Abstract
Partial resistance of cell membranes to solubilization with mild detergents and the analysis of isolated detergent-resistant membranes (DRMs) have been used operationally to define membrane domains. Given the multitude of detergents used for this purpose, we sought to investigate whether extraction with different detergents might reflect the same underlying principle of domain formation. We therefore compared the protein and lipid content of DRMs prepared with a variety of detergents from two cell lines. We found that the detergents differ considerably in their ability to selectively solubilize membrane proteins and to enrich sphingolipids and cholesterol over glycerophospholipids as well as saturated over unsaturated phosphatidylcholine. In addition, we observed cell type-dependent variations of the molecular characteristics of DRMs and the effectiveness of particular detergents. These results make it unlikely that different detergents reflect the same aspects of membrane organization and underscore both the structural complexity of cell membranes and the need for more sophisticated analytical tools to understand their architecture.
Biological membranes are composed of a puzzling variety of lipids. Such diversity would be unnecessary if lipid bilayers served only as hydrophobic barriers and homogeneous two-dimensional solvents for membrane proteins. As is now increasingly appreciated, membranes show extensive lipid-driven compartmentalization, giving rise to distinct membrane domains. These domains differ in their composition, physical properties, and biological functions.
Membranes typically exist in a fluid state characterized by unconfined diffusion of its loosely packed lipids. This state is therefore also called the liquid-disordered (ld) phase. However, studies of liposomes (1, 2) as well as model membranes (3) have shown that certain lipids have the propensity to associate with each other, thus segregating from the ld phase. These lipids are cholesterol, sphingolipids with their usually saturated fatty acids, and saturated glycerophospholipids. The structure of their hydrophobic moieties allows them to pack more tightly than the kinked unsaturated glycerophospholipids. As a result, they can establish a more ordered state, called the liquid-ordered (lo) phase (4). Most likely, lo phases also exist in cell membranes with a sufficient proportion of cholesterol and sphingolipids (5). The lo phase is thought to form discrete microdomains interspersed in the continuous ld phase. These microdomains have been termed “lipid rafts” (6) and are believed to play important roles in signal transduction and protein sorting (7–9).
A common biochemical method to analyze the domain organization of membranes is extraction with mild detergents like Triton X-100 (Triton). Although detergent treatment disrupts most lipid–lipid interactions, a minor fraction of cell membranes is preserved and can be isolated as detergent-resistant membranes (DRMs). DRMs prepared with Triton probably originate from the cholesterol- and sphingolipid-rich lo phase, which resists extraction due to its tight lipid packing (5). Detergent extraction also disrupts lipid–protein interactions, so that most membrane proteins are solubilized. Only few proteins retain their association with lipids and are recovered in DRMs. Thus, DRM association of a protein is indicative of a strong interaction with highly ordered domains in the lo phase. However, DRMs may only imperfectly reflect the distribution of membrane components between the lo and ld phases (10), and it is unknown how well the composition of DRMs correlates with the components of native lipid rafts in cell membranes.
DRMs have mainly been prepared with Triton (11) and 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) (12), but other detergents such as Brij 58 (13), Brij 96 (14), Lubrol WX (15), and Brij 98 (16) have also been used. Yet, it is unclear whether DRMs prepared with different detergents similarly reflect the same aspect of membrane organization, i.e., segregation of cholesterol- and sphingolipid-rich domains in the lo phase from the ld phase rich in unsaturated glycerophospholipids.
To facilitate comparison of results obtained with different detergents, we analyzed the protein and lipid content of DRMs isolated with various mild detergents. We used two cell lines, epithelial Madin–Darby canine kidney (MDCK) cells and human T cell leukemia cells (Jurkat cells), to test whether the characteristics of DRMs and the effectiveness of particular detergents vary between cell types. In addition, we evaluated tools to manipulate DRM association of proteins. We found that DRMs obtained with different detergents differ considerably in their protein and lipid content, and that these differences are cell type-dependent. Our results caution against equating DRMs prepared with different detergents and from different cell types.
Materials and Methods
Antibodies.
Monoclonal antibodies against human calnexin and human Src-like kinase Yes were from Transduction Laboratories (Lexington, KY), the monoclonal antibody against human transferrin receptor was from Zymed, and the polyclonal caveolin-1 antibody was from Santa Cruz Biotechnology. Monoclonal stomatin antibody was a gift from Rainer Prohaska (Institute of Biochemistry, University of Vienna, Vienna); and polyclonal rab-5 antibody was kindly provided by Marino Zerial (Max Planck Institute of Molecular Cell Biology and Genetics). Antibodies against gp114 and human placental alkaline phosphatase (PLAP) have been characterized previously (17). The VIP17/MAL antibody will be described elsewhere.
Detergents.
Detergents used were Tween 20 (Sigma), Brij 58 (Sigma), Lubrol WX (Serva), Brij 98 (Sigma), Brij 96 (Fluka), Triton X-100 (Perbio, Bonn, Germany), and CHAPS (Anatrace, Maumee, OH).
