Abstract
The Escherichia coli inner membrane protein (IMP) YidC is involved in the membrane integration of IMPs both in concert with and independently from the Sec translocase. YidC seems to be dispensable for the assembly of Sec-dependent IMPs, and so far it has been shown to be essential only for the proper Sec-independent integration of some phage coat proteins. Here, we studied the physiological consequences of YidC depletion in an effort to understand the essential function of YidC. The loss of YidC rapidly and specifically induced the Psp stress response, which is accompanied by a reduction of the proton-motive force. This reduction is due to defects in the functional assembly of cytochrome o oxidase and the F1Fo ATPase complex, which is reminiscent of the effects of mutations in the yidC homologue OXA1 in the yeast mitochondrial inner membrane. The integration of CyoA (subunit II of the cytochrome o oxidase) and Foc (membrane subunit of the F1Fo ATPase) appeared exceptionally sensitive to depletion of YidC, suggesting that these IMPs are natural substrates of a membrane integration and assembly pathway in which YidC plays an exclusive or at least a pivotal role.
Integration of inner membrane proteins (IMPs) in Escherichia coli can occur by various mechanisms. The majority of IMPs are targeted in a cotranslational manner to the Sec translocase in the inner membrane by the signal recognition particle and its receptor FtsY (reviewed in ref. 1). The Sec translocase consists of the core channel components SecY, SecE, and SecG, which constitute a heterotrimer, and the peripheral motor protein SecA. In addition, SecD, SecF, and YajC form an accessory complex that is loosely associated with the core translocase and facilitates the translocation process (reviewed in ref. 2). Recently, YidC was identified as a factor that specifically interacts with transmembrane segments (TMs) of nascent IMPs during membrane integration (3) and that is associated with the SecDFYajC subcomplex (4).
In contrast to the Sec-dependent IMPs, some small phage coat proteins, such as M13 procoat and Pf3 coat protein, were reported to insert spontaneously into the membrane, i.e., independent of the Sec translocase and the signal recognition particle targeting pathway (5, 6). Recently, this concept of unassisted integration was challenged by the observed requirement of YidC in Sec-independent IMP integration (7, 8).
YidC is an essential polytopic IMP homologous to the mitochondrial IMP Oxa1p and thylakoid membrane protein Alb3. Both Oxa1p and Alb3 have been implicated in membrane protein insertion and are essential for correct organelle function (reviewed in refs. 9 and 12). Oxa1p is needed for the proper insertion of a subset of mitochondrially encoded IMPs, such as subunits of cytochrome c oxidase and F1Fo ATPase (10–12). Because mitochondria do not possess a signal recognition particle-like targeting pathway or a Sec-like translocase, it has been suggested that Oxa1p functions in a fashion similar to YidC in the Sec-independent route in E. coli (9, 12).
The exact role of YidC in the membrane integration of IMPs has not yet been defined, but it seems to be complex. YidC has been shown to interact specifically with the TMs of Sec-dependent IMPs such as FtsQ, leader peptidase (Lep), mannitol permease, and YidC itself at various stages in the membrane integration process (3, 13–18). This suggests a role in the lateral movement of TMs from the Sec translocase into the lipid bilayer. In addition, YidC might function in the initial recognition and reception of TMs, as it contacts the TM of Lep very early in biogenesis even before it is fully exposed outside the ribosome (16). Even though all tested Sec-dependent IMPs interact with YidC during membrane insertion, YidC depletion only mildly affects the assembly of Sec-dependent IMPs (7, 13). In contrast, assembly of Sec-independent IMPs is severely impaired upon YidC depletion (7). M13 procoat seems to be able to target and bind to the inner membrane independently of YidC, but it then requires YidC to translocate its periplasmic domain across the membrane, whereupon it attains its correct topology (8).
