to eight residue segment between the C- and E-helices, in trHbO), each of the six dodecamer vertex subunits exhibits a double conformation for the main chain of amino acids 38–40; one of the two conformations brings the CD loop closer to the heme distal cavity, the other farther (Fig. 3b). An intramolecular salt bridge (between Asp-40 and ArgE10–47) stabilizes the closed CD loop conformation; a sulfate anion, peripheral to the dodecameric assembly, substitutes for Asp-40 carboxylate in the open CD conformation. On the other hand, the CD region in all of the dodecamer side subunits (green and blue in Fig. 1) displays the closed conformation only, possibly as a result of quaternary contacts around the dodecamer 3-fold axis.
Figure 2.
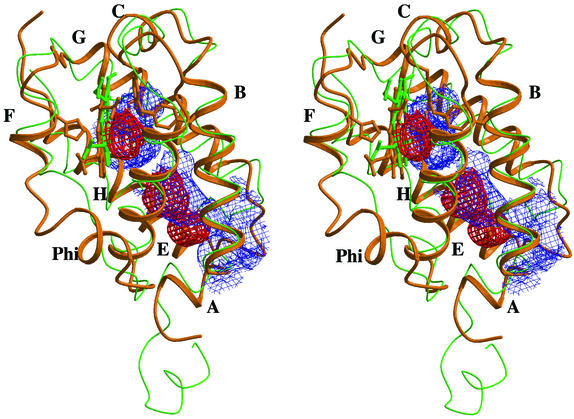
Stereo view of the trHbO subunit tertiary structure (orange ribbon) overlaid onto trHbN (green trace). Individual α-helices are labeled for trHbO. The figure includes mesh surfaces defining the trHbO protein matrix cavities (red color), and the matrix tunnel identified in trHbN (blue color).
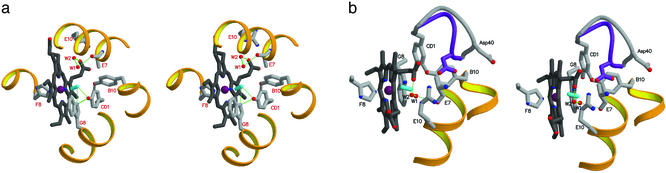
Figure 3.
(a) An exploded view of the heme distal cavity in an orientation highlighting hydrogen bonds from cyanide to TyrCD1-36 and TrpG8-88, and the TyrB10-23—TyrCD1-36 covalent bond. (b) Stereo view of the distal site cavity in cyano-met trHbO, seen from the solvent side. The CD region (Upper Right) is shown in the two observed conformations (gray and purple). Hydrogen bonds stabilizing water molecules W1 and W2 are dashed. The cyanide ligand is drawn in cyan; key residues are labeled with their topological positions.
In group I trHb, the globin fold conventional heme-proximal F-helix is substituted by an extended polypeptide segment (the “preF” segment; 11 amino acids in trHbN), preceding a one-turn F-helix bearing the proximal HisF8 residue (2, 3). In trHbO, this region hosts 15 residues, as the result of two amino acid insertions, characteristic of group II trHbs (see supporting information). The first insertion (residues 64–66 in trHbO) is reflected by the presence of a novel six-residue α-helix (the Φ-helix, residues 61–66; see Fig. 2), involved in interactions with the heme propionate D (residues Tyr-62 and Arg-66). The segment following the Φ-helix (the “ΦF-loop”) contains the group II conserved sequence motif -Gly-His-Pro-X- (where X indicates the group II specific second insertion), adopts an irregular-extended conformation, and precedes the one-turn F-helix, proximal to the heme. The sum of these structural features forces the trHbO preF region (residues 56–70) farther away from the protein core and from the heme, as compared with group I trHbs (Fig. 2) (2, 3). The deviation of the preF segment relative to group I trHbs is substantial (>7 Å at residue 68) and may be related to a significant shift of the whole heme group (about 1.3 Å with respect to trHbN) within the heme crevice of trHbO. Inspection of the trHbO structure indicates that such a shift in heme position depends on several concerted events, such as the preF loop specific conformation, reshaping of the FG interhelical region, shifting of the G-helix, a three-residue insertion (101–103) widening the GH corner, and a straight H-helix.
Heme Stabilization and the Proximal Heme Site.