Cell Culture and Metabolic Labeling.
MDCK strain II cells (MDCK cells) and MDCK cells expressing human PLAP (MDCK-PLAP cells; ref. 11) were maintained on tissue culture plates in MEM with 5% FCS/2 mM glutamine/100 units/ml penicillin/streptomycin. Jurkat cells were kept in RPMI medium 1640 with 10% FCS, glutamine, and antibiotics. For 14C- and 32P-labeling, MDCK or Jurkat cells were incubated overnight in normal medium with 10 μCi of 14C-acetate or 50 μCi of 32P-orthophosphate (Amersham Pharmacia; Ci = 37 GBq) per 10-cm dish or 107 cells, respectively.
Detergent Extraction and Flotation.
For flotation on sucrose step gradients, confluent MDCK cells from a 10-cm tissue culture plate or 107 Jurkat cells (≈1 mg of protein) were washed with homogenization buffer (HB, 250 mM sucrose in 10 mM Hepes, pH 7.4) and collected by scraping and centrifugation at 380 × g for 5 min. Cells were resuspended in 500 μl of HB with 25 μg/ml chymostatin, leupeptin, antipain, and pepstatin (CLAP) and homogenized by passing through a 25-G needle 20 times. Five hundred microliters of HB/CLAP was added containing either no detergent, 2% Tween 20, 2% Brij 58, 1% Lubrol WX, 2% Brij 98, 1% Brij 96, 2% Triton, or 8% CHAPS (all % wt/vol). Extraction was done on ice for 30 min, except for Brij 98, which was applied at 37°C for 10 min. Samples were adjusted to 42% (wt/wt) sucrose with 2 ml 56% sucrose in 10 mM Hepes, transferred into SW40 centrifuge tubes (Beckman), overlaid with 8.5 ml of 38% and 0.5 ml of 5% sucrose/Hepes, and centrifuged at 39,000 rpm (271,000 ×g) for 18 h; 2.5 ml was collected from the top as the floating fraction. For flotation on linear sucrose gradients, detergent extracts from MDCK-PLAP cells were prepared as above, except that HB was replaced by TNE (150 mM NaCl/2 mM EDTA in 50 mM Tris⋅HCl, pH 7.4). Samples were adjusted to 40% (wt/wt) sucrose with 2 ml 56% sucrose/TNE, transferred into SW40 tubes, overlaid with linear 5–35% sucrose gradients in TNE (9 ml), and centrifuged as above. Twelve fractions of 1 ml were collected from the top.
Surface Biotinylation.
Confluent MDCK cells on 10-cm tissue culture plates were biotinylated with 1 mg/ml Sulfo-NHS-LC-Biotin (Pierce) at 4°C for 30 min. After detergent extraction and flotation, equal aliquots of the floating fractions were run on 8–16% SDS/PAGE gels, blotted, and probed with peroxidase–extravidin (Sigma). For quantification, dilution series of each sample were dotted onto a nitrocellulose membrane, probed with peroxidase—extravidin, and enhanced chemiluminescent images were quantified. The background signal arising from endogenously biotinylated proteins; was negligible.
Lipid Extraction and TLC.
Lipids were extracted according to Folch et al. (18). 14C- and 32P-labeled lipids were resolved on silica high-performance TLC plates using chloroform/methanol/water 60:35:8 and chloroform/methanol/acetic acid/water 60:50:1:4, respectively, and quantified by phosphorimaging. Identification was by comigration with standards and susceptibility to mild alkaline hydrolysis.
Mass Spectrometric Lipid Analysis.
Mass spectrometric analysis was performed by using multiple precursor ion scanning (MPIS) (19). To enable quantitative analysis of different samples, each sample was spiked with an equal amount of 13C-labeled lipid extract from Pichia pastoris as internal standard. For detection of phosphatidylcholine (PC), MPIS was performed in positive ion mode with the selected characteristic fragment ion of the choline head group. To determine percent fractions, the peak area of individual PC species was divided by the sum peak area of all PC species. To determine relative concentrations, the peak area of individual PC species was divided by the sum peak area of a selected set of 13C-labeled PC species as internal standard.
Cyclodextrin, Saponin, and Sphingomyelinase Treatment.
For cyclodextrin treatment, MDCK-PLAP cells from 6-cm tissue culture plates were resuspended in 180 μl of TNE/CLAP and homogenized. One hundred sixty microliters of cell homogenate was mixed with 20 μl of 100 mM methyl-β-cyclodextrin (Sigma) and incubated at 37°C for 30 min. After cooling, samples were extracted with 20 μl of 10% Triton on ice for 30 min, adjusted to 40% iodixanol with 400 μl of Optiprep (Nycomed Pharma), transferred into TLS55 centrifuge tubes (Beckman), overlaid with 1.2 ml of 30% iodixanol/TNE, and 0.2 ml of TNE, and centrifuged at 55,000 rpm (259,000 × g) for 2 h. Two fractions of 1 ml were collected from the top. For saponin treatment, cells were treated with 0.2% saponin from Quillaja Bark (Sigma) in PBS at 4°C for 1 h. For sphingomyelinase (SMase) treatment, cells were incubated with 0.5 units/ml SMase from Staphylococcus aureus (Sigma) in MEM at 37°C for 1 h. Homogenate from saponin- or SMase-treated cells was either directly submitted to Triton extraction or first treated with cyclodextrin.