Because YidC seems to be dispensable for assembly of Sec-dependent IMPs and because known Sec-independent proteins that require YidC for proper integration are phage coat proteins, the question arises of why YidC is essential for cell viability. To identify E. coli IMPs that require YidC for proper membrane biogenesis, we analyzed the physiological consequences of YidC depletion. We observed that depletion of YidC results in a rapid and massive expression of PspA. PspA is a stress protein that responds to a variety of membrane-damaging treatments that ultimately result in a dissipation of the proton-motive force (PMF) (reviewed in ref. 19). Here, we show that the PMF is strongly reduced upon depletion of YidC and that this relates to decreased activity and defects in assembly of both cytochrome o oxidase and F1Fo ATPase.
Materials and Methods
Materials.
E. coli phospholipids were from Avanti Polar Lipids, spectinomycin sulfate was purchased from ICN, 9-amino-6-chloro-2-methoxyacridine (ACMA) was obtained from Molecular Probes, and tetraphenylphosphonium (TPP+) chloride was from Merck. All other chemicals were purchased from Sigma. YidC polyclonal antiserum was raised in rabbit against purified hisYidC (14) by AgriSera (Vännäs, Sweden). Polyclonal antisera against SecD, SecF, and SecE were raised in rabbits against the synthetic peptides IKEELSNGRTVQQAIDEGYRC (SecD), MAQEYTVEQLNHGRKC (SecF), and KGKATVAFAREARTEVRK (SecE), by AgriSera. Antiserum against SecY has been described (20). Antisera against PspA, SecA, DnaK, and DegP were kind gifts from J. Tommassen (University of Utrecht, Utrecht, The Netherlands), W. Wickner (Dartmouth Medical School, Hanover, NH), P. Geneveaux (University of Geneva, Geneva), and J. Beckwith (Harvard Medical School, Boston), respectively. Antisera against the Fob and Foc subunits of the F1Fo ATPase were a generous gift from G. Deckers-Hebestreit (University of Osnabrück, Osnabrück, Germany), and antiserum against cytochrome o oxidase was kindly provided by B. Gennis and B. Barquera (University of Illinois, Urbana).
YidC Depletion and Isolation of Inner Membrane Vesicles.
E. coli strain JS7131 (7), in which yidC is under the control of the araBAD operator/promoter, was grown overnight at 37°C in LB medium supplemented with 0.2% L-arabinose. Cells were harvested, washed with warm LB, diluted to an OD660 of 0.05, and further grown with 0.1% glucose to deplete YidC or with 0.1% L-arabinose to generate nondepleted control cells. The cultures were grown in volumes of 0.5 liter in 2-liter baffled flasks. At indicated time points, cells were harvested for Western blot analysis, PMF measurements, or the isolation of inverted inner membrane vesicles (IMVs) (21). For IMV isolation, 500 OD660 units were harvested of cultures grown in volumes of 1 liter in 2-liter baffled flasks. The harvested cultures had an OD660 of ≈0.15–0.2 after 1.5 h, ≈0.3–0.4 after 2.5 h, ≈0.4–0.5 for cultures grown on glucose for 3.5 h, and ≈0.8–0.9 for cultures grown on arabinose for 3.5 h.
PMF Measurements.
The transmembrane electrical potential (Δψ)-dependent uptake of TPP+ by intact cells was measured by using a homemade ion-selective electrode. JS7131 cells were permeabilized with 2 mM K-EDTA and 120 mM Tris, pH 8.0, at 0.1 g wet weight per ml for 5 min at 37°C (22). The cells were subsequently washed and resuspended in a buffer of 50 mM potassium phosphate, pH 8.0, containing 5 mM MgSO4 and 50 μg/ml chloramphenicol at 15 mg/ml protein. To ensure that the Δψ was the only component of the PMF, 1 μM nigericin was added, and the cells were stored on ice. Uptake of TPP+ was monitored with the electrode by using 1 mg/ml protein of cells at 25°C in the presence of 500 nM TPP+.
