Abstract
Because of their ability to inhibit proteases, protease inhibitors have generally been considered to counteract tumor progression and metastasis. However, expression of serine protease inhibitors (SPIs) in tumors is often associated with poor prognosis of cancer patients. Moreover, there is growing evidence that SPIs may even promote malignancy of cancer cells, opening new avenues for their use as biomarkers in malignancy. To isolate cancer promoting genes, we applied the suppression subtractive hybridization method to low-malignant Lewis Lung Carcinoma 3LL-S versus high-malignant 3LL-S-sc cells. This resulted in the identification of the SPI secretory leukocyte protease inhibitor (SLPI), as one of the genes whose expression was higher in 3LL-S-sc than in 3LL-S cells. By stable transfection of 3LL-S cells with mouse or human SLPI, we demonstrated that elevated levels of SLPI expression increased both the tumorigenicity and lung-colonizing potential of 3LL-S cells. Moreover, we showed that this function of SLPI depended on its protease inhibitory capacity. Our results also reveal that although SLPI enhanced the proliferation of 3LL-S cells in vitro, its promalignant activity in vivo was not solely due to its effect on cell proliferation. In this study, we report a causal role for SLPI in the malignant behavior of cancer cells, underscoring the potential malignancy-promoting activities of SPIs.
Tumor progression is generally associated with extensive tissue remodeling to provide a proper environment for tumor growth, angiogenesis, invasion, and metastasis of cancer cells (1). An impressive amount of data reveal that, among many factors, proteases expressed by cancer and/or stromal cells are key players in this process. Indeed, because of their ability to activate and release cytokines and growth factors and to degrade components of the extracellular matrix, proteases are necessary to provide optimal conditions for growth and invasion of cancer and endothelial cells. Expression of corresponding protease inhibitors in tumors is one way to control the activity of these enzymes. Protease inhibitors are therefore expected to be antimalignant (2). However, serine protease inhibitors (SPIs) are often overexpressed in different tumor types (3–7), suggesting that overexpression of these inhibitors might favor tumor progression (8). Indeed, it has been demonstrated that overexpression of a number of SPIs from the serpin and kunitz families results in enhancement of cancer cell malignancy (9–12). None of the kazal-type SPIs has yet been shown to promote malignancy of cancer cells.
Secretory leukocyte protease inhibitor (SLPI) is a member of the kazal-type SPI family. SLPI inhibits elastase, cathepsin G, trypsin, and chymotrypsin (13) and plays a significant role in protection against neutrophil proteases during massive inflammatory responses (14–17). The function of SLPI has been the subject of extensive investigation, because besides its function as an inhibitor of inflammatory proteases, SLPI exerts pleiotropic activities in different biological systems. For example, SLPI promotes wound healing (18) and in vitro cell proliferation (19, 20), inhibits HIV infection (21) and NF-κB activation (22), lyses bacteria (23), and modulates macrophage functions (24). Some of the activities of SLPI are independent of its protease inhibitory capacity toward certain proteases (21–24).
Several studies have reported a direct correlation between SLPI expression levels and tumor progression (7, 25–28). However, it has not yet been demonstrated whether SLPI contributes to the malignant phenotype of cancer cells.
In this study, we demonstrate that SLPI plays a causal role in the malignant behavior of Lewis lung carcinoma 3LL-S cells. Moreover, we show that this function of SLPI depends on its protease-inhibitory activity, but not on its ability to enhance cell proliferation.
Materials and Methods
Mice.
Six- to 8-week-old female C57BL/6 (Harlan, Horst The Netherlands) and CB17/IcrHanHsd-SCID mice (Harlan) were used in all experiments.
Cell Lines and Culture Conditions.
The 3LL-S cell line has been described elsewhere (29). The 3LL-S-sc cell line was obtained by s.c. inoculation of 2 × 106 3LL-S cells in C57BL/6 mice, followed by removal and homogenization of the resulting tumor tissue and in vitro propagation of cancer cells for at least 10 days to eliminate contaminating host cells. The human lung carcinoma cell line A549 was kindly provided by M. Mareel (Ghent University, Ghent, Belgium). All cell lines were maintained in RPMI medium 1640 supplemented with 0.3 mg/ml l-glutamine/100 units/ml penicillin/0.1 mg/ml streptomycin/10% heat-inactivated FCS (GIBCO/BRL). Cells were grown in a humidified incubator at 37°C, containing 5% CO2.
General Molecular Techniques.
Unless otherwise noted, nucleic acids were handled according to standard protocols. PCR products were purified by using the PCR Purification kit (Qiagen) as recommended by the manufacturer. Nucleotide sequences were determined by the dideoxynucleotide chain termination method. Nucleic acid homology searches were performed using the FastA program. Total RNA and mRNA were prepared using TRIzol reagent (GIBCO/BRL) and Fasttrack 2.0 kit (Invitrogen), respectively, following the suppliers' recommendations.