Results
Protein Analysis of DRMs Prepared with Different Detergents.
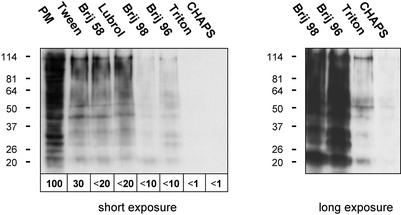
We restricted our initial analysis of membrane solubilization to plasma membrane proteins to avoid mixing proteins from different subcellular localizations and contamination with, e.g., cytoskeletal proteins. MDCK cells were surface biotinylated, extracted with 1% Tween 20, 1% Brij 58, 0.5% Lubrol WX, 1% Brij 98, 0.5% Brij 96, and 1% Triton or 4% CHAPS (65 mM), and DRMs were prepared by flotation on sucrose step gradients (Fig. 1).
Figure 1.
Protein content of plasma membrane DRMs from MDCK cells. Surface biotinylated MDCK cells were extracted with 1% Tween 20, 1% Brij 58, 0.5% Lubrol WX, 1% Brij 98, 0.5% Brij 96, and 1% Triton or 4% CHAPS. DRMs were prepared by flotation on sucrose step gradients. Equivalent aliquots of the starting material before detergent treatment, which contained the total plasma membrane protein (PM), and the various DRMs were analyzed by Western blotting using peroxidase-conjugated extravidin to reveal biotinylated proteins. Two exposures of the same membrane are shown. The amounts of biotinylated protein were quantified, setting PM to 100%.
All detergents solubilized a substantial fraction of the biotinylated plasma membrane protein. However, the different DRMs contained very different amounts of protein. For instance, Tween 20, Brij 58, and Lubrol WX produced DRMs containing >10-fold more protein than DRMs prepared with Triton and CHAPS. Therefore, these detergents were less selective in that they disrupted fewer lipid–protein interactions and allowed DRM association of a larger number of proteins.
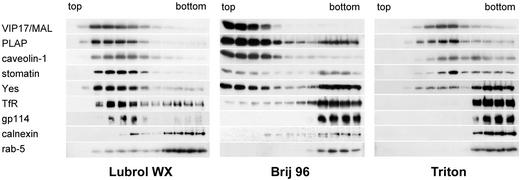
To investigate the differences between detergents in more detail, detergent resistance of particular marker proteins was evaluated by using MDCK-PLAP cells. Cell homogenate, or detergent extracts prepared with Lubrol WX, Brij 96, or Triton, were floated on linear sucrose gradients. The distribution of the tetraspan protein VIP17/MAL, GPI-anchored PLAP, caveolin-1, stomatin, the doubly acylated Src-like kinase Yes, transferrin receptor (TfR), the sialoglycoprotein gp114, calnexin, and rab-5 was analyzed by immunoblotting (Fig. 2).
Figure 2.
DRM association of marker proteins from MDCK cells. MDCK-PLAP cell homogenate was extracted with 0.5% Lubrol WX, 0.5% Brij 96, or 1% Triton, and floated on linear sucrose gradients. The distribution of marker proteins was analyzed by immunoblotting. The light fractions from the top of the gradients are on the left, the heavy bottom fractions on the right.
Flotation without detergent treatment demonstrated membrane association of the above proteins (not shown). Lubrol WX largely solubilized calnexin and rab-5 as reflected by their retention in the high-density fractions, whereas the other proteins were predominantly detergent-resistant (Fig. 2 Left). Brij 96 additionally solubilized gp114, most of TfR, and ≈50% of Yes (Fig. 2 Center), whereas Triton solubilized rab-5, calnexin, gp114, TfR, and most of Yes (Fig. 2 Right). Hence, DRM association of these marker proteins confirms the graded selectivity of the detergents observed by analysis of the plasma membrane DRM protein content.
The results with Triton agreed with earlier observations (6, 11, 20–22). Regarding Lubrol WX, calnexin had previously been found to be partly insoluble (15), consistent with our results. In contrast, TfR, which in our hands largely resisted extraction with Lubrol WX, was reported to be Lubrol-soluble in MDCK cells (15).
Therefore, analysis of the protein contents of DRMs from MDCK cells showed that the detergents differed considerably in their ability to enrich strongly lipid-associated proteins; i.e., they differed in their “DRM selectivities.”
Lipid Analysis of DRMs Prepared with Different Detergents.
