Abstract
The generic membrane trafficking signals of internal RXR and carboxyl-terminal KKXX motifs direct intracellular endoplasmic reticulum (ER) localization of the signal-bearing proteins. These signaling motifs play a critical role in partitioning proteins into designated subcellular compartments by functioning as an intracellular “zip code.” In the process of determining the potential distinctions between these two otherwise functionally identical motifs, two functional zones of these signals were revealed. The KKXX signal was effective only when it was positioned closer to the membrane surface. In contrast, under identical conditions, the internal RXR signal was functional when it was positioned distally from the membrane. Different from the C-terminal KKXX signal, the internal RXR motif may be present in multiple copies. The receptor with multivalent RXR motifs displayed similar trafficking behavior to that of the same receptor with one copy of the RXR motif. The distinctive operating ranges from their anchored membrane surface provide experimental evidence for the notion that there are functional zoning layers within which membrane protein signal motifs are active.
The diversity of vesicular membrane trafficking is the key underlying biological process responsible for the partition of membrane proteins into different compartments [see reviews by Mellman and Warren (1) and Teasdale and Jackson (2)]. Membrane vesicular trafficking is thought to be mediated largely by signal sequences of membrane-bound proteins that direct the protein-bearing vesicles to specific locations, thereby conferring functional specification of the targeted subcellular compartments. The regulation of signal sequence conferred activity may take place at multiple levels, including genetically, via alternative splicing to include and remove certain signal motifs such as those found in N-methyl-d-aspartate (NMDA) receptor (3), and biochemically, via motif masking/removal which may be induced by, for example, protein–protein interaction or proteolytic digest [see review by Ma and Jan (4)].
The sequence motifs for mediating endoplasmic reticulum (ER) residence include the carboxyl-terminal dilysine KKXX motif, which is thought to function through its interaction with coat protein I (COPI complex; refs. 1 and 5) and the internally positioned RXR motif whose interaction partners remain unknown (6). These signals have been found in a variety of membrane-bound proteins, including receptors and ion-conducting or -transporting proteins. In addition to the sorting function that codes the ER localization of many ER resident proteins, recent evidence suggests that macromolecular complexes on the cell surface, such as ion channels, use intracellular retention signals as a checkpoint to distinguish different assembly intermediates. For example, the physical accessibility of the RXR motif is correlated with different stages of assembled ATP channel complexes. Thus, a change of access to the RXR motif by an appropriate protein interaction could signal the completion of a macromolecular assembly, thereby allowing for ER exit, which often leads to surface expression (6). Similar notions were also proposed for RKR(R) of the γ-aminobutyric acid (GABA) receptor (7) and for the KKXX signal of T cell receptors (8).
Intriguingly, both KKXX and RKR motifs are positioned on the cytoplasmic side of the plasma membrane and seem to function in the same pathway for retention/retrieval of proteins for ER localization. An isoform of the COPI protein exhibits an interaction with the KKXX motif but not with the RXR motif (9), consistent with the notion that the two motifs function via distinct machinery. Furthermore, there is evidence suggesting a COPI-independent ER retrieval pathway (10). Thus, the questions concerning distinct protein machineries and/or mechanisms for the two ER localization motifs remain to be addressed. One hypothesis for the requirement of different signaling machineries for the same process is that the two systems may possess different effectiveness levels in conferring ER residence. Because both signals have been found in integral membrane proteins, we investigated the effective ranges of the two signals with reference to the plasma membrane. Two distinct functional zones were revealed and are defined by the functional effectiveness of KKXX- and RKR-mediated ER localization.
Materials and Methods
Constructs.
