Abstract
Polar residue hot spots have been observed at protein–protein binding sites. Here we show that hot spots occur predominantly at the interfaces of macromolecular complexes, distinguishing binding sites from the remainder of the surface. Consequently, hot spots can be used to define binding epitopes. We further show a correspondence between energy hot spots and structurally conserved residues. The number of structurally conserved residues, particularly of high ranking energy hot spots, increases with the binding site contact size. This finding may suggest that effectively dispersing hot spots within a large contact area, rather than compactly clustering them, may be a strategy to sustain essential key interactions while still allowing certain protein flexibility at the interface. Thus, most conserved polar residues at the binding interfaces confer rigidity to minimize the entropic cost on binding, whereas surrounding residues form a flexible cushion. Furthermore, our finding that similar residue hot spots occur across different protein families suggests that affinity and specificity are not necessarily coupled: higher affinity does not directly imply greater specificity. Conservation of Trp on the protein surface indicates a highly likely binding site. To a lesser extent, conservation of Phe and Met also imply a binding site. For all three residues, there is a significant conservation in binding sites, whereas there is no conservation on the exposed surface. A hybrid strategy, mapping sequence alignment onto a single structure illustrates the possibility of binding site identification around these three residues.
Keywords: protein–protein interfaces‖hot spots‖molecular recognition‖ binding site prediction‖residue conservation
Ringe (1) has raised the question “what makes a binding site a binding site?” Many studies have addressed this intriguing and vastly important problem. Being able to a priori predict binding sites would both limit the conformational search in drug design, facilitate the prediction of protein–protein interactions (2), and may provide leads to binding site design.
A number of studies have examined the attributes of protein-binding sites (3–5). Although binding sites on enzyme surfaces typically consist of a concave cleft shape (6, 7) and similarly small ligand binding sites on receptor surfaces (8), this is not the case for the larger protein–protein complexes (9–12). Enzyme-binding sites were shown to frequently be the largest cavities on the enzyme surface (6, 7). On the other hand, the shape of dimer-binding sites is usually quite flat (9) and practically indistinguishable from other patches on the protein surface. Native binding sites do not yield the largest possible interfaces between two protein molecules. A docking study has shown that nonnative interfaces can be larger, and bury a larger extent of total or nonpolar surface areas (13). A similar observation has been made for the number of salt bridges or hydrogen bonds (13, 14). Hence, although interfaces are frequently largely hydrophobic and bury a large extent of nonpolar surface area (15), the magnitude of the hydrophobic effect is insufficient to identify binding sites. It also does not enable distinguishing between crystal packing interfaces versus native interfaces (16). Fernandez and Scheraga (5) have made the remarkable discovery that the majority of backbone hydrogen bonds are completely wrapped intramolecularly by nonpolar groups except for a few likely to be around the binding site. The insufficiently dehydrated hydrogen bonds may be dramatically stabilized on binding.
Alanine scanning of protein–protein interfaces has shown that the binding free energy is not equally distributed at the binding interface. Rather, there are hot spots of binding energy consisting of a subset of residues at the interface (17, 18). Systematic analysis has found the hot spots to be particularly enriched in Trp, Tyr, and Arg. These were largely surrounded by hydrophobic rings, probably to occlude bulk solvent (19). Because of the significance of hot spots in protein interaction and drug discovery, unraveling hot spots in binding interfaces continues to stimulate interest, with both progress and challenges (18).
There are two major computational approaches to understand the binding hot spots: energetic evaluation and structural analysis. Computational alanine scanning using extensive molecular mechanics Poisson–Boltzmann surface area (MM-PBSA) calculations (20) as well as a simple physical model (21) reproduced successfully experimental energy changes. Monte Carlo evaluation of the energy landscape of hot spots (22) indicates that evolutionary convergent binding sites (23) correspond to the energetically most favorable states. However, there are different interpretations of the specific energy contribution. Kortemme and Baker (21) demonstrated that the hydrogen bonding term contributes significantly to the correct prediction of hot spots. In the Verkhivker et al. study (22), hydrophobic interactions were found to be critical.
Structural analysis of the protein–protein surface also provided insights into the binding hot spots. Hu et al. (24) have analyzed families of related interfaces (25, 26). Their analysis has confirmed and extended the results of Bogan and Thorn, showing that binding sites are enriched in polar residue hot spots. The Hu et al. analysis (24) sought residue conservation in structurally similar environments. Other structural analyses using conservation probability (27) or graph-spectral methods (28) also indicate the significance of conserving important interactions on homodimeric protein interfaces.
Nevertheless, none of these studies resolved the important question of whether the hot spots distinguish between the interfaces and the remainder of the surface. The main goal of the present work is to answer the question of the portion of the conserved residues in the interface over the exposed surface. We compare the conservation patterns at the binding sites versus the remainder of the surface. We are able to carry out such a study owing to MUSTA, our newly developed multiple structure comparison algorithm (29, 30).