Construction and Screening of a Subtracted cDNA Library.
A subtracted cDNA repertoire enriched for cDNA fragments up-regulated in 3LL-S-sc, as compared with 3LL-S cells, was generated using the PCR-Select cDNA Subtraction kit (CLONTECH), as instructed by the manufacturers. The cDNAs obtained from 3LL-S and 3LL-S-sc cells were used as driver and tester, respectively. The subtracted cDNA repertoire was cloned into the T/A cloning vector pCR2.1 (Invitrogen) and transformed into E. coli strain TOP10F′ (Invitrogen). Differential expression of cloned cDNA fragments was tested by Northern blot using standard protocols. Probes were generated by PCR amplification of cDNA inserts and labeled using the Rediprime II random prime labeling system (Amersham Pharmacia). The membranes were exposed to a phosphor-imaging screen and developed using the Molecular Imager system (Bio-Rad). The specific signals were quantified using the Molecular Analyst software (Bio-Rad). The signals were normalized using the house-keeping gene GAPDH.
Tumorigenicity.
A total of 2 × 106 cells were injected s.c. in the flank, and tumor length (L) and width (W) were measured at different time points by using a caliper. The tumor volume (V) was calculated as V = W × W × L × 0.4.
Evaluation of Experimental Metastatic Potential.
A total of 2 × 106 cells were injected i.v. via the tail vein. Lung-colonizing potential was measured by monitoring the lung weight and number of visible metastatic nodules after fixation in Bouin's solution (Sigma).
Transfection of 3LL-S Cells with mSLPI, hSLPI, F-hSLPI or R-hSLPI.
By using primers 5′-CGGAATTCCAGAGCTCCCCTGCCTTC-3′ and 5′-GCTCTAGACATAGAGAAATGAATGCGTTT-3′, the full-length mSLPI cDNA (including the signal peptide and the 3′ untranslated region) was obtained by RT-PCR on mRNA from 3LL-S cells. The full-length hSLPI cDNA was obtained by RT-PCR on total RNA from A549 cells using primers 5′-CGGAATTCCAGAGTCACTCCTGCCTTC-3′ and 5′-GCTCTAGACAAAGAGAAATAGGCTCGTTT-3′. By using primer pairs 5′-GAAATTGGGGGGGTTAAGCATGAAACATTGGCC-3′ and 5′-GGCCAATGTTTCATGCTTAACCCCCCCAATTTC-3′, or 5′-GGGGGTTAAGCATCCTACATTGGCCATAAGTC-3′ and 5′-GACTTATGGCCAA- TGTAGGATGCTTAACCCCC-3′, the codon for Leu-72 of the mature hSLPI protein was mutated via PCR into a codon for Phe (F-hSLPI) or Arg (R-hSLPI), respectively (the nucleotides replacements are shown in bold). PCR products were cloned into the EcoRI/XbaI sites of the pcDNA3.1(+)/Neo plasmid (Invitrogen). After sequence verification, the recombinant plasmids containing mSLPI, hSLPI, F-hSLPI, or R-hSLPI cDNA, in parallel with the empty plasmid, were electroporated into 3LL-S cells following standard protocols. Subcloning and selection in the presence of neomycin (GIBCO/BRL) resulted in the isolation of stable transfectants. mSLPI expression in transfectants was evaluated by Northern blot. Each Northern blot was repeated three times. hSLPI, F-hSLPI, or R-hSLPI secretion was evaluated by using the “human SLPI ELISA Test kit” (HyCult Biotechnology, Uden, The Netherlands). Three independent ELISAs were performed.
In Vitro Cell Proliferation Assay.
Exponentially growing cancer cells were collected, thoroughly washed in RPMI medium 1640 and incubated for 24 h in serum-free medium to synchronize the cells. The cells were collected, resuspended in serum-containing medium and seeded for 24 h in 6-fold at 104 cells per well in 96-well plates. Cell proliferation was quantified in an 18-h [3H]thymidine incorporation assay.
Statistical Analysis.
Statistical analyses were performed by the two-tailed unpaired t test.
Results
s.c. Growth of 3LL-S Cells Enhances Their Malignancy.
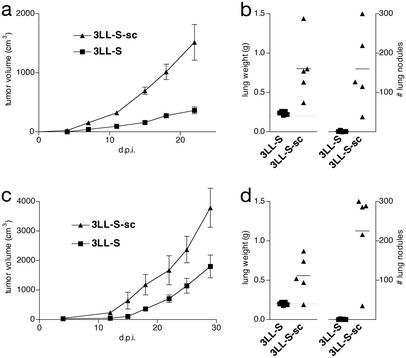
The 3LL-S cell line is a low-malignant subclone derived from the parental Lewis Lung Carcinoma (29). The low malignancy of these cells is reflected by their low tumorigenicity on s.c. inoculation (Fig. 1 a and c) and low lung-colonizing potential after i.v. injection (Fig. 1 b and d), in both syngeneic C57BL/6 mice (Fig. 1 a and b) and immune-deficient SCID mice (Fig. 1 c and d). On s.c. growth in syngeneic C57BL/6 mice, 3LL-S cells become more malignant. Indeed, as compared with the parental 3LL-S cells, cancer cells derived from s.c. 3LL-S tumors (hereafter referred to as 3LL-S-sc cells) grow significantly faster in the flank of mice (Fig. 1 a and c). In addition, 3LL-S-sc cells colonize the lung more extensively than 3LL-S cells after i.v. injection (Fig. 1 b and d). These data show that 3LL-S-sc cells are significantly more malignant than 3LL-S cells, as manifested by their increased capacity to grow at a local site and to colonize the lung.