To clone the CD4 fusion vectors, the cDNA encoding the human CD4 extracellular and transmembrane domains (amino acid 1–419) was isolated for fusion with the hemagglutinin (HA) epitope (YPYDVPDYA), and subcloned into the HindIII-BamHI sites of pCDNA3.1(+) (Invitrogen). The CD4-HA fragment was obtained by PCR using a sense primer comprising a HindIII site and 19 nucleotides corresponding to the 5′-untranslated region, together with an antisense primer comprising a BamHI site, a HA epitope sequence, and 20 nucleotides corresponding to the transmembrane region (nucleotides 1241–1260), and then ligated into pCDNA3.1(+) at HindIII-BamHI sites. The ER retention signal-containing sequences were cloned from mouse Kir6.2 and yeast wheat germ agglutinin binding protein 1 (WBP1), and fused to the BamHI-EcoRI sites of the above CD4-HA vectors to generate the complete CD4-(HA)1 vectors. The complementary oligonucleotides flanked by BamHI and EcoRI sites for sense and antisense strands, respectively, were annealed to generate the DNA fragments encoding the C-terminal 36 aa of Kir6.2 (LLDALTLASSRGPLRKRSVAVAKAKPKFSISPDSLS) and 10-aa residues of WBP1 (KKLETFKKTN), respectively, and ligated into the BamHI-EcoRI sites of the CD4-(HA)1 vectors. The mutants for these retention signals (RKR to RAA and KKTN to AATN) were created as above by using the oligonucleotides containing these mutations. The vectors with three copies of HA were created by inserting the two tandem HA epitopes flanked by BglII and BamHI into BamHI-digested CD4-(HA)1 vectors. This inactivated the original BamHI site in the CD4-(HA)1 vector and created a new BamHI site after the third HA, which enabled the construction of the CD4-(HA)5 vectors in the same way by again ligating a tandem HA repeat into the BamHI-digested CD4-(HA)3 vectors. For construction of HA-IRK1 chimeras, the HA epitope (YPYDVPDYA) was inserted at amino acid position 117 of mouse IRK1 by sequential overlap extension PCR. The C-terminal fusions were obtained by cloning Fig. 4-referenced coding sequences at PstI site engineered at the last residue. All these IRK1 chimera sequences were inserted to pCDNA3.1(+) vector at HindIII and NotI sites.
Cells and Transfection.
293T and COS-7 were cultured in a 60-mm dish in 3 ml of 50% DMEM/50% F12 medium supplemented with 10% FBS, l-glutamine, and penicillin/streptomycin. The plasmid DNA was transfected with FuGENE6 (Roche Applied Science) or by the calcium phosphate method at 50–70% confluency.
Flow Cytometry and Quantitation of Relative Surface CD4 Level.
At 48 h after transfection with 1 μg of plasmids, the 293T cells were harvested by incubation with PBS (pH 7.3) containing 0.5 mM EDTA for 10 min at 37°C and washed with Hanks' balanced salt solution (HBSS) supplemented with 5 mM Hepes (pH 7.3) and 2% FBS (staining medium). The cells were stained for surface CD4 with FITC-conjugated anti-human CD4 mAb (DAKO) at 10 μg/ml for 20 min at 4°C. After staining, the cells were washed three times with the staining medium and examined for CD4 expression with FACSCalibur (BD Biosciences) by using cellquest software (BD Biosciences). The CD4 expression was quantitated by converting the geometric mean fluorescence for each measurement to the molecules of equivalent soluble fluorochromes (MESF) by using the fluorescence quantitation kit (Bangs Laboratories, Carmel, IN). The MESF obtained from the control sample, which was transfected with pCDNA3.1(+) vector, was subtracted as a background from each test sample. The surface expression level relative to the total expression was determined by dividing the MESF values for RKR and KKXX constructs by those of RAA and AAXX constructs (with the corresponding number of HA copies), respectively. Alternatively, the relative surface expression was determined by dividing the MESF values obtained from nonpermeabilized cells by those from permeabilized cells. To achieve this, the transfected cells were fixed with 4% paraformaldehyde for 20 min at 4°C and either permeabilized or not with 0.025% Triton X-100 in PBS (pH 7.3) for 5 min at room temperature before staining with anti-CD4 antibody for 30 min at 4°C. For staining of surface IRK1 fusion proteins, the 293T cells transfected with 1 μg of plasmid DNA were incubated with rat anti-HA monoclonal antibody (Roche Applied Science) for 1 h at 4°C. On a washing step, the cells were incubated with FITC-goat anti-rat IgG antibody (Jackson ImmunoResearch) for 20 min at 4°C.
Western Blots.
The whole cell extracts were prepared by incubating a similar number of transfected 293T cells with 0.3% Triton X-100 in PBS (pH 7.3) for 15 min at 4°C in the presence of protease inhibitors and sedimenting for 20 min at 11,000 × g at 4°C. Equivalent volumes of the supernatants were resolved in 10% SDS-polyacrylamide gels, transferred to nitrocellulose membrane, and blotted with rabbit polyclonal anti-human CD4 Ab (Santa Cruz Biotechnology) and horse radish peroxidase-conjugated goat anti-rabbit IgG antibody. The immunoblots were developed with the enhanced chemiluminescence system (Amersham Pharmacia Biosciences).
Immunofluorescence and Confocal Microscopy.
The 293T and COS-7 were grown on the cover glass in a 60-mm dish and transfected with 1 μg of plasmids with FuGENE6. After 24–36 h, the cells were fixed in 4% paraformaldehyde for 20 min at 4°C, blocked with 1% FBS in PBS for 1 h at room temperature, and stained with FITC-anti-CD4 mAb for 1 h at 4°C, followed by washing twice with PBS for 10 min. For IRK1 detection, the cells were stained with anti-HA mAb for 1 h at 4°C, followed by FITC-conjugated anti-rat IgG antibody for 20 min at 4°C. Confocal images were taken with the Ultra View Confocal Imaging System (Perkin–Elmer) and processed with photoshop (Adobe Systems, Mountain View, CA).