Inspection of the electron density indicates that in all twelve trHbO subunits the heme group is rotated by 180° along the methinic α - γ meso axis, relative to (non)vertebrate globins (28). Selection of such bound heme orientation can be related to the specific location of the G- and H-helices in the heme crevice, and to specific heme/protein contacts in the regions surrounding residues TrpG8-88 and AlaH15-120. Stabilization of the bound heme is achieved through direct Fe coordination to the proximal HisF8-75 residue, electrostatic interaction of the heme propionates, and van der Waals contacts (<4.0 Å) with 21/24 protein residues (in the vertex/side subunits, respectively). Two salt bridges, to ArgE10-47 and to ArgF7-74, are provided by the heme propionates A and D, respectively. Residue E10 is strongly conserved as Arg/Lys in all three trHb groups. ArgF7-74, an invariant residue only in group II trHbs, is part of the cluster of positively charged residues surrounding the proximal site in trHbO.
The deep location of the porphyrin ring in the heme and the structural properties of the G- and H-helical regions in trHbO may be responsible for the potential solvent exposure of the heme C-pyrrole. In fact, the location of the whole pre-F segment far from the G- and H-helices, the lack of the H-helix kink observed in group I trHbs, and the residue size selection, give rise to a shallow surface depression nestled between the pre-F, G- and H-regions, on the rear of the trHbO molecule as depicted in Fig. 2. The surface depression is roughly circular (≈7-Å diameter) and primarily defined by residues PheE14-51, TyrE18-55, LeuF4-71, TyrH10-115, AlaH14-119, and LeuH18-123, all solvent exposed. Approximately at the center of the surface depression the atoms CAC and CBC of heme C-pyrrole are nearly exposed to solvent in the static crystal structure (they are accessible to a 1.2-Å radius solvent probe). This structural property, unique within the globin superfamily, might be related to a trHbO functional role, where redox processes could affect the heme Fe atom through interaction with electron donor/acceptor partners.
The heme proximal HisF8-75 residue is accommodated in a protein cavity defined by LeuF4-71, PheFG2-78, IleFG4-80, and LeuH18-123, all of which contact the porphyrin ring. The solvent side of the proximal heme pocket is sealed by the ArgF7-74–heme D propionate salt bridge; as a result, HisF8-75 is not accessible to solvent. Analysis of the stereochemical parameters describing the heme Fe coordination, on the proximal side, indicates a regular Fe—HisF8 NE2 coordination bond (2.05 ± 0.03 Å, averaged over the 12 subunits) (28), despite a systematic deviation of the HisF8 imidazole ring from heme orthogonality (the average angle between the two planes is 80.9 ± 2.7°). The average coordination bond distance between heme Fe and the heme four pyrrole N atoms is 2.04 ± 0.01 Å, the Fe atom being essentially in plane relative to these four N atoms. The proximal HisF8-75 imidazole azimuthal orientation is staggered, with a mean azimuthal angle of 33 ± 2° (relative to the heme pyrrole ND atom).
A Tyr-Tyr Covalent Bridge in the trHbO Distal Site Pocket.
All of the trHbO subunits display a cyanide anion bound to the heme Fe atom (the Fe-C coordination distances vary in the 1.77/2.02-Å range, with the exception of the K subunit displaying a Fe-C distance of 2.34 Å). The cyanide ligand is oriented roughly perpendicular to the heme plane (the Fe-C-N angle range is 164 ± 9°; Fig. 3 a and b). The spread in Fe—C coordination bond lengths suggests that, at least for the K subunit, the heme Fe may be present in the ferrous form, as a result of x-ray-induced reduction (29).
TyrCD1-36 is one of two residues involved in stabilization of the heme bound cyanide. A direct H bond (in the 2.7 ± 0.1-Å range, in the 12 subunits) connects the cyanide distal N atom to the OH group of TyrCD1-36 (Fig. 3a). Such ligand stabilization role based on the CD1 residue is totally new in the whole globin superfamily, because the CD1 site is almost invariably occupied by Phe, and the crystal structure of a wild-type globin bearing a TyrCD1 residue has never been reported. Next, the six vertex subunits display a covalent bond connecting TyrB10-23 OH and TyrCD1-36 CE2 atoms (1.43 ± 0.02 Å), resulting in a globin unprecedented posttranslational modification yielding a rigid system of two almost orthogonal aromatic rings (the angle between TyrB10-23 and TyrCD1-36 aromatic rings is 82 ± 3°; Fig. 3 a and b; for electron density maps and mass spectra details, see supporting information). In the six side subunits of the dodecamer, on the contrary, residues TyrCD1-36 and TyrB10-23 are in close contact (shortest mean distance 2.4 ± 0.1 Å), but a covalent bond is definitely absent. Within these differing distal site pictures, it should be noted that a double conformation for the CD loop (limited to amino acids 38–40) is observed only in the vertex subunits, which display the covalent link between the B10 and CD1 residues occurs.