To analyze the lipid content of DRMs, MDCK and Jurkat cells were used to label either all lipids with 14C or phospholipids with 32P. Total cell membranes (TCMs) and DRMs were obtained by flotation on sucrose step gradients. Lipids were extracted and resolved by TLC (Fig. 3). As observed for the protein content of DRMs derived from the plasma membrane, the amounts of lipid present in the DRMs varied considerably between detergents. To show qualitative changes in the lipid composition of DRMs compared with TCMs, lipids were grouped into sphingolipids, cholesterol, and glycerophospholipids. Sphingolipid/glycerophospholipid and cholesterol/glycerophospholipid ratios were calculated. The fold changes of these ratios relative to the TCMs are depicted in Fig. 4. No significant changes occurred when higher detergent concentrations were used or when DRMs where prepared by pelleting from isolated membranes instead of flotation from cell homogenate (not shown).
Figure 3.
Lipid contents of DRMs from MDCK and Jurkat cells. Lipids from TCMs and DRMs from 14C- or 32P-labeled MDCK and Jurkat cells were analyzed by TLC (Upper, 14C-labeled lipids; Lower, 32P-labeled lipids). GluC/GalC/LacC, glucosyl/galactosyl/lactosyl ceramide; SM, sphingomyelin; Gb5, pentosylglucoside; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PS, phosphatidylserine. Asterisks denote lipids not identified by comigration with lipid standards.
Figure 4.
Lipid contents of DRMs from MDCK and Jurkat cells. Lipids from the TLC plates shown in Fig. 3 were quantified and grouped into sphingolipids (GalC + GluC + LacC + SM + Gb5 for MDCK cells, GluC + SM for Jurkat cells, intensities taken from 14C-labeled lipids), cholesterol and glycerophospholipids (PE + PI + PS + PC, intensities taken from 32P-labeled lipids). Sphingolipid/glycerophospholipid and cholesterol/glycerophospholipid ratios relative to the ratios for TCMs are shown. GPL, glycerophospholipids.
Compared with TCMs, DRMs from MDCK cells prepared with Triton and CHAPS showed clear enrichment of sphingolipids and cholesterol over glycerophospholipids. The other detergents enriched these lipids less strongly. Nevertheless, their capacity for enrichment correlated with the DRM specificity with regard to proteins, such that Brij 98 and Brij 96 caused a more pronounced enrichment of sphingolipids and cholesterol than Tween 20, Brij 58, and Lubrol WX. The same trend was observed for Jurkat cells, but notable differences exist. Contrary to MDCK cells, Brij 98 and Brij 58 enriched sphingolipids and cholesterol to a similar extent, whereas Brij 96 did not.
The behaviors of Brij 58, Brij 96, and Brij 98 in MDCK and Jurkat cells illustrate that the effect of a given detergent may vary between cell types. Moreover, the enrichment of sphingolipids and cholesterol was overall less pronounced in Jurkat cells than in MDCK cells, showing that the lipid composition of DRMs prepared with a particular detergent can be cell type-dependent.
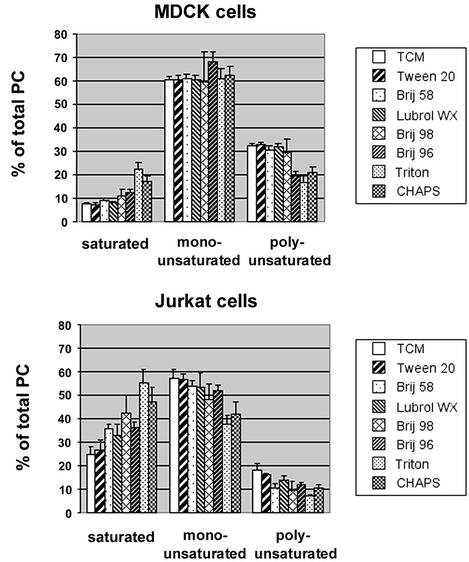
Next, we analyzed the fatty acid substitution pattern of glycerophospholipids in different DRMs. Lipids from the floating fractions of MDCK and Jurkat cells were prepared as before, and PC, the major glycerophospholipid, was analyzed by mass spectrometry. Seventeen PC species, differing in the total number of carbon atoms and/or the number of double bonds in the fatty acid moieties, were detected. For each species, its percent fraction of the total PC was calculated. In addition, the relative concentration of each species was determined to allow comparison of the amounts present in different DRMs (see Table 1, which is published as supporting information on the PNAS web site, www.pnas.org). PC species were then grouped according to the degree of saturation of their fatty acids (Fig. 5).
Figure 5.
Saturation profile of PCs in DRMs from MDCK and Jurkat cells. TCMs and DRMs were obtained as before, lipids were extracted, and PC was analyzed by mass spectrometry. Percent fractions of each PC species were determined and grouped into saturated (30:0, 31:0, 32:0, 34:0), monounsaturated (31:1, 32:1, 33:1, 34:1, 36:1), and polyunsaturated (32:2, 33:2, 34:2, 35:2, 36:3, 36:2, 38:3) PC species (in x:y, x indicates the total number of carbon atoms of the fatty acid side chains; y indicates the number of double bonds). Error bars represent standard deviations of three measurements of the same samples.