Results
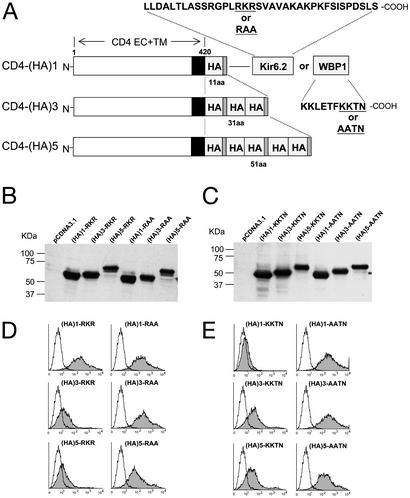
To address the question concerning the effective range, we chose to construct a chimeric type I membrane receptor system (Fig. 1A), which has a CD4 extracellular domain (CD4EC) containing the N-terminal signal peptide. The transmembrane segment is native to the CD4 antigen, consisting of 26 aa, longer than typical ER or Golgi resident membrane proteins, which often have ≈15 aa. The longer transmembrane segment reduces the possible tendency toward ER or Golgi residence as affected by the heterogeneity of lipid thickness in the ER vs. the plasma membrane (11, 12). The intracellular spacer that separates the transmembrane segment from the ER retention/retrieval signals is the tandemly positioned peptide sequence of YPYDVPDYA corresponding to the HA epitope, a viral sequence that is hydrophilic and functions as a linear peptide (13). This sequence is typically positioned extracellularly during cell entry or within the lumen during the viral biogenesis. No endogenous protein has been shown to interact with the HA epitope. Thus, the HA sequence is not likely to recruit any effectors. These presumably trafficking signal-void chimeric receptor cDNAs were expressed in the transfected cells, and the resultant fusion proteins were detected by immunoblot analysis by using an antibody specific for the CD4EC. The protein expression of different forms is comparable between RKR and RAA or KKTN and AATN, with neither degradation nor any detectable heterogeneity of protein modification that is specific for either RXR or KKXX (Fig. 1, B and C). Thus, an effect induced by the fused signals is not likely to originate from heterogeneity of protein synthesis or half-life.
Figure 1.
(A) A schematic diagram showing the coding regions of different CD4 constructs: CD4-(HA)1, CD4-(HA)3, and CD4-(HA)5. Amino acid positions are indicated at the top of the CD4-(HA)1 construct. Each of the CD4 constructs is fused with the listed protein sequences containing either the RKR motif from Kir6.2 (24) or the KKTN motif from WBP1. Specific site-directed mutations change RKR to RAA or KKTN to AATN. The number of residues that separate the transmembrane domain from the 36-aa Kir6.2 motif or WBP1 motif is indicated below each construct. (B and C) Immunoblot analyses of fusion proteins from the transfected cells. Protein lysates of transfected cells are identified by the constructs indicated at the top. The fusion protein expression was detected by anti-CD4 antibody. The molecular mass standard in kDa is indicated on the left of the gel. (D and E) FACS analyses of the surface expression are shown. The horizontal axis represents the logarithmic fluorescence intensity. The vertical axis represents the number of events (cells). Unfilled areas are signals from mock-transfected cells (negative control), and the filled areas are signals from cells transfected with the indicated CD4 constructs.
The plasma membrane expression of the chimeric receptor proteins was first determined by fluorescence-activated cell sorting (FACS). The transient expression was optimized to consistently give rise to >90% transfection, thereby allowing for improved detection sensitivity and quantification for surface expression. When cells were transfected with a construct encoding CD4-(HA)1-RKR, the specific anti-CD4 antibody binding detected a high level of surface expression that is indistinguishable from the surface expression of CD4-(HA)1-RAA, indicating that the RKR signal within this construct was not active to confer the restricted ER localization (Fig. 1D Top). However, when constructs with three or five copies of a tandemly positioned HA epitope were transfected, despite the similar protein level, the CD4-(HA)3-RKR and CD4-(HA)5-RKR chimeric receptors displayed a substantial reduction in surface expression (Fig. 1D Left). In contrast, a specific mutation of RKR to RAA restored the surface expression and eliminated the variation induced by various copies of HA inserts (Fig. 1D Right). These results indicate that the intracellular localization was RKR dependent. Furthermore, the RKR-mediated intracellular localization was ineffective when the RKR signal was separated with one copy of the HA epitope. The effectiveness was progressively restored when the RKR signal was separated by three or five copies of HA epitopes.