We find that structurally conserved residues distinguish between binding sites and exposed protein surfaces. For three residues (Trp, Phe, and Met), there is a significant conservation in binding sites, whereas there is no conservation on the exposed surface.
Methods
The initial dataset of interfaces with 1,629 two-chain interface entries in the Protein Data Base (25, 31) has been clustered into 351 families, by using the Geometric Hashing, sequence order-independent structural comparison algorithm. A threshold of <90% sequence identity was imposed. A representative was taken from each cluster, and the process was repeated with a 30–90% sequence similarity range for the next clustering level. This procedure resulted in 10 families (with at least four members), totalling 86 entries (Table 1). All are included in the study of Hu et al. (24). The data set has a good balance of interface type, including obligate dimers (hemoglobin and immnunoglobulins), proteinase inhibitors, antigens, protease complexes, and hormones. Therefore, we expect no significant bias of a specific family.
Table 1.
List of 10 protein interface families used in the present study
| No. | Family representative | Members in the family | Protein classification | ||||
|---|---|---|---|---|---|---|---|
| 1 | 1babAD | 1hdsAD | 1bbbBC | 1bbbAD | 1hdsBC | Hemoglobin | |
| 2 | 1bbbAB | 1hdsCD | 1hdaCD | 1pbxAB | 2mhbAB | 2dhbAB | |
| 3 | 1choEI | 3sgbEI | 1ppeEI | 4tpiZI | 1brcEI | 1ppfEI | Serine proteinase inhibitor |
| 1tabEI | 1tgsZI | 2kaiBI | |||||
| 4 | 1cseEI | 1sibEI | 1meeAI | 1sbnEI | 2sniEI | 5sicEI | |
| 5 | 1hhjAB | 2mhaAB | 1hocAB | 1hsaAB | 2vaaAB | 1hsaDE | Histocompatibility antigen |
| 6 | 1hilCD | 2cgrLH | 2fb4LH | 1bafLH | 1bbdLH | 1mfbLH | Immunoglobulin |
| 1dbjLH | 1figLH | 1gigLH | 1ifhLH | 2mcpLH | |||
| 1mamLH | 2fbjLH | 2fgwLH | 1cbvLH | 1ggiLH | |||
| 1igfMJ | 1igiLH | 1indLH | 7fabLH | 1ncbLH | |||
| 1tetLH | 2iffLH | 3hfmLH | 1dfbLH | 1faiLH | |||
| 1fvdAB | 1gcLH | ||||||
| 7 | 1vfaAB | 1fvcAB | 1reiAB | 1fgvLH | 1fvbLH | 1igmLH | |
| 1jhlLH | 1migLH | 2fvwLH | |||||
| 8 | 1hviAB | 2rspAB | 1hvpAB | 2mipAB | 1sivAB | Protease complexe | |
| 9 | 2gstAB | 1glqAB | 1guhAB | 1gssAB | 1hncAB | Glutathione transferase | |
| 10 | 1izaAB | 1trzAB | 6rlxAB | 1izaCD | 6rlxCD | Hormone | |
The data set of studied proteins.
Multiple structure comparisons with the structural coordinates being the only input, disregarding sequence and motif information, is a difficult problem. Multiple structure alignment (MUSTA) solves the problem while avoiding the expensive full conformational space search (29, 30). The input consists of an ensemble of N molecules represented by its Cα-atoms coordinates. The algorithm consists of three major stages: (i) detection of seed matches and of candidate multidimensional transformations; (ii) clustering of the transformations in each of the multidimensional transformation components and extension of the seed matches corresponding to the cluster prototypes; and (iii) computation of the highest scoring multidimensional transformations. These induce the largest cores. Lower ranked transformations can be obtained as well. These lead to the smaller substructural motifs.
The surface residues have been identified by using ACCESS (32). For each residue the surface area was calculated and compared with that of the residue in Gly–X–Gly (33). Surface residue was defined when its accessible surface area was >20% of the residue in the extended conformation. The calculation was carried out on the complex. Hence, below, the term “surface” is exposed surface and does not include residues that are contact residues.
Contact residues were defined as in Tsai et al. (25). Briefly, two residues are considered to be in contact across the interface if there is at least a pair of atoms, one from each residue, at a distance smaller than the sum of their vdW radii plus a threshold of 0.5 Å. The general propensities are calculated as follows:
 |
where ni is the number of conserved residues of type i at the contact interface, si is the number of conserved residues of type i on the surface, Ni is the total number of residues i in the protein, and N is the total number of amino acids in the protein. p1 and p2 measure the propensity of the amino acid to be conserved on the interface and exposed surface, respectively. [There was a typo in the earlier publication of Hu et al. (24), and the sum(ni) was mistakenly reported as total number of interface residues, whereas it should be the total number of conserved residues, as used in the computation.] All of the above numbers are normalized (averaged) in each family to make them independent of the entries in each family. Therefore, all protein families are equally weighted.
Results and Discussion
Structural Conservation on the Binding Site and the Exposed Surface.