Figure 1.
Malignant potential of 3LL-S and 3LL-S-sc cells. (a) s.c. growth of 3LL-S and 3LL-S-sc cells in C57BL/6 mice (P = 0.0056 at 22 days postinoculation, d.p.i.). (b) Lung-colonizing potential of 3LL-S and 3LL-S-sc cells in C57BL/6 mice at 32 d.p.i. (P = 0.013 and 0.0081 for lung weight and number of lung nodules, respectively). (c) s.c. growth of 3LL-S and 3LL-S-sc cells in SCID mice (P = 0.032 at 29 d.p.i.). (d) Lung-colonizing potential of 3LL-S and 3LL-S-sc cells in SCID mice at 21 d.p.i. (P = 0.016 and 0.0020 for lung weight and number of lung nodules, respectively).
Mouse SLPI Expression Is Up-Regulated During s.c. Growth of 3LL-S Cells.
To identify genes whose expression is modulated during s.c. growth of 3LL-S cells, the suppression subtractive hybridization approach was adopted. This approach led to the identification of a 480-bp cDNA fragment corresponding to the 3′ fragment of the mouse SLPI (mSLPI) mRNA (13).
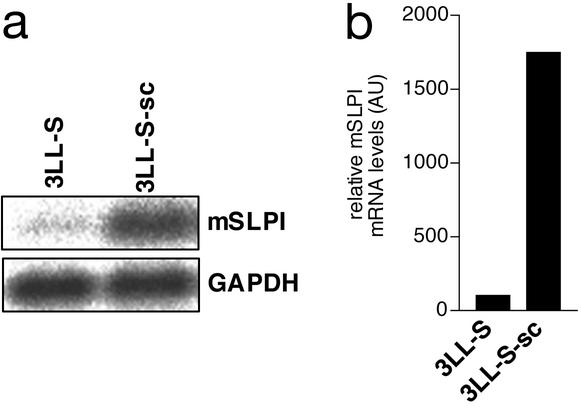
The up-regulation of mSLPI expression on s.c. growth of 3LL-S cells was further validated by Northern blot. These Northern blot experiments (Fig. 2a) and subsequent normalization with the housekeeping gene GAPDH, revealed that the mSLPI mRNA level was ≈15-fold higher in 3LL-S-sc cells as compared with 3LL-S cells (Fig. 2b).
Figure 2.
mSLPI expression in 3LL-S and 3LL-S-sc cells. (a) Northern blot analysis of expression of mSLPI and GAPDH. (b) Normalized mSLPI mRNA levels. The relative quantities of mSLPI mRNA were determined by densitometry and normalized by using GAPDH.
Mouse SLPI Overexpression Enhances the Malignancy of 3LL-S Cells.
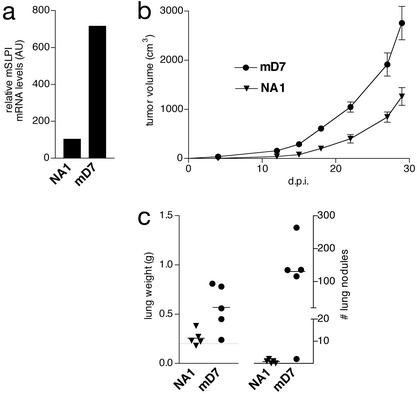
The above experiments revealed a direct correlation between mSLPI expression levels and the malignant behavior of 3LL-S and 3LL-S-sc cells. We next investigated whether elevated levels of mSLPI expression enhanced the tumorigenicity and/or lung-colonizing potential of 3LL-S cells. To this end, 3LL-S cells were transfected with a plasmid expressing mSLPI. As negative control-transfectant, the empty plasmid was introduced into 3LL-S cells. The stable mSLPI-transfectant mD7, in which the mSLPI mRNA level was ≈7-fold higher than that in 3LL-S cells, was selected for further analysis (Fig. 3a). The control-transfectant clone NA1, with mSLPI mRNA levels similar to that of 3LL-S, was used as negative control.