The transmembrane pH gradient (ΔpH) in IMVs was monitored by using the pH-sensitive fluorescent dye ACMA (23). Reaction mixtures at 30°C contained 12.5 μg/ml IMVs and 4 μM ACMA in 50 mM Hepes-KOH, pH 8.0/50 mM KCl/2 mM MgCl2/1 mM DTT/0.2 mg/ml BSA (buffer A). Reactions were started by the addition of 1.25 mM NADH or 1 mM ATP, and ACMA fluorescence was measured in a Perkin–Elmer LS50B luminescence spectrophotometer at excitation and emission wavelengths of 409 and 474 nm, respectively.
Enzymatic Assays.
ATPase activity of IMVs was measured at 37°C as described (24) by using malachite green. NADH consumption was determined at 25°C with 10 μg/ml IMVs in buffer A. Reactions were started by the addition of 200 μM NADH and monitored photometrically at 340 nm by using a Cary–Varian spectrophotometer. Activities were calculated by using a molar extinction coefficient ɛNADH of 6.3 × 103 liter⋅mol−1⋅cm−1.
NADH dehydrogenase activity was determined at 25°C by using 25 μg/ml IMVs and 50 μM of the electron acceptor 2,6-dichloroindophenol (DCIP) in buffer A containing 1% (wt/vol) Triton X-100. Reactions were initiated by the addition of 400 μM NADH. DCIP reduction was monitored photometrically at 600 nm. Activities were calculated by using a molar extinction coefficient ɛDCIP of 1.5 × 104 liter⋅mol−1⋅cm−1.
Cytochrome o oxidase activity was measured with an oxygen electrode (Yellow Springs Instruments) at 25°C (25). Reaction mixtures contained 100 mM potassium phosphate at pH 6.6, 5 mM MgCl2, 0.2 mg/ml E. coli phospholipids, 1.25% (wt/vol) octyl glucoside, 100 μg/ml IMVs, and 2.5 mM potassium ascorbate. Reactions were started by adding 10 μM of the artificial electron donor phenazine methosulfate (PMS).
Spectral Analysis.
Cytochrome absorption spectra of IMVs were determined by using a Cary–Varian spectrophotometer. Dithionite-reduced and air-oxidized absorption spectra of IMVs were recorded at 25°C with 0.5 mg/ml IMVs in 10 mM Hepes–KOH, pH 7.5.
Results
Depletion of YidC Induces a Strong and Specific PspA Stress Response.
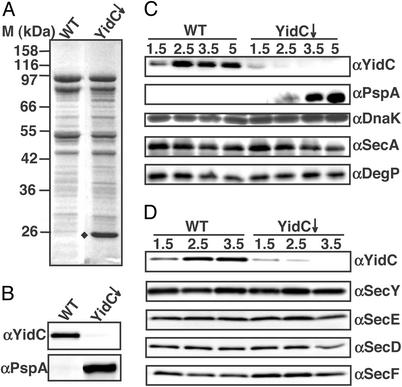
To study why YidC is essential for cell viability, we analyzed the physiological consequences of YidC depletion. First, the effect of YidC depletion on the protein composition of the inner membrane was examined. IMVs were purified from strain JS7131, in which YidC expression is under control of the araBAD operator/promoter (7). Cells were grown for 3.5 h in the presence of 0.1% arabinose (control cells, WT) or glucose (YidC-depleted cells), and used for the isolation of IMVs. As shown by immunoblotting, the amount of YidC in the IMVs decreased to an undetectable level when cells were grown under depleting conditions (Fig. 1B Upper). Analysis of the IMVs by SDS/PAGE and Coomassie brilliant blue staining revealed no dramatic changes in the overall protein profile except for the appearance of a protein of ≈25 kDa in the YidC-depleted IMVs (Fig. 1A). This protein was identified by immunoblotting as phage shock protein A (PspA; Fig. 1B Lower), a peripheral IMP with a predicted molecular mass of 25.6 kDa (reviewed in ref. 19). Because PspA has been characterized as a stress protein that is induced under conditions that affect the integrity of the inner membrane, we examined the PspA levels at various time points during YidC depletion to determine whether PspA induction is an early effect of YidC depletion or perhaps a late effect attributable to accumulative membrane damage. Cells were harvested after 1.5, 2.5, 3.5, and 5 h of depletion, and whole cell samples were analyzed by immunoblotting to determine the YidC and PspA content (Fig. 1C). YidC was still detectable after 1.5 h of depletion, but its signal decreased over time. PspA became well detectable after 2.5 h, when YidC was barely detectable. This observation indicates that PspA expression responds promptly to YidC depletion before significant effects on growth and cell morphology are observed (data not shown).