Although there has been a number of multiple sequence alignment algorithms, to date there have been very few approaches to multiple structure alignment and detection of a recurring substructural motif. Among these, very few perform both multiple structure comparison and motif detection simultaneously, considering all structures at the same time, rather than initiating from a pairwise superimposed molecular seed, which can lead to a bias. These include MUSTA (29, 30), multiprot (34), and MASS (multiple alignment of order-independent secondary structures; O. Dror, H. Bengamini, R.N., and H.W., unpublished data). MUSTA is state of-the-art in its capabilities. Given an ensemble of protein structures, the algorithm automatically finds the largest common substructure (core) of Cα-atoms that appears in all of the molecules in the ensemble. The detection of the core and the structural alignment are performed simultaneously. Additional structural alignments are also obtained and ranked by the sizes of the substructural motifs that are present in the entire ensemble. Because it is independent of amino acid sequence order, it can be applied to protein surfaces, protein–protein interfaces, and protein cores to find the optimally and suboptimally spatially recurring substructural motifs.
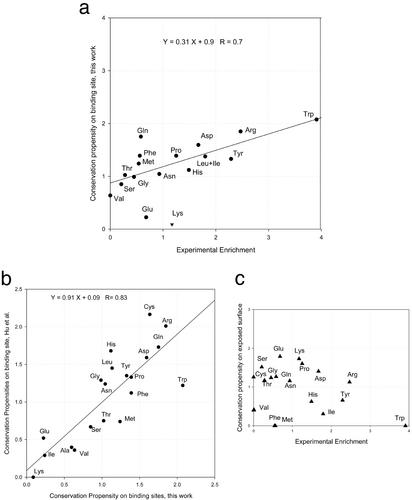
We used MUSTA to structurally align the members of each family. For each family, when >80% family members have an identical residue aligned within 1.0 Å, we define the residue as conserved. Finally, the conservation propensities are computed. The results of the conservation propensities on the surface and binding sites are reported in Table 2. Cross comparisons of the MUSTA alignments with the experimental hot spots enrichment and computed propensities from the previous pairwise alignment are necessary to validate the new propensities we have obtained. As may be seen in Fig. 1, there are very good correlations between the current binding conservation and both the experimental data of hot spot enrichment and the previously published binding conservation values (24). The correlation coefficient between the current work and our earlier work is 0.82, with an average deviation of 0.25. The largest difference comes from Trp (2.07 in this work and 1.22 in ref. 24). It seems that our earlier work underestimated the conservation of Trp significantly (experimental enrichment 3.91).
Table 2.
Conservation propensities and change of conservation propensities on the binding sites exposed surfaces
| Residue | P_2 | P_1 | A | B |
|---|---|---|---|---|
| ALA | 0.90 | 0.63 | 0.94 | 1.14 |
| VAL | 0.36 | 0.70 | 0.88 | 1.09 |
| PHE | 0 | 1.35 | 0 | 0.87 |
| PRO | 1.64 | 1.37 | 1.23 | 1.14 |
| MET | 0 | 1.21 | 0 | 0.80 |
| GLY | 1.15 | 0.96 | 1.13 | 1.05 |
| ILE | 0.31 | 0.23 | 0.79 | 0.26 |
| LEU | 0.41 | 1.12 | 0.90 | 1.08 |
| ASP | 1.37 | 1.55 | 1.06 | 1.36 |
| GLU | 1.83 | 0.29 | 1.13 | 0.40 |
| LYS | 1.71 | 0.14 | 0.97 | 0.30 |
| ARG | 1.15 | 1.80 | 0.92 | 1.13 |
| SER | 1.55 | 0.83 | 1.03 | 1.10 |
| THR | 1.20 | 1.07 | 0.93 | 1.10 |
| TYR | 0.67 | 1.29 | 1.30 | 0.80 |
| HIS | 0.64 | 1.09 | 0.67 | 0.87 |
| CYS | 1.28 | 1.59 | 3.18 | 1.05 |
| ASN | 1.18 | 1.10 | 0.95 | 1.10 |
| GLN | 1.29 | 1.70 | 0.95 | 1.10 |
| TRP | 0 | 2.02 | 0 | 1.74 |
P_1, binding sites; P_2, exposed surfaces; A, [conservation propensities on P_2]/[propensity on P_2]; B, [conservation propensities on P_1]/[propensity on P_1].
Figure 1.
(a) Correlation of new binding-site conservation propensities with experimental enrichment of hot spots. (b) Correlation of new binding-site conservation propensities with the previous work of Hu et al. (c) No correlation of exposed surface conservation propensity with experimental hot spots.
In general, our current work correlates with the experimental hot spot enrichment much better than our previous work (24) did. Following the earlier work, we use combined propensity of Leu and Ile. Three outliers were identified previously (Val, Lys, and Trp). However, only Lys was identified as an outlier in the current correlation (Fig. 1a). The overall correlation with experimental hot spots is 0.7, comparable with the previous study. As may be seen from Fig. 1a, Glu, Gln, and Lys deviate most from the regression line. If we were to exclude these three residues, the correlation of the current work with the experimental hot spot enrichment would be 0.87. As expected, there is no correlation between the surface conservation propensity and the experimental hot spots enrichment. This noncorrelation contrasts with the high correlation between the binding sites conservation and the experimental hot spot enrichment, and validates our approach. This is particularly important for our studies of surfaces.