It is not known whether the RXR signal is different from the well known KKXX recognition signal, which interacts with COPI and mediates ER residence. Current evidence comes from a yeast two-hybrid analysis where an KKXX but not RKR peptide showed interaction with one of the COPI isoforms (6). To test the relationship of the ER residence ability to the length of spacer, similarly spaced constructs were made by using the ER residence signal, KKTN, from the WBP1 (ref. 14; Fig. 1A). In contrast to RKR, the single-copy HA construct resulted in no surface expression of CD4-(HA)1-KKTN. The surface expression was restored when KKTN was mutated to AATN (Fig. 1E), indicating the KKTN sequence-dependent intracellular localization. This result is consistent with the notion that KKTN is fully effective in conferring ER localization. Contrasting with the CD4-RKR chimera, an incremental increase of spacing between the transmembrane segment and KKTN by insertions of additional HA epitopes progressively reduced the effectiveness of intracellular localization, thereby allowing for the surface expression of the CD4-(HA)3-KKTN and CD4-(HA)5-KKTN chimeric receptors (Fig. 1E Left). These results show that the effectiveness of KKTN-mediated ER residence critically depends on the length of spacing peptide separating it from the transmembrane segment. Furthermore, when compared with that of the RKR signal, the KKTN displays contrasting sensitivity to the spacing between the signal sequence and plasma membrane. This contrasting effect on RKR and KKTN signals by identical sets of HA spacers is consistent with the notion that the length dependence was not caused by a novel sequence as a result of the HA–HA fusion.
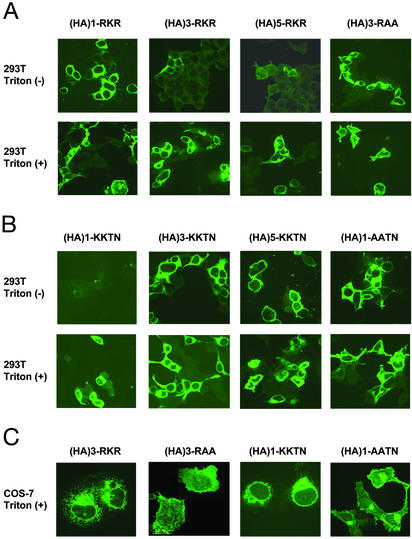
Fig. 2 shows the immunolocalization of the chimeric receptors that detect the surface expression by using transiently transfected 293T cells. The positive expression was detected in detergent-permeablized cells transfected with CD4 constructs with different copies of HA epitopes (Fig. 2 A and B Lower). In the absence of detergent treatment, the CD4 antibody could detect protein surface expression from cells transfected with the CD4-(HA)1-RKR construct but not from cells transfected with the CD4-(HA)3-RKR or CD4-(HA)5-RKR construct, indicative of differential effectiveness of retention (Fig. 2A Upper). Consistent with the RKR-mediated ER localization, specific mutation of RKR to RAA abolished the effect (Fig. 2A Right most). Using constructs of CD4-(HA)n-KKTN (n = 1, 3, and 5) in the absence of detergent (Fig. 2B Upper), anti-CD4 antibody detected no signal from CD4-(HA)1-KKTN, indicative of fully effective retention. For cells transfected with CD4-(HA)3-KKTN or CD4-(HA)5-KKTN, anti-CD4 antibody could readily detect surface expression. The differential retention effectiveness is KKTN-specific because CD4-(HA)1-AATN could be detected in the absence of detergent (Fig. 2B Right most). To better visualize the subcellular staining signal and rule out the observed effect being 293T cell specific, COS-7 cells transfected with either CD4-(HA)3-RKR/RAA or CD4-(HA)1-KKTN/AATN were stained with anti-CD4 antibody and imaged similarly with confocal microscopy. The HA-spacer-dependent surface expression could be readily observed. The intracellular staining of permeabilized COS-7 cells of either RKR or KKTN showed indistinguishable patterns consistent with ER residence (Fig. 2C).
Figure 2.
Immunocytochemistry analyses of surface expression chimeric CD4 receptors by confocal microscopy. (A) Immunostaining of 293T cells that were transiently transfected with the indicated RKR constructs (indicated on the top). Cells were stained before (Upper) and after (Lower) detergent treatments. (B) Immunostaining of 293T cells that were transiently transfected with the indicated KKTN constructs (indicated on the top). Cells were stained before (Upper) and after (Lower) detergent-mediated membrane permeation. (C) Immunostaining of COS-7 cells that were transiently transfected with the indicated RKR/RAA and KKTN/AATN constructs (indicated on the top). Cells were stained after detergent treatments.