In addition to TyrB10-23 and TyrCD1-36, the trHbO distal site pocket is characterized by residues AlaE7-44 and TrpG8-88, which can also be defined as quite unusual for the heme distal pocket, both in trHbs and generally in 3-on-3 folded globins (18, 19). AlaE7-44 substitutes for a residue (either His or Gln), often providing the main H bonding capabilities to the heme distal ligand in Hbs and Mbs. The small size and apolarity of the E7 residue may be required in trHbO to grant ligand access to the heme. In fact, in the cyano-met trHbO structure the heme distal pocket is connected to the external solvent space through a path defined by the AlaE7-44 ArgE10-47 residues, and the porphyrin plane; along this path two protein bound water molecules are found in the crystal structure (Fig. 3 a and b). One of these (W1), at the entrance of the distal site, is present in all of the dodecamer subunits, being hydrogen bonded to the carbonyl O atom of AlaE7-44. The other (W2), in all of the vertex subunits, is located between the heme propionates, at hydrogen bonding distance from ArgE10-47, W1 and the heme propionate A (Fig. 3 a and b).
At the dead end of the distal pocket, TrpG8-88, conserved in trHb group II, fills the inner part of the heme distal site, preventing farther diffusion of ligands into the distal pocket. TrpG8-88 indole ring is parallel and in contact with the heme plane (partially contacting the B and C pyrrole rings; Fig. 3 a and b). The location of the indole NE1 atom allows establishment of a bifurcated hydrogen bond connecting TrpG8-88 to the bound cyanide (3.20 ± 0.10 Å) and to TyrCD1-36 OH atom (3.31 ± 0.06 Å). A distal site tight hydrogen bonded network is thus established, linking two globin very unusual residues, i.e., TyrCD1-36 and TrpG8-88, to the heme ligand. Further rigidity to such structural arrangement is conferred, in the vertex subunits, by the B10—CD1 covalent bond. As a result of the combined presence of the TyrB10/TyrCD1 pair, the small AlaE7(44), and the bulky TrpG8-88 side chains, the trHbO distal cavity is slightly shrunk relative to trHbN (49 ± 7 Å3 vs. 62 Å3, respectively), the room available for heme ligand diffusion being mostly localized in the surroundings of the AlaE7-44—LeuE11-48 pair (Fig. 2).
Protein Matrix Cavities in trHbO.
The group I trHb protein matrix tunnel, typically stretching from the AB/GH corner region to the heme distal site, and branching to a second surface aperture between the H and G helices (1, 3), is dramatically restricted in trHbO. Such a change in the inner organization of the two proteins results from different relative orientations of the G and H helices and from six amino acid substitutions in trHbO (AlaB1→Thr, ValB5→Ile, ValG8→Trp, AlaG9→Leu, LeuG12→Met, and AlaH12→Glu). The changes, on average increasing the volume of the substituted residues, partly fill the protein matrix tunnel space, as defined in trHbN (Fig. 2). In particular, most of the trHbN short tunnel branch and the deeper part of the distal site pocket are occupied by TrpG8-88 in trHbO. The long tunnel branch, about 20 Å long in trHbN, retains only two cavities in trHbO (cavity 1, 37 ± 6 Å3 in the vertex subunits, and 33 ± 3 Å3 in the side subunits, respectively; cavity 2, 21 ± 3 Å3 in vertex subunits, and 31 ± 4 Å3 in side subunits, respectively), both fully shielded from solvent contact. The volume displayed by each cavity is comparable to the volume of the Xe-4 cavity observed in sperm whale Mb (11).
Discussion
The cyano-met trHbO crystal structure shows aggregation of the protein in a compact dodecameric assembly. Six of the sulfate anions bound to the dodecamer fall in a strongly positively charged protein region, roughly matching the preF protein loop in the inner region of the assembled dodecamer. It is therefore possible that electrostatic compensation of several Arg residues on the heme proximal side by an anionic species, such as sulfate, is required for trHbO to achieve a dodecameric quaternary assembly.