Figure 1.
YidC depletion induces PspA expression. IMVs were isolated from JS7131 cells depleted of YidC (YidC↓) for 3.5 h or grown under control conditions (WT) as described in Materials and Methods. IMVs (10 μg) were analyzed by SDS/PAGE and stained with Coomassie brilliant blue (A) or immunostained with anti- (α)YidC (B Upper) or αPspA (B Lower). (A, ♦) The ≈25-kDa protein PspA. (C) Effects of YidC depletion on the expression of stress-related proteins. Cells were depleted for 1.5, 2.5, 3.5, and 5 h or grown under control conditions; 0.05 OD660 unit of cells was analyzed by immunoblotting using antibodies against YidC, PspA, DnaK, SecA, and DegP. (D) Effects of YidC depletion on the expression of translocase components. IMVs were isolated from cells depleted of YidC for 1.5, 2.5, and 3.5 h; 10 μg of protein was analyzed by SDS/PAGE and immunoblotting with antibodies against YidC, SecY, SecE, SecD, and SecF.
Next, we analyzed whether the response is specific for PspA or part of a more general stress response. Cellular levels of three stress-related proteins were examined by immunoblotting: the cytosolic chaperone DnaK, the translocase component SecA, and the periplasmic chaperone/protease DegP. DnaK is a heat shock protein (26), SecA expression is induced upon secretion stress (27), and DegP expression responds to cell envelope stress (28). The levels of DnaK, DegP, and SecA did not change significantly upon YidC depletion (Fig. 1C). Taken together, the results indicate that YidC depletion results in a rapid, strong, and specific increase in the expression of PspA.
Among the stimuli that induce the PspA response are conditional mutations in secD, secF, and secA (29) and depletion of SecE (E. N. G. Houben and J.L., unpublished observation). Because part of YidC is physically associated with the Sec translocase (3, 4), we investigated whether YidC depletion influences the composition of the Sec translocase, which might indirectly stimulate PspA expression. IMVs derived from YidC-depleted and control cells were analyzed by immunoblotting to monitor the levels of the various translocase subunits (Fig. 1D). The levels of SecY, SecE, SecD, and SecF were hardly affected at 2.5 h when the PspA response became detectable. After prolonged depletion of YidC, the levels of SecE, SecD, and SecF did decrease slightly. These data strongly suggest that the induction of the PspA response is not caused by reduced levels of translocon components.
Depletion of YidC Affects the PMF.
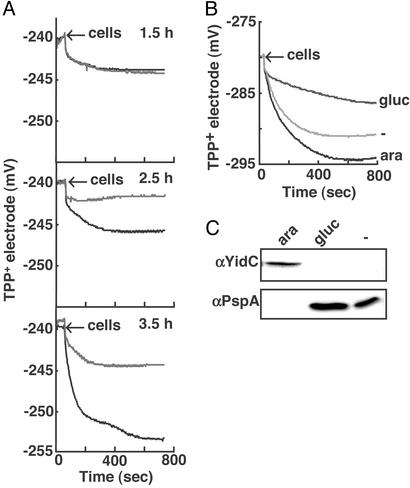
The primary signal for the PspA response is considered to be a reduction of the PMF (19). PspA expression responds to uncoupling agents (19), and a pspA mutant strain was shown to be affected in maintenance of the PMF under PspA-inducing stress conditions (30). Therefore, it was of interest to investigate the effect of YidC depletion on the PMF. For this purpose, Δψ was measured in intact cells by using a TPP+-selective electrode. Measurements were performed in the presence of nigericin (a K+/H+ antiporter) and potassium to ensure that Δψ is the only component of the PMF. Cells were depleted for YidC for 1.5, 2.5, and 3.5 h, and the outer membrane was permeabilized with EDTA. After 1.5 h of growth, control and YidC-depleted cells show a similar, low uptake of TPP+ (Fig. 2A). After 2.5 and 3.5 h of growth, cells grown in the presence of arabinose (WT) showed an increased TPP+ uptake as compared with the YidC-depleted cells, indicating that the PMF in the control cells is higher than in the YidC-depleted cells (Fig. 2A). The reduction of the PMF in YidC-depleted cells coincides with the elevated PspA expression at 2.5 h (Fig. 1C).