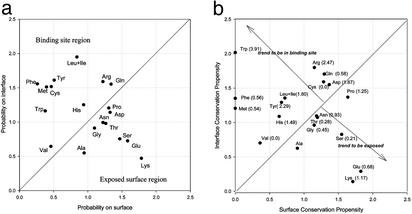
We compare the binding-site properties with those of the surfaces. Such a comparison should be useful in distinguishing between binding sites and the nonbinding surface. Fig. 2a compares the propensity of amino acids to be on the binding interface and on the surface. Fig. 2b compares the conservation propensity of amino acids on the two surface types. In Fig. 2b, the numbers in parentheses after the residue names are the experimental hot spot enrichment by Bogan and Thorn (19).
Figure 2.
Propensity map of the residues to be in the binding-site contact layer versus exposed surface (a) and conservation propensities in the binding-site contact layer versus exposed surface (b).
As expected, inspection of Fig. 2a shows an overall dominant appearance of hydrophobic residues at the interface (15). Gly and Pro are slightly preferred on the surface. This tendency reduces in the conservation patterns (Fig. 2b). The propensities of Phe, Met, and Leu+Ile all drop significantly. The general propensity of Gly and Val at the interface is similar to their conservation at the interface. However, Pro is slightly preferred to be conserved, which is understandable because of its unique conformation.
Val is reminiscent of the aberrant (low frequency) behavior of Leu in the Ala scanning data (19). In Hu et al. (24) the frequency of Leu was high and that of Ile was low. However, the sums of both Leu and Ile in the two studies correlated well. The same situation was observed in the current study. Val shows a slight tendency to be at the interface (Fig. 2a). Hu et al. also observed a low Val propensity at the interface. Experimentally, its hot spot enrichment is zero (19). Hence, it appears that Val tends to be buried (either at the interface or in the protein core) with no specific role in binding.
The general occurrence of polar hot spots, as compared with specific polar and nonpolar residue conservation at binding sites such as observed in the proteases, argues to their contribution to the binding free energy, rather than to specific binding. A similar conclusion has been reached by Kleanthous et al. (35–37). Through studies of DNase binding by the immunity proteins they have shown that conserved residue hot spots (from helix III) act as a binding site anchor, whereas the variable residues (from helix II) define specificity. Further, they find Tyr to be conserved.
Several polar residues move significantly in the binding sites: exposed surface maps (Fig. 2). Three top ranking hot spots (Trp, Arg, Tyr) are very interesting. Trp has only a moderate propensity on the interface. However, its conservation propensity jumps to the very top, corresponding to its highest ranking in experimental hot spot enrichment. Thus, it appears that the role played by Trp is unique, probably owing to its large size and aromatic nature (38). Additionally, it is possible that the Trp/Ala mutation creates a large cavity, due to the significant difference in sizes. After Trp, Arg also gets a moderate increase in the conservation propensity. It has the second highest conservation propensity on binding sites, also consistent with experimental data. Tyr, however, shows an opposite trend. It has a very high propensity to be on the interface, and only a moderate conservation propensity. Even though in Fig. 2b it still locates in the preferred binding region, the trend deserves further investigation. A careful examination of individual propensities for each family reveals that Tyr is the most abundant residue on the interface for the 1hilCD and 1vfaAB families, both being immunoglobulins. As noted in our previous study (24), the antibody shows a random surface distribution and tends to be less conserved. This may explain the decrease in conservation propensity of Tyr. Asp is the only residue that switchs from the region of exposed surface (Fig. 2a) to the binding region (Fig. 2b). Note that it also has a high value in experimental enrichments.
Three residues (Lys, Glu, and Ser) strongly and two (Thr and Asn) moderately prefer to be on the surface. These five residues do not change the propensity patterns from Fig. 2a to Fig. 2b. With regard to Lys, an aberrant behavior was also observed in the Hu et al. study, also with very low contact layer conservation. Lys is more frequent on the surface than in the interface or in the contact region. Given its high flexibility, and the fact that we multiply align the entire chains, its deviation distance in the superposition might have exceeded the (2.0 Å) distance threshold set in the alignment.
Overall, Fig. 2b clearly indicates that structurally conserved residues do distinguish between binding sites and exposed protein surfaces. Trp, the most important residue in terms of both conservation and free energy change in Ala scanning is remarkably conserved only in binding sites. Thus, the conservation of Trp on the protein surface has a very high probability to be around a binding site. To a lesser extent, conservation of Phe and Met also imply a binding site. This proposition needs, however, extensive benchmark trials. For Phe and Met, there is a significant conservation in binding sites while there is no conservation on the surface. Trp, Phe and Tyr may be conserved to ensure the structural stability on folding and binding. One such example is Trp's role in protecting fragile hydrogen bonds from water (39).