Figure 3.
mSLPI overexpression enhances the malignancy of 3LL-S cells (a) Normalized mSLPI mRNA levels in the mock-transfectant NA1 and mSLPI-transfectant mD7. The relative quantities of mSLPI mRNA were determined by densitometry and normalized by using GAPDH. (b) s.c. growth of NA1 and mD7 in SCID mice (P = 0.0011 at 27 d.p.i.). (c) Lung-colonizing potential of NA1 and mD7 in SCID mice at 36 d.p.i. (P = 0.023 and 0.014 for lung weight and number of lung nodules, respectively).
The role of mSLPI in increasing malignancy of 3LL-S cells was tested by measuring the tumorigenicity and lung-colonizing potential of the mSLPI overexpressing clone mD7 and the control mock-transfectant clone NA1. As shown in Fig. 3, a 7-fold mSLPI overexpression significantly enhanced tumor growth (Fig. 3b) and lung-colonizing potential (Fig. 3c) of 3LL-S cells injected s.c. or i.v., respectively.
Human SLPI (hSLPI) Expression in 3LL-S Cells Enhances Their Malignancy.
Although mSLPI and hSLPI exhibit only 58% identity at the amino acid level, the proteases they inhibit are similar (30). Besides, it has been shown that, similar to mSLPI, hSLPI is up-regulated during cancer progression (25, 27). Hence, we investigated whether, similar to mSLPI, hSLPI also promotes the malignancy of 3LL-S cells.
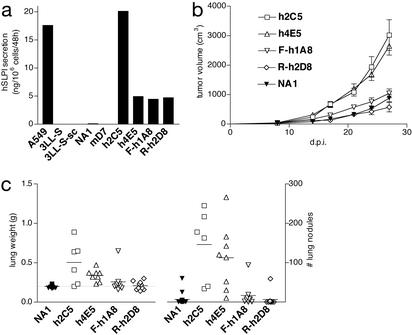
To assess the malignancy-promoting activity of hSLPI, 3LL-S cells were transfected with a plasmid expressing hSLPI. Based on ELISA, two stable hSLPI-transfectants, clones h2C5 and h4E5, secreting ≈20 and 5 ng of hSLPI per 106 cells per 48 h, respectively, were selected for further analysis. Conditioned medium from the human lung cancer cell line A549 was used as a positive control in ELISA. 3LL-S, 3LL-S-sc cells, mSLPI-transfectant clone mD7 and the control-transfectant clone NA1 did not yield any ELISA signal in these experiments, demonstrating that ELISA signals obtained with hSLPI-transfectants were specific for hSLPI (Fig. 4a).
Figure 4.
The promalignant effect of hSLPI depends on its protease inhibitory activity. (a) Secretion levels of hSLPI, F- or R-hSLPI by A549, 3LL-S, and 3LL-S-sc cells, 3LL-S mock-transfectant NA1, mSLPI-transfectant mD7, hSLPI-transfectants h2C5 and h4E5, F-hSLPI-transfectant F-h1A8, and R-hSLPI-transfectant R-h2D8. (b) s.c. growth of NA1, h2C5, h4E5, F-h1A8, and R-h2D8 in SCID mice (P = 0.0003 and 0.0001 for h2C5 and h4E5, respectively, as compared with NA1. P = 0.0063 and 0.0012 for F-h1A8 and R-h2D8, respectively, as compared with h4E5). P values were calculated from the data at 27 d.p.i. (c) Lung-colonizing potential of NA1, h2C5, h4E5, F-h1A8, and R-h2D8 in SCID mice at 36 d.p.i. (lung weight: P < 0.0001 for h2C5 and h4E5, as compared with NA1. P = 0.19 and 0.0007 for F-h1A8 and R-h2D8, respectively, as compared with h4E5. Number of lung nodules: P < 0.0001 for h2C5 and h4E5, as compared with NA1. P = 0.0054 and 0.0012 for F-h1A8 and R-h2D8, respectively, as compared with h4E5).
The effect of hSLPI on the malignancy of 3LL-S cells was then tested by measuring the tumorigenicity and lung-colonizing potential of the hSLPI-expressing clones h2C5 and h4E5 and the control mock-transfectant clone NA1. Similar to the mSLPI-transfectant mD7, the hSLPI-transfectants h2C5 and h4E5 grew much faster in the flank of mice than the mock-transfectant NA1 (Fig. 4b). As measured by both the number of lung nodules and lung weight, both hSLPI-transfectants exhibited a significantly higher lung-colonizing potential as compared with the mock-transfectant clone NA1 (Fig. 4c). A hSLPI secretion level of ≈5 ng per 106 cells per 48 h was sufficient to enhance the malignancy of 3LL-S cells; indeed, although clones h2C5 and h4E5 differed ≈4-fold in their hSLPI secretion levels, they did not differ significantly in their tumorigenicity (P = 0.52) and lung-colonizing potential (P = 0.12 and 0.48 for lung weight and number of lung nodules, respectively). Therefore, despite the differences in their amino acid sequences, both mouse and human SLPIs enhance the malignant potential of 3LL-S cells.