In MDCK cells, ≈60% of the total cell membrane PC was monounsaturated, <10% was fully saturated, and ≈30% possessed more than one double bond. With increasing detergent strength, increasing enrichment of saturated PCs at the expense of polyunsaturated PCs was observed. A similar trend was found for Jurkat cells, which additionally showed depletion of monounsaturated PCs with increasing solubilization efficiency of the detergents.
Interestingly, PCs from Jurkat cells contained a larger fraction of saturated PCs (25% of the total cell membrane PC compared with <10% in MDCK cells). This could explain the less pronounced enrichment of sphingolipids and cholesterol in DRMs from these cells. Saturated glycerophospholipids preferentially partition into the lo phase, so that liquid-ordered domains in Jurkat cell membranes may contain a comparatively high proportion of glycerophospholipids. As a result, enrichment of these domains by DRM isolation does not cause the marked depletion of glycerophospholipids characteristic of MDCK cells. These results again underscore differences between DRMs from different cell types.
Taken together, the lipid analysis showed quantitative as well as qualitative differences between DRMs prepared with different detergents. DRM selectivities of the detergents, as defined by their ability to enrich a subset of proteins during DRM preparation, correlated well with their ability to enrich sphingolipids and cholesterol over glycerophospholipids and saturated over unsaturated glycerophospholipids. Thus, the term “DRM selectivity” can be extended to comprise characteristic changes in the lipid composition of DRMs. Trends were similar in MDCK and Jurkat cells, but significant cell type-dependent differences with regard to the composition of DRM lipids were observed.
Tools for Manipulating DRM Association.
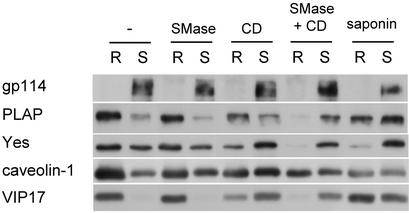
Finally, tools to manipulate DRM association of proteins were evaluated. Methyl-β-cyclodextrin (CD) extracts cholesterol from membranes and disrupts domains whose integrity depends on cholesterol. Saponin complexes cholesterol and is thought to sequester it away from other interactions. We also tested SMase, which we speculated might disturb detergent-resistant domains if their integrity depended on sphingomyelin. MDCK-PLAP cells were subjected to these treatments. Effects on the lipid composition of the samples were monitored by TLC (not shown). After extraction with 1% Triton, soluble and insoluble material was separated by flotation on Optiprep step gradients and the distribution of gp114, PLAP, Yes, caveolin-1, and VIP17/MAL was analyzed by immunoblotting (Fig. 6). Without detergent extraction, none of the treatments disrupted membrane association of the above proteins, except for CD, which solubilized a minor portion of Yes (not shown).
Figure 6.
Manipulation of DRM association. MDCK cells were left untreated or treated with SMase, CD, saponin, or first SMase and then CD. After extraction with 1% Triton and flotation on Optiprep step gradients, two fractions containing the detergent-resistant (R, resistant) and detergent-soluble (S, soluble) material were collected and analyzed by immunoblotting.
When CD was used on intact MDCK cells, DRM association of all proteins tested was essentially unaffected, even when cholesterol was depleted by >70% (not shown). Only when used on cell homogenate, as in Fig. 6, was CD effective. It depleted cholesterol by ≈50% without affecting other lipids, while reducing DRM association of PLAP, Yes, caveolin-1, and VIP17/MAL. Saponin, which caused no lipid extraction, had similar effects as CD. SMase treatment converted <30% of sphingomyelin into ceramide, but on its own did not reduce DRM association of the above proteins. However, SMase treatment augmented the effects of CD, indicating that resistance to Triton extraction depends on both cholesterol and sphingomyelin. No synergy was observed between saponin and SMase (not shown).
Discussion
We have demonstrated that DRMs prepared with a variety of mild detergents show considerable quantitative, as well as qualitative differences in their protein and lipid content. First, the amount of proteins and lipids recovered in the DRM fractions differed dramatically between detergents (Figs. 1 and 3). Second, the detergents differentially solubilized a set of marker proteins (Fig. 2). Third, the detergents differed in their ability to enrich cholesterol and sphingolipids over glycerophospholipids (Fig. 4) and saturated over unsaturated phosphatidylcholine (Fig. 5). Hence, the DRM selectivities of the detergents, as defined by their ability to enrich a subset of membrane proteins, cholesterol, sphingolipids, and saturated phosphatidylcholine, varied widely. These observations make it unlikely that the biophysical basis for how biological membranes resist different detergents is by being the same.
In addition, we observed significant cell type-dependent variations in the composition of DRMs and in the DRM selectivities of some detergents. In MDCK cells, the approximate order of DRM selectivity was CHAPS ≈ Triton > Brij 98 ≈ Brij 96 > Lubrol WX ≈ Brij 58 > Tween 20. In Jurkat cells, it was CHAPS ≈ Triton > Brij 98 ≈ Brij 58 > Lubrol WX ≈ Brij 96 > Tween 20. In other cell types, further deviations from these trends have been observed. Brij 96, for instance, was found to be at least as selective as Triton in neurons and the myelin membrane (14, 23). In addition, some proteins may not follow the general order of DRM selectivity. In macrophages, for example, CD11b was Triton-insoluble but Lubrol-soluble, even though Triton was generally a more effective solubilizer in those cells as judged by the DRM lipid content (24).