Other studies also indicate the importance of aromatic residues in protein interactions (28), consistent with our findings of the conservation of Trp and Phe in binding sites. However, it is interesting to see that Met also prefers binding sites over the surface. This may be due to the fact that Met, although hydrophobic, still has the ability to form weak hydrogen bonds. Also, a sulfur atom has a large polarizability, which may be useful for electrostatic interactions around the binding sites. Verkhivker et al. (22) found that Met residues play a significant role in the human Ig binding. However, in that case, Met residues are critical for local conformational changes during binding.
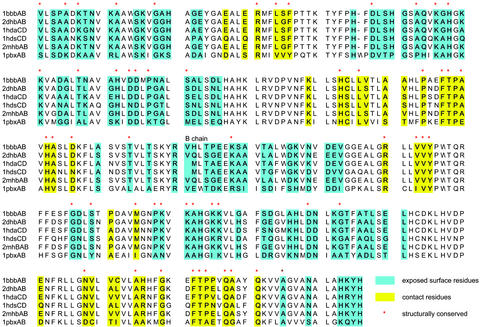
The current results indicate that multiple structural alignment could be a valuable tool to identify possible binding sites on protein surfaces when several related protein structures are available. However, what if only one structure is available, along with homologous sequences? Here we test a hybrid alignment, mapping a sequence alignment onto a single structure. As an example, let us assume that 1bbbAB is the only structure available in the hemoglobin family. Fig. 3 shows alignment of six sequences from the 1bbbAB family whose overall sequence identity is 37%. We identify binding site residues (yellow) and exposed residues (green) on the 1bbbAB structure. The structurally conserved residues are marked with red stars. Three observations may be made from the hybrid alignment (1). Most structurally conserved residues around the binding site are sequentially conserved (25 of 29), whereas less sequentially conserved residues observed to be structurally conserved (28 of 54) (2). Neither Phe nor Met are sequentially conserved on the exposed surface (3). Phe and Met are conserved in both sequence and structural alignment around the binding site. Thus, in principle, we expect that hybrid alignment may be used to predict binding sites combined with conservation around Trp, Phe, and Met.
Figure 3.
Comparison of structural alignment and sequential alignment of 1bbbAB interfaces.
In a similar approach, evolutionary trace (ET) studies (40, 41) have demonstrated that spatial clustering of evolutionarily important residues is a general phenomenon (41). Friedberg and Margalit (42) also found that the structural/functional key residues from structural alignment show conservation in the respective sequences. Madabushi et al. (41) used ET to rank residues in a protein sequence by evolutionary importance and map top ranked residues or evolutionarily privileged clusters onto a representative structure. Good identification of binding sites is achieved in the ET study. However, no specific information about the nature of the amino acids was reported. A combination of ET with a focus on Trp, Phe, and Met might be helpful in future binding site predictions.
Properties of Binding Epitopes: Structurally Conserved Residues and Contact Area.
Intermolecular binding cannot be too strong to obstruct biological function. Yet, binding interfaces can be quite large. A similar observation has been made for small molecule binding (43). The question then arises as to how, despite the large interface size, the total binding free energy does not exceed a maximal value, set by protein function. In principle, many factors may be involved. Here we examine the possibility that if a large fraction of the binding free energy derives from conserved (hot spot) residues, then their number should be limited despite the overall large interface area. This evolutionary strategy would keep a lid on maximal intermolecular binding affinity.
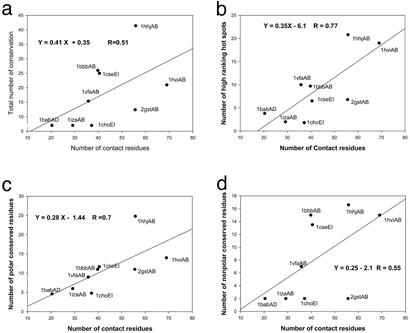
Assuming that structurally conserved residues are energy hot spots, our results are inconsistent with such a solution. As may be seen in Fig. 4, there is an apparent correlation between the number of structurally conserved residues and the contact surface size, measured by the number of contact residues. The correlation is very strong when we only count the high ranking hot spot residues (Trp, Arg, Tyr, Leu+Ile, Asp, His, Pro, and Lys; Fig. 4b, R = 0.77). The correlation of polar residues (Fig. 4c) is stronger than the correlation of hydrophobic residues (Fig. 4d). Thus, larger binding interfaces entail an increase in the number of high ranking (mostly polar) residue hot spots. How then to reconcile this apparent contradiction? First, we note that it is possible that the interface size included in our study is not large enough to reach the plateau yet. We do see some indication of such a plateau for the 1hilCD family (not shown), which has an average of 166 contact residues and only 25 conserved residues. Nevertheless, this sole point does not offset the trend observed with the other nine protein families. Here we favor an alternative explanation: when the interface is large, packing is not as optimal as when it is significantly smaller and backbone and side-chain contacts may move to optimize their interactions. This explanation is consistent with the absence of a correlation between the binding energy and the change of side-chain contact surface for individual residues (19). Furthermore, when a ligand interacts, specific contacts and desolvation must be balanced to favor ligand binding. Hence, hot spots cannot be simply clustered together in a small contact area. Because most of the residues at the interface are well known to be hydrophobic (15), the increase in the number of residue hot spots with the interface size is further consistent with the proposition made by Bogan and Thorn (19) that polar hot spots are surrounded by hydrophobic rings. Water exclusion may lead to a better interacting environment of the energetically hot residues rather than provide thermodynamic stability directly.