The Protease Inhibitory Activity of hSLPI Is Involved in Its Malignancy-Promoting Capacity.
To assess the role of the protease-inhibitory activity of hSLPI in its capacity to promote malignancy of 3LL-S cells, two mutant hSLPIs were generated. In these mutants, Leu-72 of the mature wild-type hSLPI protein was replaced by Phe (in F-hSLPI) or Arg (in R-hSLPI). These mutations have already been shown to result in a drastic alteration in the inhibitory activity of hSLPI toward serine proteases (31).
Plasmids expressing each of these mutants were used to transfect 3LL-S cells. Two transgenic cell lines, F-hSLPI-transfectant F-h1A8 and R-hSLPI-transfectant R-h2D8, having expression levels similar to that of the WT hSLPI-transfectant h4E5, were selected for further study (Fig. 4a). The transfectants F-h1A8 and R-h2D8 were compared with the mock-transfectant NA1 and the hSLPI-transfectant h4E5 for their capacity to colonize the lung and to grow locally. As depicted in Fig. 4b, both mutant hSLPI-transfectants grew significantly slower than the hSLPI-transfectant h4E5 and exhibited growth curves similar to the mock-transfectant NA1. Moreover, after i.v. injection, both mutant hSLPI-transfectants F-h1A8 and R-h2D8 colonized the lung less efficiently than the WT hSLPI-transfectant h4E5. This was reflected by both a decreased number of lung nodules and a lower lung weight, the latter to a lesser extent (Fig. 4c). These experiments demonstrate that the protease inhibitory activity of hSLPI is involved in the promotion of 3LL-S malignancy.
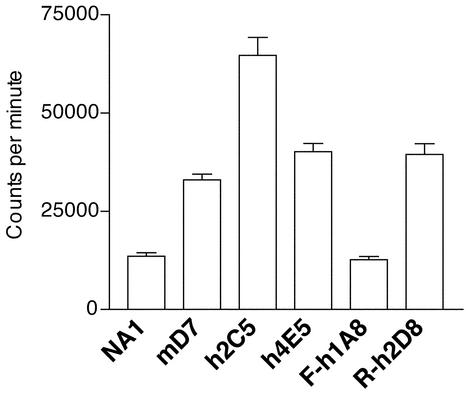
The Promalignant Activity of SLPI Is Not Mediated by Its Effect on in Vitro Cell Proliferation.
In view of two recent reports linking SLPI expression with in vitro proliferation rates of human endometrial cells (19, 20), the influence of SLPI on 3LL-S cell proliferation was tested. To this end, in vitro proliferation rates of mock and SLPI transfectants were compared. SLPI-transfectant clones mD7, h2C5, and h4E5 proliferated, respectively, 2.4, 4.8, and 3.0 times faster than the mock-transfectant NA1, demonstrating that SLPI indeed promotes the proliferation of 3LL-S cells in vitro (Fig. 5). When Leu-72 was mutated to Phe, the in vitro growth-stimulating effect of hSLPI was abrogated: transfectant F-h1A8 proliferated significantly slower than the hSLPI-transfectant h4E5 and exhibited the same proliferation rate as the mock-transfectant NA1. However, replacement of Leu-72 by Arg did not change the effect of hSLPI on the proliferation of 3LL-S cells; indeed, transfectant R-h2D8 proliferated as fast as the hSLPI-transfectant h4E5 and significantly faster than the mock-transfectant NA1 (Fig. 5). Taking together these data and the in vivo malignancy of these cells, there is not always a direct correlation between the in vitro proliferation rate of these cells and their in vivo malignant behavior. Therefore, the promalignant activity of SLPI cannot be explained solely by its effect on in vitro cell proliferation.
Figure 5.
Effect of SLPI expression on the in vitro cell proliferation of 3LL-S cells. Cell proliferation rates of transfected 3LL-S cells were measured by [3H]thymidine uptake. The data shown are representative of five independent experiments. P < 0.0001 for mD7, h2C5, h4E5, and R-h2D8 and P = 0.4922 for F-h1A8, as compared with NA1. P < 0.0001 for F-h1A8 and P = 0.8381 for R-h2D8, as compared with h4E5.
Discussion
As reported for other SPIs (8), several studies have demonstrated that SLPI is up-regulated in the course of cancer development: (i) SLPI has been identified as highly up-regulated in ovarian carcinomas as compared with nontransformed ovarian epithelia (7, 25, 26). (ii) It has been shown that patients with nonsmall cell lung carcinoma have higher serum SLPI levels than healthy individuals, with concomitant high hSLPI expression in tumor tissue. Furthermore, serum SLPI levels in these patients correlated with tumor stage and response to therapy (27). (iii) It has been reported that mouse SLPI mRNA levels directly correlate with the metastatic potential of liver carcinoma cells (28). Although these studies showed that SLPI is overexpressed in tumor tissue or in malignant cancer cells, the significance of SLPI up-regulation in tumor progression and cancer cell malignancy has not yet been reported.