The comparison of different detergents clearly shows the limitations of DRMs with regard to providing insight into the presence of distinct membrane domains. Isolation of DRMs with one or more detergents cannot yield information about the spatial organization of membranes. DRM association of proteins and lipids reflects only the end result of the extraction process. Different types of detergent-resistant domains may exist (14, 15), but the presence of proteins or lipids in DRMs obtained with a particular detergent does not necessarily indicate association with the same domains in the native membrane. These considerations are illustrated by studies of polarized cells that show readily discernible separation of different membrane domains. In migrating T lymphocytes, several proteins and the glycolipids GM1 and GM3, all of which associate with DRMs, localize to different regions of the cells, the leading edge, and the uropod (25). Similarly, during yeast mating, a number of DRM-associated proteins relocalize to the mating projection of the cells. However, DRM-associated proteins are also found outside the mating projection (26). Hence, separate detergent-resistant domains exist in those cells, but they are indistinguishable by the DRM criterion. Detergent treatment leads to coalescence of detergent-resistant cholesterol-sphingolipid micodomains, which come together to form aggregates or fuse into continuous membrane sheets (27, 28). It has also been demonstrated that detergent intercalation into model membranes can promote the formation of DRMs (29). On the other hand, proteins that are present in lipid domains may be solubilized by the detergent due to low affinity for the surrounding lipids or to due to the specifics of the protein–lipid interface. In these cases, antibody cross-linking can stabilize or promote partitioning into the lipid domains. For instance, the T cell receptor in Jurkat cells is Triton-soluble. When it was cross-linked by an antibody, it acquired resistance to Triton extraction (30). The T cell receptor may be constitutively present in detergent-resistant domains, but its interaction with such domains could be too weak to survive Triton extraction. However, it is also possible that oligomerization of the receptor with antibodies or by other means shifts the receptor into detergent-resistant domains. Therefore, on its own, differential association of proteins or lipids with different DRMs is insufficient to define distinct membrane domains. For cholesterol–sphingolipid microdomains, the problem is compounded by the small size of individual rafts, each raft containing only a limited number of components (8). Rafts can be visualized only by morphological methods after raft clustering, e.g., by antibody cross-linking (31).
As observed previously (14, 23) and confirmed here, DRMs prepared with different detergents have different densities (Fig. 2). In addition to proteins and lipids, intercalated detergent molecules influence DRM density, and removal of weakly associated proteins and lipids by detergent may alter the protein/lipid ratio of detergent-resistant domains. Therefore, different DRM densities with different detergents do not necessarily indicate the isolation of different types of domains. However, if two proteins that reside in the same membrane float to different densities during DRM preparation, it can be concluded that they had different lipid environments before detergent treatment (15).
This leads to the question of how the DRM criterion should best be used to test the association of a particular protein with specialized membrane domains. Given the relatively low DRM selectivities of some detergents, we suggest to first use a rather strong detergent. If the chosen detergent disrupts lipid association of the protein of interest, a weaker detergent may be tried or the detergent concentration may be lowered. For instance, some gp114 and TfR was insoluble in 20 mM CHAPS, whereas they were fully solubilized by 65 mM CHAPS (unpublished data). The mass ratio of detergent to protein also determines to what extent DRM proteins are solubilized (32). This probably explains the discrepancy between Fig. 2, which shows that the majority of Yes was solubilized by Triton, and Fig. 6, where >50% of Yes was insoluble, because the detergent/protein mass ratio was about 10:1 in Fig. 2 but only 5:1 in Fig. 6. Regardless of the conditions for detergent treatment, a completely soluble membrane protein localized at the same membrane as the protein of interest should be included in the analysis. This is an essential negative control for incomplete solubilization of bulk membrane. In addition, lipid analysis of DRMs should complement the analysis of DRM protein contents. There are no universally applicable conditions for DRM isolation. Instead, optimization will be required in each case.
If a protein is found to be DRM-associated, treatment with cyclodextrin, saponin, and sphingomyelinase may be applied to test whether DRM association depends on cholesterol and sphingomyelin. We show that in MDCK cells, cyclodextrin and saponin reduce DRM association of the plasma membrane proteins PLAP, Yes, caveolin-1, and VIP17/MAL (Fig. 5). Remarkably, cholesterol depletion of intact MDCK cells by >70% did not disturb DRM association of these proteins, but cholesterol depletion of cell homogenate by 50% was sufficient. It is not clear why cyclodextrin was more effective on cell homogenate. Possibly cholesterol is difficult to extract from the apical plasma membrane of epithelial cells, which has long been known to be unusually stable because of its high concentration of glycosphingolipids (33). As a result, DRM association of proteins at the apical plasma membrane may be particularly resistant to cyclodextrin unless the apical membrane is mechanically disrupted first. Alternatively, living MDCK cells may be able to maintain detergent-resistant domains under conditions of severe cholesterol depletion by redistributing the remaining cholesterol.