Figure 2.
YidC depletion affects the PMF. (A) Cells were grown as described in Materials and Methods. After 1.5, 2.5, or 3.5 h of depletion, cells were harvested and pretreated, and TPP+ uptake was measured as described in Materials and Methods. The WT signals are shown in black and the YidC↓ signals are shown in gray. (B) Cells were grown overnight in the presence of 0.2% arabinose, washed, and diluted in LB medium containing no sugars, 0.1% glucose, or arabinose. After 3.5 h, the cells were harvested and treated as described above. (C) Cell samples of B were taken and analyzed by SDS/PAGE and Western blotting with antibodies against YidC and PspA.
Cells that are depleted for YidC are grown in the presence of glucose instead of arabinose. Addition of glucose to aerobically grown cells has been shown to alter the expression of certain proteins involved in respiration, such as succinate dehydrogenase (31). To exclude the possibility that the diminished PMF in YidC-depleted cells is due to an effect of glucose on the expression of respiratory chain components rather than a direct consequence of the loss of YidC, the TPP+ uptake measurements were repeated with JS7131 grown for 3.5 h in the absence of sugars and in the presence of glucose or arabinose. The TPP+ uptake of cells grown in the absence of sugars is reduced compared with growth on arabinose, but not as dramatically as in YidC-depleted cells grown on glucose (Fig. 2B). Cells grown in the absence of sugars are depleted for YidC and show enhanced levels of PspA (Fig. 2C), albeit less pronounced than cells grown in the presence of glucose. In conclusion, the PspA response and concomitantly the reduction of the PMF seem to be directly related to the reduced levels of YidC present in YidC-depleted JS7131.
Depletion of YidC Interferes with the Membrane Assembly of Energy-Transducing Enzymes.
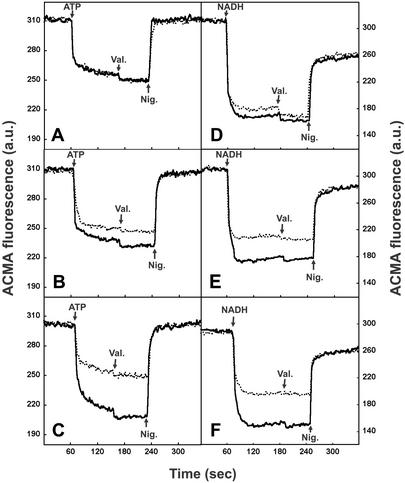
To examine the effects of YidC depletion on the PMF in more detail, we compared IMVs purified from control and YidC-depleted cells with respect to their ability to generate a PMF in vitro. IMVs were energized with either ATP or NADH, and the generation of a PMF was monitored with the pH-sensitive fluorescent dye ACMA. To ensure that the ΔpH is the only component of the PMF, the generated Δψ was converted to a ΔpH by the addition of valinomycin (a K+ ionophore) in the presence of K+ ions. Upon depletion of YidC, a clear defect in PMF generation with either ATP or NADH was observed, which progressed during depletion (Fig. 3). These results are consistent with the in vivo measurements and demonstrate that YidC-depleted IMVs have a reduced capacity to generate a PMF.