Figure 4.
Correlations of the number of all conserved residues with the number of contact residues (a), the number of high ranking hot spot (Trp, Arg, Tyr, Leu+Ile, Asp, His, Pro, and Lys) residues with the number of contact residues (b), the number of all conserved polar residues with the number of contact residues (c), and the number of all conserved hydrophobic residues with the number of contact residues (d).
Although here we focus on (largely polar) conserved residue hot spots, it behooves us to note that a second important determinant of protein-binding sites is flexibility (23, 44–46). Analyzing interactions between biological molecules cannot be reduced to a static description of molecular structures (47). Rather, the binding partner should be considered, as well as the time component of the interaction (47). The fact that multiple different molecules bind at the same, presumably specific sites, argues for binding site dynamics (45, 46). Effectively dispersing hot spots within a large contact area rather than clustering them in a compact site, may be a strategy to retain essential interactions while still allowing certain flexibility. This may rationallize the increase in the number of conserved residues with the increase in the contact areas. In such a mechanism, most conserved polar residues at binding interfaces confer rigidity to minimize the entropic cost on binding, whereas the residues surrounding the conserved residues form a flexible cushion. One way to test such a mechanism is through studies of fluctuation frequencies. Demirel et al. (48) used the Gaussian network model to show that key residues in folding/function have the highest frequencies and are more rigid. Our preliminary results suggest that structurally conserved residues have the highest frequencies (T. Haliloglu, O. Keskin, and R.N., unpublished results). Cole and Warwicker (49) also found that a protein–protein interface is less flexible than the surface, consistent with more conserved residues on the interfaces than on surfaces.
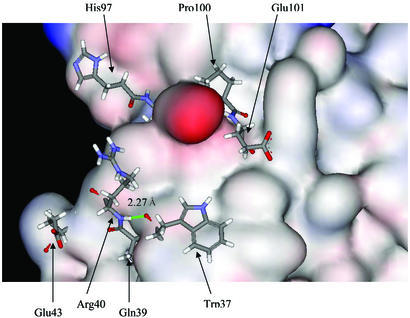
Recently, Fernandez and Scheraga (5) have made the remarkable observation that the hydrophobic residues in binding sites complete the dehydration shell of binding partners. They proposed that the under-dehydrated hydrogen bonds (UDHB) can contribute significantly to the binding energy. We have not systematically examined this new concept to see whether the conserved hot spots correspond to the UDHB. However, an examination of the hot spots from the interface of the D chain of 1babAD does indicate such a possible relationship. As shown in Fig. 5, the structurally conserved residues have exposed backbone carbonyl atoms and there is one hydrogen bond between Trp-37 and Arg-40. It will be very interesting to investigate interacting partners and backbone hydrogen bonds involving conserved hot spots.
Figure 5.
The conserved hot spots on the D chain of 1babAD interface. The hot spots are illustrated as sticks, and for the remaining residues only the surface is shown. Note that the carbonyl backbone atoms of the hot spots are exposed. There is a hydrogen bond between Trp-37 and Arg-40. This arrangement is consistent with the UDHB (under-dehydrated hybrogen bonds; see ref. 5).
Conclusions
The role played by hot spots in protein–protein associations and their contribution to the interaction energy has already been documented (17, 50). Here we show that hot spots are likely to be conserved and that these structurally conserved residues differentiate between binding sites and the remainder of the molecular surface.
Addressing the problem of residue conservation on surfaces and in interfaces in members of protein families necessitates a structural comparison method that is amino acid sequence order-independent. Furthermore, to uniquely derive the interchanges (and conservation) within an ensemble of molecules necessitates a multiple structure comparison method. Initiating from a pair-alignment seed may cause a bias. Hence, MUSTA (29, 30) is well suited to such a task, simultaneously comparing all molecules.
We find that the number of conserved polar residue hot spots is a function of the interface size. This could either show that in our data set this number has not yet reached a plateau or, alternatively, it may reflect a compromise between protein interactions and protein flexibility. The most conserved polar residues at binding interfaces confer rigidity to minimize the entropic cost on binding, whereas the surrounding residues provide a flexible cushion.