In the present study, we demonstrated that as compared with 3LL-S cells, 3LL-S-sc cells have a higher capacity to grow locally and to colonize the lung. This increased malignancy of 3LL-S-sc cells was shown to be associated with elevated levels of mSLPI in these cells. A comparable correlation between mSLPI expression and malignant behavior of liver carcinoma cells has also been made by Morita et al. (28). They reported that besides mSLPI, described by Zitnik et al. (13) and also identified in our study, a differentially spliced variant of mSLPI, mSLPI-β, coding for a protein with a hydrophilic signal peptide, is expressed in liver carcinoma cells. However, RT-PCR and RACE experiments revealed that mSLPI-β is not expressed in our tumor model (our unpublished data).
We used SLPI-transfectants to demonstrate that overexpression of either mSLPI or hSLPI was sufficient to enhance the tumorigenicity and lung-colonizing potential of 3LL-S cells. These experiments revealed that SLPI plays a causal role in the malignant phenotype of 3LL-S cells, a function of SLPI that has previously not been reported. Our results also show that although mouse and human SLPI share only 58% amino acid identity (13), both orthologues increase s.c. tumor growth and experimental metastasis of transfected 3LL-S cells.
As compared with 3LL-S-sc cells, SLPI-transfectants metastasize to the lung and grow locally at a slower rate. The differences in the kinetics of metastasis formation and tumor growth between SLPI-transfectants and 3LL-S-sc cells can be explained by the fact that the expression of genes other than mSLPI is also affected when 3LL-S cells grow s.c.; for example, S100A4, which has already been demonstrated to be a metastasis-promoting gene in 3LL cells (32), was among several genes isolated from our suppression subtractive hybridization library and showed elevated mRNA levels in 3LL-S-sc versus 3LL-S cells (data not shown).
SLPI is a boomerang-shaped molecule belonging to the kazal-family of SPIs and consists of two homologous whey acidic protein domains of similar size (33). Crystallographic and enzyme kinetics analyses have located the protease inhibitory activity of hSLPI to its C-terminal domain (33, 34). Mutation of Leu-72 in the C-terminal half of the hSLPI protein to Phe (F-hSLPI) or Arg (R-hSLPI) alters the Ki toward different serine proteases (31). However, these mutants do not differ from the wild-type hSLPI in their ability to inhibit the activation of NF-κB (35), modulate the phenotype of macrophages (24), or inhibit HIV infectivity of monocytes (21), suggesting that the serine proteases toward which the inhibitory activity is affected in these hSLPI mutants do not play a role in the above-mentioned biological activities. In our model, replacement of Leu-72 by Phe or Arg had a dramatic effect on the malignancy-promoting activity of hSLPI. Indeed, although hSLPI expression in 3LL-S cells enhances the capacity of these cells to grow locally and to metastasize to distant organs, F- and R-hSLPI expression does not significantly influence the phenotype of 3LL-S cells. Hence, the promalignant activity of hSLPI is related to its capacity to inhibit serine proteases.
It has been reported that exogenous human or porcine SLPI and endogenous human SLPI promote the in vitro proliferation of endometrial epithelial cells (19, 20). Similar to these studies, we observed that both wild-type mouse and wild-type human SLPI enhanced the proliferation of 3LL-S cells in vitro. Moreover, in vitro proliferation rates of WT SLPI-transfectants and the mutant SLPI-transfectant F-h1A8 directly correlated with their in vivo malignant behavior. This suggested that the promalignant activity of SLPI could be due to its effects on cell proliferation. However, although clones R-h2D8 and h4E5, expressing similar levels of hSLPI, did not differ in their in vitro proliferation rates, they did differ in their tumorigenicity and lung-colonizing potential. On the other hand, although mock-transfectant NA1 and mutant hSLPI-transfectants F-h1A8 and R-h2D8 exhibited a similar low tumorigenicity and lung-colonizing potential, clone R-h2D8 proliferated faster than clone NA1 and F-h1A8. Therefore, the promalignant activity of SLPI cannot be accounted for by its effect on cell-proliferation. The differences in the effects of these mutations on in vitro growth-stimulating properties of SLPI may be caused by the differences in the extent to which these mutations influence SLPI inhibitory activity (31).
The exact mechanism by which SLPI promotes malignancy is not yet known, but at least one hypothesis can be advanced: it is possible that the enhancement of malignancy by SLPI is due to its effects on angiogenesis. Endostatin, a potent antiangiogenic factor (36), is generated by elastase (37). Because both mSLPI and hSLPI are potent inhibitors of elastase (30), and as compared with wild-type hSLPI, the mutants F- and R-hSLPI have, respectively, a 500- and 15,000-fold increase in Ki toward elastase (31), it is tempting to speculate that SLPI might enhance malignancy by preventing the formation of the angiogenesis inhibitor endostatin. In line with this hypothesis is the finding that endostatin exhibits potent antitumor activities in the 3LL tumor model (36).