Figure 3.
YidC-depleted IMVs are defective in PMF generation. JS7131 IMVs were prepared from cultures grown for 1.5 (A and D), 2.5 (B and E), or 3.5 (C and F) h on 0.1% glucose (YidC↓, dotted line) or arabinose (WT, continuous line). The generation of ΔpH was determined by monitoring the fluorescence quenching of ACMA. Where indicated, 1 mM ATP (A–C) or 1.25 mM NADH (D–F) was added to the IMVs to generate a PMF. Valinomycin was used at 0.75 μM to convert the generated Δψ into a ΔpH. Subsequently, the ΔpH was dissipated by the addition of 1.5 μM nigericin.
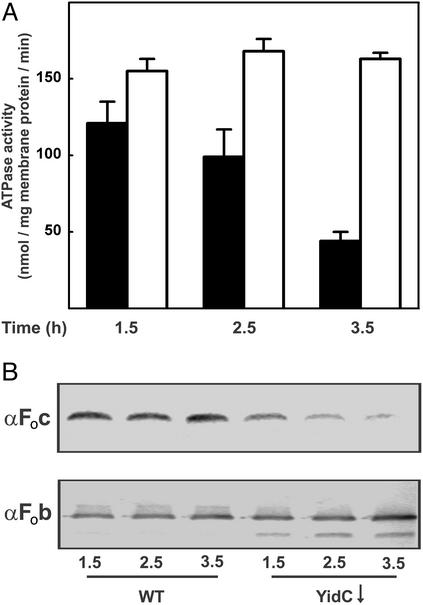
The effect of YidC depletion on the PMF could be caused by an increased proton permeability of the membrane but, alternatively, could be caused by reduced levels of energy-transducing enzymes, such as F1Fo ATPase and respiratory chain components. Therefore, control and YidC-depleted IMVs were tested for their total ATPase activity. In WT IMVs, 95% of this activity can be attributed to F1Fo ATPase (M.v.d.L., unpublished data). The ATPase activity of YidC-depleted IMVs gradually decreased as compared with the control, resulting in a residual activity of only 26% after 3.5 h (Fig. 4A). This decrease correlated with a drastically reduced amount of the small, ring-forming Foc subunit of F1Fo ATPase in the membrane as detected by immunoblotting (Fig. 4B). The total amount of the Fob subunit was unchanged, but this subunit seemed more susceptible to proteolytic degradation, because an additional lower molecular weight Fob fragment was observed in the YidC-depleted IMVs (Fig. 4B). Depletion of SecDFYajC, which associates with YidC (4), had no effect on the Foc and Fob levels (data not shown).
Figure 4.
Depletion of YidC leads to a decreased amount of functional F1Fo ATPase complex. JS7131 IMVs were prepared from cultures grown for 1.5, 2.5, or 3.5 h on 0.1% glucose (black bars, YidC↓) or arabinose (white bars, WT). (A) Total ATPase activity of IMVs. The data points are the average of three independent measurements. (B) SDS/PAGE and immunoblot analysis of the Fob and Foc subunit levels in IMVs.