Although here we have analyzed a single site per protein on each of the molecules, in vivo each protein interacts with other proteins/ligands/cofactors on its surface. A protein should be considered in terms of its multiple-molecule interactions. Because the secondary sites are unknown, here we distinguished between the primary site, defined in the crystal structure and the remainder of the molecular surface. Our analysis did not yield particular constellations of conserved residues with respect to each other, which could be used as templates to search for additional sites. Nevertheless, multiple structural superpositions yielding conserved residues on the protein surfaces may suggest the existence of such sites. This especially holds if one observes the conservation of Trp. From Fig. 2b, the conservation of Trp on the protein surface indicates a highly possible binding site. To a lesser extent, conservation of Phe and Met also imply a binding site. For all three residues, there is a significant conservation in binding sites, whereas there is no conservation on the exposed surface. Furthermore, our finding that similar residue hot spots occur across different protein families suggests that affinity and specificity are not necessarily coupled.
Hence, to conclude, structurally conserved residues distinguish between binding sites and exposed protein surfaces. This finding may have profound implications for binding-site prediction and for drug design. Drugs can be designed not only to bind at the predicted site, but to specifically target hot spot residues.
Acknowledgments
We thank members of the Nussinov–Wolfson structural bioinformatics group at Tel Aviv University. We thank Drs. C.-J. Tsai and S. Kumar for their help. We thank Dr. Jacob V. Maizel for discussions and encouragement. The research of H.W. and R.N. in Israel has been supported in part by the “Center of Excellence in Geometric Computing and Its Applications” funded by the Israel Science Foundation (administered by the Israel Academy of Sciences). The research of H.W. is partially supported by the Hermann Minkowski–Minerva Center for Geometry at Tel Aviv University. This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. NO1-CO-12400.
Abbreviation
- MUSTA
multiple structure alignment
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Ringe D. Curr Opin Struct Biol. 1995;5:825–829. doi: 10.1016/0959-440x(95)80017-4. [DOI] [PubMed] [Google Scholar]
- 2.Halperin I, Ma B, Wolfson H J, Nussinov R. Proteins. 2002;47:409–443. doi: 10.1002/prot.10115. [DOI] [PubMed] [Google Scholar]
- 3.Schmitt S, Kuhn D, Klebe G. J Mol Biol. 2002;323:387–406. doi: 10.1016/s0022-2836(02)00811-2. [DOI] [PubMed] [Google Scholar]
- 4.Ofran Y, Rost B. J Mol Biol. 2003;325:377–387. doi: 10.1016/s0022-2836(02)01223-8. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez A, Scheraga H A. Proc Natl Acad Sci USA. 2003;100:113–118. doi: 10.1073/pnas.0136888100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laskowski R A, Luscombe N M, Swindells M B, Thornton J M. Protein Sci. 1996;5:2438–2452. doi: 10.1002/pro.5560051206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peters K P, Fauck J, Frommel C. J Mol Biol. 1996;256:201–213. doi: 10.1006/jmbi.1996.0077. [DOI] [PubMed] [Google Scholar]
- 8.Pettit F K, Bowie J U. J Mol Biol. 1999;285:1377–1382. doi: 10.1006/jmbi.1998.2411. [DOI] [PubMed] [Google Scholar]
- 9.Connolly M. Biopolymers. 1986;25:1229–1247. doi: 10.1002/bip.360250705. [DOI] [PubMed] [Google Scholar]
- 10.Jones S, Thornton J M. Proc Natl Acad Sci USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones S, Thornton J M. J Mol Biol. 1997;272:121–132. doi: 10.1006/jmbi.1997.1234. [DOI] [PubMed] [Google Scholar]
- 12.Jones S, Thornton J M. J Mol Biol. 1997;272:133–143. doi: 10.1006/jmbi.1997.1233. [DOI] [PubMed] [Google Scholar]
- 13.Norel R, Petrey D, Wolfson H, Nussinov R. Proteins. 1999;36:307–317. [PubMed] [Google Scholar]
- 14.Xu D, Lin S L, Nussinov R. J Mol Biol. 1997;265:68–84. doi: 10.1006/jmbi.1996.0712. [DOI] [PubMed] [Google Scholar]
- 15.Tsai C J, Lin S L, Wolfson H J, Nussinov R. Protein Sci. 1997;6:53–64. doi: 10.1002/pro.5560060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsai C J, Nussinov R. Protein Sci. 1997;6:1426–1437. doi: 10.1002/pro.5560060707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clackson T, Wells J A. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 18.DeLano W L. Curr Opin Struct Biol. 2002;12:14–20. doi: 10.1016/s0959-440x(02)00283-x. [DOI] [PubMed] [Google Scholar]