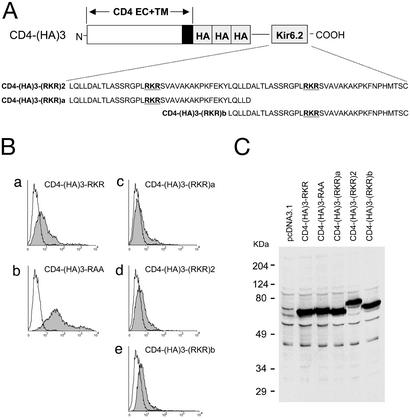
Unlike internal signals, C-terminal sequence motifs are often exposed as found in many proteins of known structure (see review by Chung et al. in ref. 15). Another important distinction between a terminal sequence signal and an internal sequence signal is that there may be multiple copies of internal signals for a given protein. Thus, there can be only one terminal signal sequence per protein, such as KKTN. Would an additional copy of RKR signal confer higher stringency of retention efficiency? To test this hypothesis, we used a CD4-(HA)3 backbone, which gave detectable retention (Fig. 1D). Thus, three different fusion receptor constructs were made, which included two forms of single-copy RKR sequences (originated from Kir6.2), CD4-(HA)3-(RKR)a and CD4-(HA)3-(RKR)b, and a tandemly linked form of two copies of RKR, CD4-(HA)3-(RKR)2 (Fig. 3A). The protein expression of these constructs was similar based on immunoblot analyses (Fig. 3C). When the two single-copy constructs were expressed, the residual surface expression was found at a level similar to that of other constructs of three HA spacer, i.e., the retention persists (Fig. 3 Ba–c and Be). When testing the two-copy RKR construct, the resultant fusion receptor was expressed (Fig. 3C). The level of surface expression was essentially identical to that of either of the single-copy constructs, suggesting that the two copies of RKR, at least under the sequence context of the referenced receptor system, did not confer additional effectiveness for receptor retention.
Figure 3.
(A) A schematic diagram of chimeric CD4 receptor with one or two copies of the Kir6.2 RKR signal. (B) FACS analyses of the surface expression are shown. The horizontal axis represents the logarithmic florescence intensity. The vertical axis represents the number of events (cells). Unfilled areas are signals from mock-transfected cells (negative control), and the filled areas are signals from cells transfected with the indicated CD4 constructs. (C) Immunoblot analyses of fusion proteins from the transfected cells. Protein lysates of transfected cells are identified by the constructs indicated at the top. The fusion protein expression was detected by anti-CD4 antibody. An overexposed blot is shown to demonstrate the absence of construct-specific degradation. The molecular mass standard in kDa is indicated on the left of the gel.
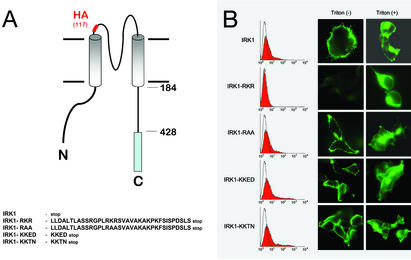
To test the effects of these spacing effects on a native protein, we chose an inward rectifier (mIRK1, Kir2.1; ref. 16). In heterologous expression systems, IRK1 is constitutively trafficked to cell surface. We inserted an HA tag at the extracellular loop between M1 and pore region, which has been previously shown to have no effect on channel function (6, 17). At the C terminus of IRK1, we fused either RKR or KKXX signals (Fig. 4A), which confer the dominant ER residence when positioned in appropriate spacing in the CD4 receptor system. The resultant IRK1 fusion constructs were then tested for the cell surface expression detected by either FACS analyses (Fig. 4B Left) or by immunocytochemistry coupled with confocal imaging (Fig. 4B Right). Whereas all constructs have comparable expression, they displayed differential surface expression. The RKR but not KKTN or KKED (18) signal was able to prevent IRK trafficking to cell surface. Mutation of RKR to RAA abolished the effects, indicating that the effect is sequence specific. According to the structure of cytoplasmic domain of a highly homologous protein, G-protein-coupled inward rectifier potassium channels (19), the extended physical length of five copies of the HA linkers would be comparable to that of structure (≈32 Å). Based on the structure and spacing used in CD4 constructs, the reported result is in complete agreement with the prediction that retention signal of RKR but not KKED or KKTN is active when positioned ≈32 Å from cytoplasmic leaflet of membrane.
Figure 4.
(A) A schematic diagram of the five IRK1 chimeric constructs. The site of the HA epitope insertion is as indicated. The peptide sequences fused to the C terminus are shown (Lower). (B) Different chimeric constructs were expressed and indicated (Left). The surface expression was detected by anti-HA antibody followed by FACS analyses (Left). The horizontal axis represents the logarithmic fluorescence intensity. The vertical axis represents the number of events (cells). Unfilled areas are signals from mock-transfected cells (negative control). Filled areas are the anti-HA signal. (Right) Immunofluorescence microscopy to detect the localization of indicated chimera in transfected cells by anti-HA under either permeabilized (Triton +) or nonpermeabilized (Triton −) condition.