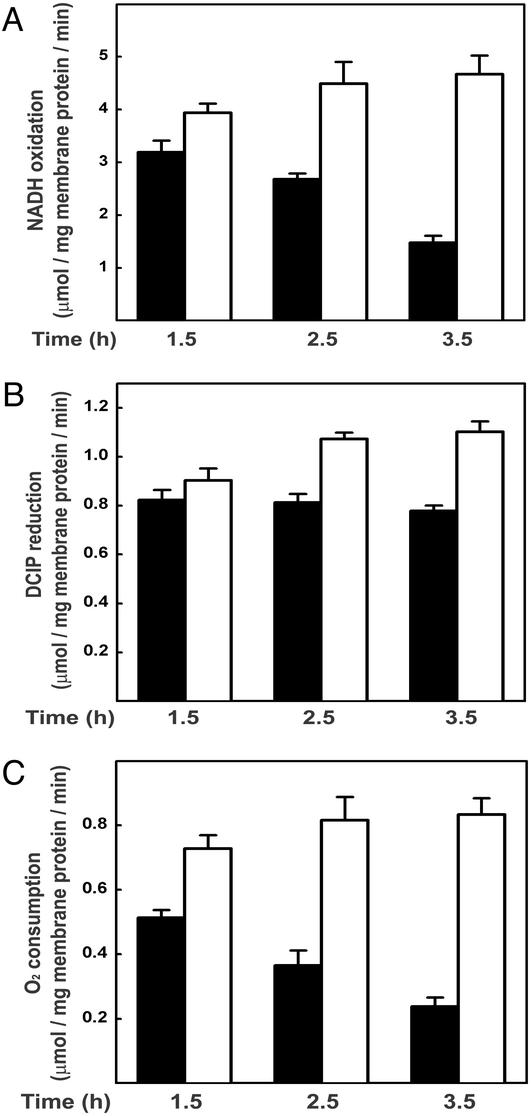
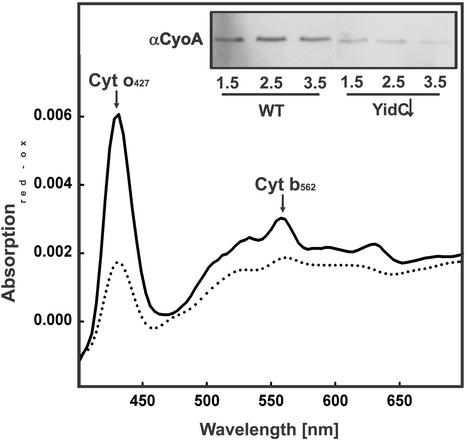
As described above, YidC depletion also affected PMF generation with NADH. Therefore, we measured the NADH consumption of YidC-depleted and control IMVs as a measure of the total respiratory activity. NADH oxidation activity of YidC-depleted IMVs was ≈32% compared with control IMVs after 3.5 h of depletion (Fig. 5A). This result suggests that YidC depletion also causes a defect in the respiratory electron transport chain. In these experiments, electron transport from NADH to oxygen is measured. This process involves two multisubunit membrane protein complexes, i.e., NADH dehydrogenase and cytochrome o oxidase, that are connected by the small electron carrier ubiquinol. To further specify the defect, activities of NADH dehydrogenase and cytochrome o oxidase were measured separately. For this purpose, the NADH dehydrogenase activity of detergent-solubilized IMVs was measured through the NADH-dependent reduction of the artificial electron acceptor DCIP. Remarkably, this activity was hardly affected by the YidC depletion (Fig. 5B), suggesting the presence of a functional NADH dehydrogenase complex. On the other hand, measurements of the cytochrome o oxidase activity by using the artificial electron donor phenazine methosulfate revealed a strong reduction upon YidC depletion (Fig. 5C). Reduced versus oxidized absorption spectra of the IMVs demonstrate that the loss of cytochrome o oxidase activity correlates with a reduced amount of the cofactors cytochrome o and cytochrome b562 (Fig. 6), which are bound to the CyoB subunit (subunit I). As shown by immunoblotting, the amount of the ubiquinol-binding subunit CyoA (subunit II) is also decreased upon depletion of YidC (Fig. 6 Inset). Strikingly, the reduction of cytochrome o oxidase activity quantitatively resembles the effect of YidC depletion on NADH consumption, indicating that the reduced electron transport activity is mainly caused by a defect in cytochrome o oxidase assembly. Taken together, these results show that depletion of YidC has a dramatic effect on the functional assembly of F1Fo ATPase and cytochrome o oxidase, which are the major energy-transducing membrane protein complexes in E. coli grown under aerobic conditions.
Figure 5.