- 19.Bogan A A, Thorn K S. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 20.Massova I, Kollman P A. J Am Chem Soc. 1999;121:8133–8143. [Google Scholar]
- 21.Kortemme T, Baker D. Proc Natl Acad Sci USA. 2002;99:14116–14121. doi: 10.1073/pnas.202485799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verkhivker G M, Bouzida D, Gehlhaar D K, Rejto P A, Freer S T, Rose P W. Proteins. 2002;48:539–557. doi: 10.1002/prot.10164. [DOI] [PubMed] [Google Scholar]
- 23.DeLano W L, Ultsch M H, de Vos A M, Wells J A. Science. 2000;287:1279–1283. doi: 10.1126/science.287.5456.1279. [DOI] [PubMed] [Google Scholar]
- 24.Hu Z, Ma B, Wolfson W, Nussinov R. Proteins. 2000;39:331–342. [PubMed] [Google Scholar]
- 25.Tsai C J, Lin S L, Wolfson H, Nussinov R. J Mol Biol. 1996;260:604–620. doi: 10.1006/jmbi.1996.0424. [DOI] [PubMed] [Google Scholar]
- 26.Nussinov R, Wolfson H J. Proc Natl Acad Sci USA. 1991;88:10495–10499. doi: 10.1073/pnas.88.23.10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valdar W S J, Thornton J M. Proteins. 1991;42:108–124. [PubMed] [Google Scholar]
- 28.Brinda K, Kannan N, Vishveshwara S. Protein Eng. 2002;15:265–277. doi: 10.1093/protein/15.4.265. [DOI] [PubMed] [Google Scholar]
- 29.Leibowitz N, Fligelman Z, Nussinov R, Wolfson H. Proteins. 2001;43:235–245. doi: 10.1002/prot.1034. [DOI] [PubMed] [Google Scholar]
- 30.Leibowitz N, Nussinov R, Wolfson H. J Comp Biol. 2001;8:93–121. doi: 10.1089/106652701300312896. [DOI] [PubMed] [Google Scholar]
- 31.Bernstein F C, Koetzle T F, Williams G J B, Meyer E F, Jr, Brice M D, Rodgers J R, Kennard O, Shimanouchi T, Tasumi M. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 32.Lee B, Richards F M. J Mol Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 33.Chotia C. J Mol Biol. 1975;105:1–14. [Google Scholar]
- 34.Shatsky M, Nussinov R, Wolfson H. Algorithms in Bioinformatics, Lecture Notes In Computer Science. Vol. 2452. Berlin: Springer; 2002. pp. 235–250. [Google Scholar]
- 35.Kuhlmann U C, Pommer A J, Moore G R, James R, Kleanthous C. J Mol Biol. 2000;301:1163–1178. doi: 10.1006/jmbi.2000.3945. [DOI] [PubMed] [Google Scholar]
- 36.Li W, Hamill S J, Hemmings A M, Moore G R, James R, Kleanthous C. Biochemistry. 1998;37:11771–11779. doi: 10.1021/bi9808621. [DOI] [PubMed] [Google Scholar]
- 37.Wallis R, Leung K Y, Osborne M, Moore G R, James R, Kleanthous C. Biochemistry. 1998;37:476–485. doi: 10.1021/bi971884a. [DOI] [PubMed] [Google Scholar]
- 38.Samanta U, Pal D, Chakrabarti P. Proteins. 2000;38:288–300. [PubMed] [Google Scholar]
- 39.Fernandez A. Protein Eng. 2002;15:1–16. doi: 10.1093/protein/15.1.1. [DOI] [PubMed] [Google Scholar]
- 40.Lichtarge O, Bourne H R, Cohen F E. J Mol Biol. 1996;257:342–358. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- 41.Madabushi S, Yao H, Marsh M, Kristensen D M, Philippi A, Sowa M E, Lichtarge O. J Mol Biol. 2002;316:139–154. doi: 10.1006/jmbi.2001.5327. [DOI] [PubMed] [Google Scholar]
- 42.Friedberg I, Margalit H. Protein Sci. 2002;11:350–360. doi: 10.1110/ps.18602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuntz I D, Chen K, Sharp K A, Kollman P A. Proc Natl Acad Sci USA. 1999;96:9997–10002. doi: 10.1073/pnas.96.18.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sundberg E J, Mariuzza R A. Structure (London) 2000;8:R137–R142. doi: 10.1016/s0969-2126(00)00167-2. [DOI] [PubMed] [Google Scholar]
- 45.Ma B, Wolfson H, Nussinov R. Curr Opin Struct Biol. 2001;11:364–369. doi: 10.1016/s0959-440x(00)00216-5. [DOI] [PubMed] [Google Scholar]
- 46.Ma B, Shatsky M, Wolfson H, Nussinov R. Protein Sci. 2002;11:184–197. doi: 10.1110/ps.21302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Regenmortel M H V. J Mol Recognit. 1999;12:1–2. doi: 10.1002/(SICI)1099-1352(199901/02)12:1<1::AID-JMR449>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 48.Demirel M C, Atilgan A R, Jernigan R L, Erman B, Bahar I. Protein Sci. 1998;7:2522–2532. doi: 10.1002/pro.5560071205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cole C, Warwicker J. Protein Sci. 2002;11:2860–2870. doi: 10.1110/ps.0222702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cunningham B C, Wells J A. J Mol Biol. 1993;234:554–563. doi: 10.1006/jmbi.1993.1611. [DOI] [PubMed] [Google Scholar]