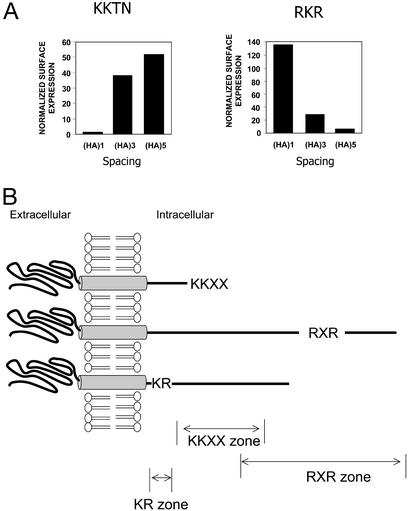
To quantify the surface expression over intracellular localization, two approaches were taken. In the first approach, surface expression for RKR constructs was normalized to measurements obtained from RAA constructs with the same number of copies of the HA epitope (Fig. 5A). Second, surface expression in nonpermeabilized transfectants was normalized to the measurements of total protein expression from permeabilized cells for each construct (data not shown). Both approaches essentially yielded identical results. The quantification suggests that the spacer length progressively alters the effectiveness. This result argues that the differential surface expression reported here was independent of the transfection efficiency, although one would predict that a very high expression of protein per cell could eventually saturate the retention machinery. Fig. 5B shows contrasting sensitivity to spacing for the RKR- and KKTN-mediated ER localization. The effectiveness of the two recognition signals covers both a proximal membrane zone by a KKXX-directed process and a distal membrane zone by the RXR-directed machinery.
Figure 5.
(A) Histogram of quantification of surface expression of CD4 chimeric receptor as a function of spacing. The normalized surface expression of CD4-KKTN (Left) and RKR (Right) was determined by dividing the MESF values for RKR and KKTN constructs by those of RAA and AATN constructs (with the corresponding number of HA copies), respectively (see Materials and Methods). (B) A schematic diagram to show that the KKTN and RKR signals are active in proximal and distal zones. Repositioning the signals into a different zone by a posttranslational event including modification, folding, and protein interaction may cause the receptor to alter its subcellular localization. The membrane stop transfer signal (KR zone) immediately followed transmembrane segment is also shown.
Discussion
In this series of experiments, we used the same reporter system to compare the spacing requirements of two different ER localization signals. The evidence revealed the presence of activity zones linearly layered in reference to the cytoplasmic side of the membrane surface. In other receptor signaling systems, such as functional interactions of a G-protein with its coupled receptors, the laterally placed zone defined by a protein complex is relatively common, where one or a cluster of receptor molecules through its/their ability to associate with other membrane-bound proteins defines/define a radius of activity zone within which there is functional coupling among different players. Recent examples, such as other potassium channels (6, 20), indicate that multiple transport signal sequences may be found in one receptor. The linearly organized zoning layers of different localization sequences reported here invite speculation that multiple functional domains within an intracellular domain of a membrane receptor (or ion channel) may define multiple layers of independent signaling events that may not be restricted to the protein localization signals.
The evidence reported here shows that the machinery that mediates KKXX- or RXR-dependent retention possesses strict sensitivity to the length of spacer that separates the trafficking signal motifs and the receptor-anchored membrane. When the RXR signal was first identified, questions were raised concerning whether the two signals (both are positively charged) interact with the same retention system mediated by COPI (6). The contrasting sensitivity of their retention/retrieval activity to the spacing identified in this report provides further evidence that the two signals confer protein residence in ER via different mechanisms and that their full activities were observed in different zones with respect to the intracellular side of the membrane surface. Because of the progressive effects by the spacer length, a possible overlap area of RKR and KKTN is suggested (Fig. 5B). This area seems not to be fully effective by either signal. It is curious as to whether there is yet another signal that is fully effective to cover this zone. Interestingly, the positively charged RXR signal also resembles the stop transfer signal positioned immediately after a membrane-spanning segment found in many transmembrane proteins (21). For example, a KKR/HRR motif is present immediately after the M2 transmembrane segment of inward rectifier potassium channels including a G-protein-gated inward rectifier (mGIRK1, Kir3.1; ref. 22), human renal outer medullary inward rectifier (hROMK, Kir1.1; ref. 23), mouse inward rectifier (IRK1, Kir2.1; ref. 16), and ATP-gated inward rectifier (Kir6.2; ref. 24). One possibility supported by the experimental evidence here is that the machinery for RKR, to serve as an ER residence signal, must be positioned with appropriate spacing from the cytoplasmic side of a transmembrane segment. In this way, the retention machinery would be able to distinguish a stop transfer signal from an RXR–ER residence signal. Similarly, whereas the KKXX signal is more active when positioned proximally to the cytoplasmic side of membrane leaflet, it is conceivable that it would require a minimal distance to allow assembly of the recognition protein complex.