There are more precedents of proteins whose DRM association is unaffected by cyclodextrin treatment. In BHK cells, DRM association of the bacterial toxin aerolysin is not significantly reduced by cyclodextrin, even though the integrity of the domains it interacts with was shown to depend on cholesterol by using saponin (34). In microvillar vesicles derived from the small intestinal brush border membrane, galectin-4 remains detergent-resistant despite cholesterol depletion by >70% (35). In agreement with studies of liposomes (1), cholesterol might even be dispensable for the formation of detergent-resistant domains in membranes such as the brush border membrane, which is extremely rich in sphingolipids. We found that sphingomyelinase treatment augments the effect of cyclodextrin, consistent with the idea that stability of detergent-resistant domains requires both cholesterol and sphingomyelin. The cleavage product of sphingomyelin, ceramide, appears to be unable to substitute for the cleaved sphingomyelin, even though it strongly supports the formation of lo domains in model membranes (36). However, sphingomyelinase on its own did not reduce DRM association of the proteins tested, nor did it enhance the effect of saponin. On the other hand, this treatment may be more effective in cells like fibroblasts, where >90% of the sphingomyelin is susceptible to sphingomyelinase (37).
These tools to manipulate DRM association of proteins, like DRM association itself, constitute “inclusive” criteria. In the case of a positive result, they can support a hypothesis, but a negative result does not necessarily provide contradicting evidence. For instance, reduction of DRM association of a protein by cholesterol depletion suggests interaction with a domain whose integrity depends on cholesterol. If no reduction is found, no conclusions can be drawn, because the remaining cholesterol may be sufficient to keep such domains intact. As discussed above, detergent insolubility of a protein indicates association with detergent-resistant domains, but detergent solubility does not exclude interaction with these domains, because the interaction may not be strong enough to be preserved during DRM preparation.
In summary, isolation of DRMs will remain a valuable tool for the analysis of biological membranes. DRMs are a most useful starting point for defining membrane subdomains, including cholesterol–sphingolipid rafts. Triton and CHAPS are the most reliable detergents for analyzing possible raft association. Applying a variety of detergents may reveal subtle differences in lipid–protein interactions or changes in the lipid environment of proteins during processes like T cell activation. However, caution is needed when drawing conclusions from DRM experiments and when comparing results obtained with different detergents and from different cell types. To avoid the pitfalls of our current relatively crude methodology, analyses should be done as comprehensively and with as many experimental tools as possible. The complexity of membranes is just beginning to be recognized, so that awareness of methodological limitations is important to prevent misinterpretations.
Supplementary Material
Acknowledgments
We thank Deborah Brown for critically reading the manuscript. We also thank Denis Corbeil, Wieland Huttner, and Joachim Füllekrug for discussion and suggestions. M.H. was partially supported by the Uehara Memorial Foundation.
Abbreviations
- DRMs
detergent-resistant membranes
- MDCK
Madin–Darby canine kidney
- TCMs
total cell membranes
- CHAPS
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
- ld
liquid-disordered
- lo
liquid-ordered
- PC
phosphatidylcholine
- SMase
sphingomyelinase
- TfR
transferrin receptor
- CD
methyl-β-cyclodextrin
References
- 1.Schroeder R, London E, Brown D A. Proc Natl Acad Sci USA. 1994;91:12130–12134. doi: 10.1073/pnas.91.25.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed S N, Brown D A, London E. Biochemistry. 1997;36:10944–10953. doi: 10.1021/bi971167g. [DOI] [PubMed] [Google Scholar]
- 3.Dietrich C, Bagatolli L A, Volovyk Z N, Thompson N L, Levi M, Jacobson K, Gratton E. Biophys J. 2001;80:1417–1428. doi: 10.1016/S0006-3495(01)76114-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ipsen J H, Karlstrom G, Mouritsen O G, Wennerstrom H, Zuckermann M J. Biochim Biophys Acta. 1987;905:162–172. doi: 10.1016/0005-2736(87)90020-4. [DOI] [PubMed] [Google Scholar]
- 5.Brown D A, London E. J Membr Biol. 1998;164:103–114. doi: 10.1007/s002329900397. [DOI] [PubMed] [Google Scholar]