Within the trHbO dodecamer, the typical 2-on-2 trHb fold is promptly recognized. Accordingly, all of the dodecamer subunits show an extended preF loop, but this is structurally modified relative to group I trHbs by group II-specific residue insertions. The trHbO dodecamer subunits can be divided into two classes. The vertex subunits display two conformations for residues 38–40 in their CD loops, together with a globin unprecedented covalent bond linking TyrB10-23 phenolic O atom to TyrCD1-36 aromatic ring. It must be stressed that, despite the spatial location of both Tyr residues next to the CD region, the covalent isodityrosine moiety displays no signs of alternate conformations. On the contrary, the trHbO side subunits neither host covalent modifications nor display double conformation in their CD loops.
The systematic occurrence of the posttranslational covalent modification in the vertex subunits of the assembled dodecamer (orange and gray in Fig. 1) and the observation of the Tyr—Tyr covalent bond in mass spectra of the native protein exclude that such a bond is the result of x-ray-induced chemical processes. Indeed, the crystal structure shows that TyrB10-23 and TyrCD1-36 are closely located, even in the absence of a covalent link (side subunits), likely due to aromatic-electrostatic interactions between the hydroxyl O atom of TyrB10-23 and the aromatic H atoms of TyrCD1-36 (30). Such close location may be a key factor lowering the activation energy for formation of the B10—CD1 covalent bond. It should also be mentioned that subtle structural differences between the vertex and side trHbO subunits, other than the alternate CD loop conformations and the B10—CD1 covalent bond, are observed throughout the heme cavity and the protein structure (for brevity, further details will be reported elsewhere).
Evidence of a posttranslational modification introducing a covalent bond at Tyr residues is present in cytochrome c oxidase (31, 32), where a covalent link between His NE2 and Tyr CE2 atoms is required to establish a functional environment in the catalytic site (33). An ether link connecting two Tyr residues, identical to that observed in trHbO, has been reported as an oxidative modification of the plant cell wall extensins, where formation of isodityrosine may support insolubilization of these structural proteins (34, 35). Moreover, it has long been known that in H2O2-treated Mb the Fe-heme triggers formation of Tyr-phenoxyl radicals, whose reactivity can affect residues distantly located within the protein scaffold (38–40). Similar heme-linked reactivity, leading to Tyr-His covalent modification, has been reported for catalase (41). Taken together, the above observations suggest that formation of the TyrB10—TyrCD1 covalent bond in trHbO is a process likely occurring in solution, before assembly of the dodecameric species observed in the crystal, via encounter of the hemeprotein with an oxidative species. In the absence of complementary information, it is, however, premature to draw conclusions on the functional role played by isodityrosine, or its formation, in trHbO.
Accessibility of diatomic ligands (such as O2, CO, and NO) to the trHbO distal site is favored by the small-apolar AlaE7 residue, which does not obstruct entrance to the heme distal cavity. In cyano-met trHbO, differently from any other trHb or globin known, the cyanide ligand is stabilized by a hydrogen-bonded network based on residues TyrCD1 and TrpG8, but not on the trHb classical TyrB10 distal residue, which is hydrogen bonded to the ligand in Chlamydomonas eugametos cyano-met trHb (2) and in cyano-met trHbN (M. Milani, unpublished results). We note that in the vertex subunits, further rigidity to the distal site hydrogen-bonded network is provided by the TyrCD1—TyrB10 isodityrosine moiety.
A rigid/tight hydrogen-bonding network stabilizing the heme ligand through TyrCD1-36 and TrpG8-88 is in keeping with resonance Raman and mutational data, with the trHbO very low O2 and CO dissociation rate constants, and with the observation that the TyrB10-23→Phe mutation has little effect on the ligand dissociation rates (13, 15). The reported small effect on O2 dissociation rate related to the TyrCD1-36 →Phe mutation may result from contained structural reorganization within the distal site, favoring the onset of a compensatory ligand—TyrB10-23) hydrogen bond.
The present data show that specific selection of bulky residues in trHbO (TrpG8 being a notable example) convert the trHbN protein matrix long tunnel branch (connecting the solvent space to the distal site) into two small cavities, along the same path. The two matrix cavities, therefore, present a further overall modulation of the trHb architecture that is likely to be characteristic of group II trHbs, as judged by analysis of residues conserved at sites B1, B5, G8, G9, G12, and H12 (see supporting information) defining the cavities. The general presence of a small distal site E7 residue in group II trHbs would then be mirrored by a restricted protein matrix tunnel, which is instead essential in group I trHbs, where access to the heme via the E7 gate appears to be prevented by GlnE7 or LeuE7 residues. Nevertheless, the presence of two protein matrix cavities, comparable in size but not in location to the Xe-4 site of sperm whale Mb (11), might still indicate that they may be required as ligand docking stations in the yet unknown functional or dynamic behavior of the protein. In this respect, the very modest sequence conservation relative to group I trHbs and the innovative structural organization presented by trHbO in subunit assembly, distal site structure, ligand recognition strategy, and within the protein matrix strongly suggest that trHbO and trHbN are involved in quite different functional roles in M. tuberculosis.