Applying the zoning effects reported here to native receptor requires consideration in several aspects. First, many membrane-bound proteins may have more than one type and/or copy of localization signal. Through oligomerization, additional signals may be recruited. Second, some receptors may undergo proteolytic processing, a mechanism which may remove or expose localization signal(s). Third, a linearly long spacer on folding may bring a “distal” signal into proximity to cytoplasmic leaflet of membrane. These factors may complicate the conclusion when comparing evidence with different reporter systems. One example is that different variants of KKXX signal behave differently within the same protein context (9). When comparing results from deletion experiments, KKXX displayed quite differently in two different receptors: the simian immunodeficiency virus (SIV) envelope (Env) protein and UDP-glucuronosyl transferase (25, 26). One way to resolve this is to use the approach taken in this paper, which compares different localization signals with the same reporter systems.
Whereas it is difficult to precisely determine the distance thereby defining the physical boundary of the zones, when comparing numbers of residues spacing between the transmembrane segment of CD4 and the 36-aa Kir6.2 RKR peptide in various constructs (Fig. 1A; also see Materials and Methods), our evidence suggests that a minimum of functional spacing would be somewhere between 16 and 46 Å (assuming that the HA epitope could adapt a more compact alpha-helix fold). This finding is in agreement with the structure of the cytoplasmic domain of the G-protein-gated inward rectifier potassium channel, which would place the Kir6.2 RKR-peptide in this study more than 32 Å from the cytoplasmic side of the plasma membrane (19). The results from the IRK1 chimera experiments (Fig. 4) provided direct evidence further supporting the notion.
Linear spacing sensitivity is often found in the context of two signals, which provides an effective means of allowing for proper activities such as those conferred by two physically connected but functionally distinct protein domains. In nucleic acids, differential spacing of two DNA binding sites allows the sequence motifs to be phased either on the same side or on different sides of DNA molecules. Concerning the RXR or KKXX signal, the zone-specific effects may reflect the structural features of the two protein machineries. Perhaps the membrane surface serves as a reference point that allows for protein machinery to measure the “distance.” Thus, protein folding or interaction with other proteins of the intracellular domain may bring about changes that position the signals at either active or inactive zones. For example, the distally effective RXR signal may be brought to closer proximity to the membrane via mechanisms such as protein interaction, resulting in exit of protein from ER. Conversely, a distantly positioned KKXX signal may be brought closer to the membrane by posttranslational events such as lipidation, thereby conferring ER retention. Thus, in addition to direct physical masking of a protein localization signal, placing the signal into the appropriate zone may be the basis for conferring a posttranslational checkpoint for determining protein surface expression or differential intracellular localization.
Acknowledgments
We thank Dr. Lily Jan for providing Kir6.2 cDNA constructs and insightful comments, the members of the Li laboratory, and Drs. Carolyn Machamer and Jeremy Nathans for helpful comments on this manuscript. This work is supported by grants from the National Institutes of Health (to M.L.), the Cystic Fibrosis Foundation (to M.L.), and an Established Investigator Award from the American Heart Association (to M.L.).
Abbreviations
- ER
endoplasmic reticulum
- COPI
coat protein I
- HA
hemagglutinin
- MESF
molecules of equivalent soluble fluorochromes
- FACS
fluorescence-activated cell sorting
- WBP1
wheat germ agglutinin binding protein 1
Note Added in Proof.
After this paper's acceptance, a report by Yuan et al. (27) has shown that the RXR motif can bind to the 143-3 protein.