In conclusion, in this report we could unequivocally attribute a previously undescribed activity, the promotion of cancer cell malignancy, to SLPI. In addition, we demonstrated that the promalignant activity of hSLPI is related to its capacity to inhibit proteases, but not to its proliferation-stimulating properties. Because SLPI overexpression has been shown to be associated with progression of tumors of different histological origin, it is interesting to assess the contribution of SLPI to the malignant behavior of different tumor types. These experiments will reveal whether the promalignant activity of SLPI we observed in Lewis Lung Carcinoma can be generalized for other tumors.
Acknowledgments
We thank Mr. E. Vercauteren, Miss M. Gobert, and Mrs. E. Omasta for their excellent technical assistance. We are grateful to Dr. M. Mareel for providing the A549 cell line. N.D. is a fellow of the Fonds voor Wetenschappelijk Onderzoek. This project was in part supported by Sportvereniging Tegen Kanker, Stichting Emmanuel van der Schueren, and Krediet aan Navorsers from Fonds voor Wetenschappelijk Onderzoek.
Abbreviations
- d.p.i.
days postinoculation
- SLPI
secretory leukocyte protease inhibitor
- hSLPI
human SLPI
- mSLPI
mouse SLPI
- SPI
serine protease inhibitor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Johnsen M, Lund L R, Romer J, Almholt K, Dano K. Curr Opin Cell Biol. 1998;10:667–671. doi: 10.1016/s0955-0674(98)80044-6. [DOI] [PubMed] [Google Scholar]
- 2.Liotta L A, Kohn E C. Nature. 2001;411:375–379. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- 3.Karashima S, Kataoka H, Itoh H, Maruyama R, Koono M. Int J Cancer. 1990;45:244–250. doi: 10.1002/ijc.2910450207. [DOI] [PubMed] [Google Scholar]
- 4.Higashiyama M, Doi O, Yokouchi H, Kodama K, Nakamori S, Tateishi R. Cancer. 1995;76:1368–1376. doi: 10.1002/1097-0142(19951015)76:8<1368::aid-cncr2820760812>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 5.Grondahl-Hansen J, Christensen I J, Rosenquist C, Brunner N, Mouridsen H T, Dano K, Blichert-Toft M. Cancer Res. 1993;53:2513–2521. [PubMed] [Google Scholar]
- 6.Muller-Pillasch F, Wallrapp C, Bartels K, Varga G, Friess H, Buchler M, Adler G, Gress T M. Biochim Biophys Acta. 1998;1395:88–95. doi: 10.1016/s0167-4781(97)00129-2. [DOI] [PubMed] [Google Scholar]
- 7.Hough C D, Sherman-Baust C A, Pizer E S, Montz F J, Im D D, Rosenshein N B, Cho K R, Riggins G J, Morin P J. Cancer Res. 2000;60:6281–6287. [PubMed] [Google Scholar]
- 8.Kataoka H, Itoh H, Koono M. Pathol Int. 2002;52:89–102. doi: 10.1046/j.1440-1827.2002.01320.x. [DOI] [PubMed] [Google Scholar]
- 9.Kataoka H, Uchino H, Iwamura T, Seiki M, Nabeshima K, Koono M. Am J Pathol. 1999;154:457–468. doi: 10.1016/s0002-9440(10)65292-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsuchiya H, Katsuo S, Matsuda E, Sunayama C, Tomita K, Ueda Y, Binder B R. Gen Diagn Pathol. 1995;141:41–48. [PubMed] [Google Scholar]
- 11.Suminami Y, Nagashima S, Murakami A, Nawata S, Gondo T, Hirakawa H, Numa F, Silverman G A, Kato H. Cancer Res. 2001;61:1776–1780. [PubMed] [Google Scholar]
- 12.Meng J Y, Kataoka H, Itoh H, Koono M. Int J Cancer. 2001;92:31–39. [PubMed] [Google Scholar]
- 13.Zitnik R J, Zhang J, Kashem M A, Kohno T, Lyons D E, Wright C D, Rosen E, Goldberg I, Hayday A C. Biochem Biophys Res Commun. 1997;232:687–697. doi: 10.1006/bbrc.1997.6358. [DOI] [PubMed] [Google Scholar]
- 14.McElvaney N G, Nakamura H, Birrer P, Hebert C A, Wong W L, Alphonso M, Baker J B, Catalano M A, Crystal R G. J Clin Invest. 1992;90:1296–1301. doi: 10.1172/JCI115994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song X, Zeng L, Jin W, Thompson J, Mizel D E, Lei K, Billinghurst R C, Poole A R, Wahl S M. J Exp Med. 1999;190:535–542. doi: 10.1084/jem.190.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lentsch A B, Yoshidome H, Warner R L, Ward P A, Edwards M J. Gastroenterology. 1999;117:953–961. doi: 10.1016/s0016-5085(99)70355-0. [DOI] [PubMed] [Google Scholar]