Supplementary Material
Acknowledgments
We are grateful to Prof. Paolo Visca for helpful discussions. This work was supported by Agenzia Spaziale Italiana Grants I/R/294/02 and Consiglio Nazionale delle Ricerche Functional Genomics. M.G. was supported by Natural Sciences and Engineering Research Council of Canada Grant 46306-01. M.M. and M.B. are grateful to H. G. Gaslini (Genoa, Italy) for continuous support.
Abbreviations
- Hb
hemoglobin
- Mb
myoglobin
- trHb
truncated Hb
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates reported in this paper have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1NGK).
trHb residues are identified by their three-letter codes, by their globin topological position (28), and by sequence numbers.
References
- 1.Wittenberg J B, Bolognesi M, Wittenberg B A, Guertin M. J Biol Chem. 2002;277:871–874. doi: 10.1074/jbc.R100058200. [DOI] [PubMed] [Google Scholar]
- 2.Pesce A, Couture M, Dewilde S, Guertin M, Yamauchi K, Ascenzi P, Moens L, Bolognesi M. EMBO J. 2000;19:2424–2434. doi: 10.1093/emboj/19.11.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Milani M, Pesce A, Ouellet Y, Ascenzi P, Guertin M, Bolognesi M. EMBO J. 2001;20:3902–3909. doi: 10.1093/emboj/20.15.3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ermler U, Siddiqui R A, Cramm R, Friedrich B. EMBO J. 1995;14:6067–6077. doi: 10.1002/j.1460-2075.1995.tb00297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolognesi M, Bordo D, Rizzi M, Tarricone C, Ascenzi P. Prog Biophys Mol Biol. 1997;68:29–68. doi: 10.1016/s0079-6107(97)00017-5. [DOI] [PubMed] [Google Scholar]
- 6.Tarricone C, Galizzi A, Coda A, Ascenzi P, Bolognesi M. Structure (London) 1997;5:497–507. doi: 10.1016/s0969-2126(97)00206-2. [DOI] [PubMed] [Google Scholar]
- 7.Hargrove M S, Brucker E A, Stec B, Sarath G, Arredondo-Peter R, Klucas R V, Olson J S, Phillips G N., Jr Structure Folding Des. 2000;8:1005–1014. doi: 10.1016/s0969-2126(00)00194-5. [DOI] [PubMed] [Google Scholar]
- 8.Goodman M D, Hargrove M S. J Biol Chem. 2001;276:6834–6839. doi: 10.1074/jbc.M009254200. [DOI] [PubMed] [Google Scholar]
- 9.Pesce A, Dewilde S, Kiger L, Milani M, Ascenzi P, Marden M C, Van Hauwaert M L, Vanfleteren J, Moens L, Bolognesi M. J Mol Biol. 2001;309:1153–1164. doi: 10.1006/jmbi.2001.4731. [DOI] [PubMed] [Google Scholar]
- 10.Ilari A, Bonamore A, Farina A, Johnson K A, Boffi A. J Biol Chem. 2002;277:23725–23732. doi: 10.1074/jbc.M202228200. [DOI] [PubMed] [Google Scholar]
- 11.Brunori M, Gibson Q H. EMBO Rep. 2001;2:674–679. doi: 10.1093/embo-reports/kve159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scott E E, Gibson Q H, Olson J S. J Biol Chem. 2001;276:5177–5188. doi: 10.1074/jbc.M008282200. [DOI] [PubMed] [Google Scholar]
- 13.Mukai M, Savard P Y, Ouellet H, Guertin M, Yeh S R. Biochemistry. 2002;41:3897–3905. doi: 10.1021/bi0156409. [DOI] [PubMed] [Google Scholar]
- 14.Pathania R, Navani N K, Gardner A M, Gardner P R, Dikshit K L. Mol Microbiol. 2002;45:1303–1314. doi: 10.1046/j.1365-2958.2002.03095.x. [DOI] [PubMed] [Google Scholar]
- 15. Ouellet, H., Juszczak, L., Savard, P. Y., Wittenberg, J. B., Wittenberg, B. A., Friedmann, J. L. & Guertin, M. (2003) Biochemistry, in press.