References
- 1.Mellman I, Warren G. Cell. 2000;100:99–112. doi: 10.1016/s0092-8674(00)81687-6. [DOI] [PubMed] [Google Scholar]
- 2.Teasdale R D, Jackson M R. Annu Rev Cell Dev Biol. 1996;12:27–54. doi: 10.1146/annurev.cellbio.12.1.27. [DOI] [PubMed] [Google Scholar]
- 3.Standley S, Roche K W, McCallum J, Sans N, Wenthold R J. Neuron. 2000;28:887–898. doi: 10.1016/s0896-6273(00)00161-6. [DOI] [PubMed] [Google Scholar]
- 4.Ma D, Jan L Y. Curr Opin Neurobiol. 2002;12:287–292. doi: 10.1016/s0959-4388(02)00319-7. [DOI] [PubMed] [Google Scholar]
- 5.Nilsson T, Jackson M, Peterson P A. Cell. 1989;58:707–718. doi: 10.1016/0092-8674(89)90105-0. [DOI] [PubMed] [Google Scholar]
- 6.Zerangue N, Schwappach B, Jan Y N, Jan L Y. Neuron. 1999;22:537–548. doi: 10.1016/s0896-6273(00)80708-4. [DOI] [PubMed] [Google Scholar]
- 7.Margeta-Mitrovic M, Jan Y N, Jan L Y. Neuron. 2000;27:97–106. doi: 10.1016/s0896-6273(00)00012-x. [DOI] [PubMed] [Google Scholar]
- 8.Klausner R D, Lippincott-Schwartz J, Bonifacino J S. Annu Rev Cell Biol. 1990;6:403–431. doi: 10.1146/annurev.cb.06.110190.002155. [DOI] [PubMed] [Google Scholar]
- 9.Zerangue N, Malan M J, Fried S R, Dazin P F, Jan Y N, Jan L Y, Schwappach B. Proc Natl Acad Sci USA. 2001;98:2431–2436. doi: 10.1073/pnas.051630198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Girod A, Storrie B, Simpson J C, Johannes L, Goud B, Roberts L M, Lord J M, Nilsson T, Pepperkok R. Nat Cell Biol. 1999;1:423–430. doi: 10.1038/15658. [DOI] [PubMed] [Google Scholar]
- 11.Masibay A S, Balaji P V, Boeggeman E E, Qasba P K. J Biol Chem. 1993;268:9908–9916. [PubMed] [Google Scholar]
- 12.Bretscher M S, Munro S. Science. 1993;261:1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- 13.Schulze-Gahmen U, Rini J, Arevalo J, Stura E, Kenten J, Wilson I. J Biol Chem. 1988;263(32):17100–17105. [PubMed] [Google Scholar]
- 14.te Heesen S, Janetzky B, Lehle L, Aebi M. EMBO J. 1992;11:2071–2075. doi: 10.1002/j.1460-2075.1992.tb05265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung J J, Shikano S, Hanyu Y, Li M. Trends Cell Biol. 2002;12:146–150. doi: 10.1016/s0962-8924(01)02241-3. [DOI] [PubMed] [Google Scholar]
- 16.Kubo Y, Baldwin T J, Jan Y N, Jan L Y. Nature. 1993;362:127–133. doi: 10.1038/362127a0. [DOI] [PubMed] [Google Scholar]
- 17.Ma D, Zerangue N, Lin Y F, Collins A, Yu M, Jan Y N, Jan L Y. Science. 2001;291:316–319. doi: 10.1126/science.291.5502.316. [DOI] [PubMed] [Google Scholar]
- 18.Hill K J, Stevens T H. Mol Biol Cell. 1994;5:1039–1050. doi: 10.1091/mbc.5.9.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishida M, MacKinnon R. Cell. 2002;111:957–965. doi: 10.1016/s0092-8674(02)01227-8. [DOI] [PubMed] [Google Scholar]
- 20.O'Kelly I, Butler M H, Zilberberg N, Goldstein S A. Cell. 2002;111:577–588. doi: 10.1016/s0092-8674(02)01040-1. [DOI] [PubMed] [Google Scholar]
- 21.Kuroiwa T, Sakaguchi M, Mihara K, Omura T. J Biol Chem. 1991;266:9251–9255. [PubMed] [Google Scholar]
- 22.Kubo Y, Reuveny E, Slesinger P A, Jan Y N, Jan L Y. Nature. 1993;364:802–806. doi: 10.1038/364802a0. [DOI] [PubMed] [Google Scholar]
- 23.Ho K, Nichols C G, Lederer W J, Lytton J, Vassilev P M, Kanazirska M V, Hebert S C. Nature. 1993;362:31–38. doi: 10.1038/362031a0. [DOI] [PubMed] [Google Scholar]
- 24.Inagaki N, Gonoi T, Clement J P t, Namba N, Inazawa J, Gonzalez G, Aguilar-Bryan L, Seino S, Bryan J. Science. 1995;270:1166–1170. doi: 10.1126/science.270.5239.1166. [DOI] [PubMed] [Google Scholar]
- 25.Kinosaki M, Masuko T, Sogawa K, Iyanagi T, Yamamoto T, Hashimoto Y, Fujii-Kuriyama Y. Cell Struct Funct. 1993;18:41–51. doi: 10.1247/csf.18.41. [DOI] [PubMed] [Google Scholar]
- 26.Vincent M J, Martin A S, Compans R W. J Biol Chem. 1998;273:950–956. doi: 10.1074/jbc.273.2.950. [DOI] [PubMed] [Google Scholar]
- 27.Yuan H, Michelsen K, Schwappach B. Curr Biol. 2003;13:638–646. doi: 10.1016/s0960-9822(03)00208-2. [DOI] [PubMed] [Google Scholar]