- 17.Gipson T S, Bless N M, Shanley T P, Crouch L D, Bleavins M R, Younkin E M, Sarma V, Gibbs D F, Tefera W, McConnell P C, et al. J Immunol. 1999;162:3653–3662. [PubMed] [Google Scholar]
- 18.Ashcroft G S, Lei K, Jin W, Longenecker G, Kulkarni A B, Greenwell-Wild T, Hale-Donze H, McGrady G, Song X Y, Wahl S M. Nat Med. 2000;6:1147–1153. doi: 10.1038/80489. [DOI] [PubMed] [Google Scholar]
- 19.Zhang D, Simmen R C, Michel F J, Zhao G, Vale-Cruz D, Simmen F A. J Biol Chem. 2002;277:29999–30009. doi: 10.1074/jbc.M203503200. [DOI] [PubMed] [Google Scholar]
- 20.Badinga L, Michel F J, Simmen R C. Biol Reprod. 1999;61:380–387. doi: 10.1095/biolreprod61.2.380. [DOI] [PubMed] [Google Scholar]
- 21.McNeely T B, Shugars D C, Rosendahl M, Tucker C, Eisenberg S P, Wahl S M. Blood. 1997;90:1141–1149. [PubMed] [Google Scholar]
- 22.Lentsch A B, Jordan J A, Czermak B J, Diehl K M, Younkin E M, Sarma V, Ward P A. Am J Pathol. 1999;154:239–247. doi: 10.1016/s0002-9440(10)65270-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiemstra P S, Maassen R J, Stolk J, Heinzel-Wieland R, Steffens G J, Dijkman J H. Infect Immun. 1996;64:4520–4524. doi: 10.1128/iai.64.11.4520-4524.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, DeWitt D L, McNeely T B, Wahl S M, Wahl L M. J Clin Invest. 1997;99:894–900. doi: 10.1172/JCI119254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hough C D, Cho K R, Zonderman A B, Schwartz D R, Morin P J. Cancer Res. 2001;61:3869–3876. [PubMed] [Google Scholar]
- 26.Shigemasa K, Tanimoto H, Underwood L J, Parmley T H, Arihiro K, Ohama K, O'Brien T J. Int J Gynecol Cancer. 2001;11:454–461. doi: 10.1046/j.1525-1438.2001.01062.x. [DOI] [PubMed] [Google Scholar]
- 27.Ameshima S, Ishizaki T, Demura Y, Imamura Y, Miyamori I, Mitsuhashi H. Cancer. 2000;89:1448–1456. doi: 10.1002/1097-0142(20001001)89:7<1448::aid-cncr6>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 28.Morita M, Arakawa H, Nishimura S. Adv Enzyme Regul. 1999;39:341–355. doi: 10.1016/s0065-2571(98)00020-x. [DOI] [PubMed] [Google Scholar]
- 29.Remels L M, De Baetselier P C. Int J Cancer. 1987;39:343–352. doi: 10.1002/ijc.2910390313. [DOI] [PubMed] [Google Scholar]
- 30.Wright C D, Kennedy J A, Zitnik R J, Kashem M A. Biochem Biophys Res Commun. 1999;254:614–617. doi: 10.1006/bbrc.1998.0108. [DOI] [PubMed] [Google Scholar]
- 31.Eisenberg S P, Hale K K, Heimdal P, Thompson R C. J Biol Chem. 1990;265:7976–7981. [PubMed] [Google Scholar]
- 32.Takenaga K, Nakamura Y, Sakiyama S. Oncogene. 1997;14:331–337. doi: 10.1038/sj.onc.1200820. [DOI] [PubMed] [Google Scholar]
- 33.Grutter M G, Fendrich G, Huber R, Bode W. EMBO J. 1988;7:345–351. doi: 10.1002/j.1460-2075.1988.tb02819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van-Seuningen I, Davril M. Biochem Biophys Res Commun. 1991;179:1587–1592. doi: 10.1016/0006-291x(91)91755-2. [DOI] [PubMed] [Google Scholar]
- 35.Mulligan M S, Lentsch A B, Huber-Lang M, Guo R F, Sarma V, Wright C D, Ulich T R, Ward P A. Am J Pathol. 2000;156:1033–1039. doi: 10.1016/S0002-9440(10)64971-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Reilly M S, Boehm T, Shing Y, Fukai N, Vasios G, Lane W S, Flynn E, Birkhead J R, Olsen B R, Folkman J. Cell. 1997;88:277–285. doi: 10.1016/s0092-8674(00)81848-6. [DOI] [PubMed] [Google Scholar]
- 37.Wen W, Moses M A, Wiederschain D, Arbiser J L, Folkman J. Cancer Res. 1999;59:6052–6056. [PubMed] [Google Scholar]