- 16.Pathania R, Navani N K, Rajamohan G, Dikshit K L. J Biol Chem. 2002;277:15293–15302. doi: 10.1074/jbc.M111478200. [DOI] [PubMed] [Google Scholar]
- 17.Couture M, Yeh S R, Wittenberg B A, Wittenberg J B, Ouellet Y, Rousseau D L, Guertin M. Proc Natl Acad Sci USA. 1999;96:11223–11228. doi: 10.1073/pnas.96.20.11223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bashford D, Chothia C, Lesk A M. J Mol Biol. 1987;196:199–216. doi: 10.1016/0022-2836(87)90521-3. [DOI] [PubMed] [Google Scholar]
- 19.Kapp O H, Moens L, Vanfleteren J, Trotman C N, Suzuki T, Vinogradov S N. Protein Sci. 1995;4:2179–2190. doi: 10.1002/pro.5560041024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poole R K, Hughes M N. Mol Microbiol. 2000;36:775–783. doi: 10.1046/j.1365-2958.2000.01889.x. [DOI] [PubMed] [Google Scholar]
- 21.Uson I, Sheldrick G M. Curr Opin Struct Biol. 1999;9:643–648. doi: 10.1016/s0959-440x(99)00020-2. [DOI] [PubMed] [Google Scholar]
- 22.Terwilliger T C, Berendzen J. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morris R J, Perrakis A, Lamzin V. Acta Crystallogr D. 2002;58:968–975. doi: 10.1107/s0907444902005462. [DOI] [PubMed] [Google Scholar]
- 24.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 25.Murshudov G N, Lebedev A, Vagin A A, Wilson K S, Dodson E J. Acta Crystallogr D. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 26.Brunger A T, Adams P D, Clore G M, Delano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 27.Berman H M, Westbrook J, Feng Z, Gilliland G, Bhat T N, Weissig H, Shindyalov I N, Bourne P E. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perutz M F. Annu Rev Biochem. 1979;48:327–386. doi: 10.1146/annurev.bi.48.070179.001551. [DOI] [PubMed] [Google Scholar]
- 29.Bolognesi M, Rosano C, Losso R, Borassi A, Rizzi M, Wittenberg J B, Boffi A, Ascenzi P. Biophys J. 1999;77:1093–1099. doi: 10.1016/S0006-3495(99)76959-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burley S K, Petsko G A. Science. 1985;229:23–28. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- 31.Ostermeier C, Harrenga A, Ermler U, Michel H. Proc Natl Acad Sci USA. 1997;94:10547–10553. doi: 10.1073/pnas.94.20.10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshikawa S, Shinzawa-Itoh K, Nakashima R, Yaono R, Yamashita E, Inoue N, Yao M, Fei M J, Libeu C P, Mizushima T, et al. Science. 1998;280:1723–1729. doi: 10.1126/science.280.5370.1723. [DOI] [PubMed] [Google Scholar]
- 33.Pinakoulaki E, Pfitzner U, Ludwig B, Varotsis C. J Biol Chem. 2002;277:13563–13568. doi: 10.1074/jbc.M112200200. [DOI] [PubMed] [Google Scholar]
- 34.Brady J D, Sadler I H, Fry S C. Biochem J. 1996;315:323–327. doi: 10.1042/bj3150323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Epstein L, Lamport D T A. Phytochemistry. 1984;23:1241–1246. [Google Scholar]
- 36.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 37.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 38.Tew D, Ortiz de Montellano P R. J Biol Chem. 1988;263:17880–17886. [PubMed] [Google Scholar]
- 39.Catalano C E, Choe Y S, Ortiz de Montellano P. J Biol Chem. 1989;264:10534–10541. [PubMed] [Google Scholar]
- 40.Witting P K, Mauk A G. J Biol Chem. 2001;276:16540–16547. doi: 10.1074/jbc.M011707200. [DOI] [PubMed] [Google Scholar]
- 41.Matè M J, Sevinc M S, Hu B, Bujons J, Bravo J, Switala J, Ens W, Loewen P C, Fita I. J Biol Chem. 1999;274:27717–27725. doi: 10.1074/jbc.274.39.27717. [DOI] [PubMed] [Google Scholar]
- 42.Ouellet H, Ouellet Y, Richard C, Labarre M, Wittenberg B A, Wittenberg J B, Guertin M. Proc Natl Acad Sci USA. 2002;99:5902–5907. doi: 10.1073/pnas.092017799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.