- 6.Simons K, Ikonen E. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 7.Brown D A, London E. Annu Rev Cell Dev Biol. 1998;14:111–136. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- 8.Simons K, Toomre D. Nat Rev Mol Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 9.Ikonen E. Curr Opin Cell Biol. 2001;13:470–477. doi: 10.1016/s0955-0674(00)00238-6. [DOI] [PubMed] [Google Scholar]
- 10.London E, Brown D A. Biochim Biophys Acta. 2000;1508:182–195. doi: 10.1016/s0304-4157(00)00007-1. [DOI] [PubMed] [Google Scholar]
- 11.Brown D A, Rose J K. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- 12.Kurzchalia T V, Dupree P, Parton R G, Kellner R, Virta H, Lehnert M, Simons K. J Cell Biol. 1992;118:1003–1014. doi: 10.1083/jcb.118.5.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bohuslav J, Cinek T, Horejsi V. Eur J Immunol. 1993;23:825–831. doi: 10.1002/eji.1830230409. [DOI] [PubMed] [Google Scholar]
- 14.Madore N, Smith K L, Graham C H, Jen A, Brady K, Hall S, Morris R. EMBO J. 1999;18:6917–6926. doi: 10.1093/emboj/18.24.6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Röper K, Corbeil D, Huttner W B. Nat Cell Biol. 2000;2:582–592. doi: 10.1038/35023524. [DOI] [PubMed] [Google Scholar]
- 16.Drevot P, Langlet C, Guo X J, Bernard A M, Colard O, Chauvin J P, Lasserre R, He H T. EMBO J. 2002;21:1899–1908. doi: 10.1093/emboj/21.8.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verkade P, Harder T, Lafont F, Simons K. J Cell Biol. 1999;148:727–739. doi: 10.1083/jcb.148.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Folch J, Lees M, Stanley G H S. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 19.Ekroos K, Chernushevich I V, Simons K, Shevchenko A. Anal Chem. 2002;74:941–949. doi: 10.1021/ac015655c. [DOI] [PubMed] [Google Scholar]
- 20.Puertollano R, Martín-Belmonte F, Millán J, del Carmen de Marco M, Albar J P, Kremer L, Alonso M A. J Cell Biol. 1999;145:141–151. doi: 10.1083/jcb.145.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snyers L, Umlauf E, Prohaska R. Eur J Cell Biol. 1999;78:802–812. doi: 10.1016/S0171-9335(99)80031-4. [DOI] [PubMed] [Google Scholar]
- 22.Lafont F, Lecat S, Verkade P, Simons K. J Cell Biol. 1998;142:1413–1427. doi: 10.1083/jcb.142.6.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor C M, Coetzee T, Pfeiffer S E. J Neurochem. 2002;81:993–1004. doi: 10.1046/j.1471-4159.2002.00884.x. [DOI] [PubMed] [Google Scholar]
- 24.Drobnik W, Borsukova H, Bottcher A, Pfeiffer A, Liebisch G, Schutz G J, Schindler H, Schmitz G. Traffic. 2002;3:268–278. doi: 10.1034/j.1600-0854.2002.030404.x. [DOI] [PubMed] [Google Scholar]
- 25.Gómez-Moutón C, Abad L J, Mira E, Lacalle R A, Gallardo E, Jiménez Baranda S, Illa I, Manez S, Martínez-A C. Proc Natl Acad Sci USA. 2001;98:9642–9647. doi: 10.1073/pnas.171160298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bagnat M, Simons K. Proc Natl Acad Sci USA. 2002;99:14183–14188. doi: 10.1073/pnas.172517799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurzchalia T V, Hartmann E, Dupree P. Trends Cell Biol. 1995;5:187–189. doi: 10.1016/s0962-8924(00)88990-4. [DOI] [PubMed] [Google Scholar]
- 28.Mayor S, Maxfield F R. Mol Biol Cell. 1995;6:929–944. doi: 10.1091/mbc.6.7.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heerklotz H. Biophys J. 2002;83:2693–26701. doi: 10.1016/S0006-3495(02)75278-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janes P W, Steven C L, Magee A I. J Cell Biol. 1999;147:447–461. doi: 10.1083/jcb.147.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harder T, Scheiffele P, Verkade P, Simons K. J Cell Biol. 1998;141:929–942. doi: 10.1083/jcb.141.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ostermeyer A G, Beckrich B T, Ivarson K A, Grove K E, Brown D A. J Biol Chem. 1999;274:34459–34466. doi: 10.1074/jbc.274.48.34459. [DOI] [PubMed] [Google Scholar]
- 33.Simons K, van Meer G. Biochemistry. 1988;27:6197–6202. doi: 10.1021/bi00417a001. [DOI] [PubMed] [Google Scholar]
- 34.Abrami L, van der Goot G. J Cell Biol. 1999;147:175–184. doi: 10.1083/jcb.147.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansen G H, Immerdal L, Thorsen E, Niels-Christiansen L-L, Nystrom B T, Demant E J F, Danielsen E M. J Biol Chem. 2001;276:32338–32344. doi: 10.1074/jbc.M102667200. [DOI] [PubMed] [Google Scholar]
- 36.Xu X, Bittman R, Duportail G, Heissler D, Vilcheze C, London E. J Biol Chem. 2001;276:33540–33546. doi: 10.1074/jbc.M104776200. [DOI] [PubMed] [Google Scholar]
- 37.Slotte J P, Bierman E L. Biochem J. 1988;250:653–658. doi: 10.1042/bj2500653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.