Depletion of YidC leads to a decreased activity of the respiratory chain. JS7131 IMVs were prepared from cultures grown for the indicated times on 0.1% glucose (YidC↓, black bars) or arabinose (WT, white bars). (A) NADH consumption was monitored photometrically as decrease of absorption at 340 nm. (B) NADH dehydrogenase activities were determined by using Triton X-100-solubilized IMVs and the artificial electron acceptor DCIP. NADH-dependent reduction of DCIP was monitored photometrically as decrease of absorption at 600 nm. (C) Cytochrome o oxidase activities were determined by using the artificial electron donor system phenazine methosulfate/ascorbate. Phenazine methosulfate/ascorbate-dependent O2 consumption was measured by using an oxygen electrode. The data points are the average of three independent measurements.
Figure 6.
Depletion of YidC leads to a decreased amount of functional cytochrome o oxidase complex. JS7131 IMVs were prepared from cultures grown for 1.5, 2.5, or 3.5 h on 0.1% glucose (YidC↓) or arabinose (WT). Difference spectra (dithionite-reduced minus air-oxidized) of YidC-depleted (3.5 h on 0.1% glucose, dotted line) or WT (3.5 h on 0.1% arabinose, continuous line) IMVs were recorded. (Inset) IMVs were analyzed by SDS/PAGE and immunostaining with antibodies against cytochrome o oxidase.
Discussion
YidC is involved in the biogenesis of IMPs in E. coli. The precise function(s) and the essential nature of YidC, however, have remained elusive. In the present study, we show that depletion of YidC rapidly and specifically induces the expression of PspA, a stress response that was previously suggested to be triggered by a dissipation of the PMF (19). Consistently, YidC depletion was shown to affect the PMF, which could be attributed to an early and drastic effect on the functional assembly of cytochrome o oxidase and F1Fo ATPase complexes.
The induction of PspA expression upon depletion of YidC proved to be a reliable indicator of a reduced PMF in line with previous suggestions that PspA senses membrane damage and/or a reduction of the PMF (19, 30). Both in vivo and in vitro measurements demonstrated a decrease in the PMF that coincides with PspA induction. Under aerobic conditions, the PMF is generated mainly by the respiratory chain membrane protein complexes NADH dehydrogenase and cytochrome o oxidase (32). The overall respiratory activity is reflected by the NADH oxidation rate of IMVs, which is drastically reduced in YidC-depleted IMVs. When the functional link between NADH dehydrogenase and cytochrome o oxidase is interrupted by solubilization of the membrane and the addition of an artificial electron acceptor or acceptor, the activities of both enzyme complexes can be determined separately. Depletion of YidC leads to a strong reduction of cytochrome o oxidase activity, whereas the NADH dehydrogenase activity is hardly affected. However, if cytochrome o oxidase activity is lowered, electron transfer will be reduced as well, resulting in reduced NADH oxidation by NADH dehydrogenase. Examination of the content of YidC-depleted inner membranes revealed a reduction in the levels of subunit II (CyoA) of cytochrome o oxidase and of the subunit I (CyoB)-associated cytochromes o and b562. Interestingly, cytochrome o oxidase is homologous to the cytochrome c oxidase in mitochondria, which depends on the YidC homologue Oxa1p for proper insertion and assembly of its various constituents (11, 12). Oxa1p facilitates the insertion of a variety of mitochondrially encoded proteins, but of all of the substrates tested, the CyoA homologue, Cox2p, displays the strictest dependency on Oxa1p for membrane insertion (11).
Saccharomyces cerevisiae mutated in OXA1 shows respiratory defects not only because of a reduced assembly of the terminal oxidase but also because the F1Fo ATPase seems to be compromised (12). Likewise, we observed that in YidC-depleted inner membranes, the ATPase activity was severely reduced. In E. coli, the proton-conducting domain, Fo, consists of three subunits (a–c) (reviewed in ref. 33). Depletion of YidC leads to a strong reduction of the amount of the Foc subunit in IMVs, whereas the Fob subunit is normally inserted but seems to be subject to degradation. This degradation of Fob might be caused by the reduction of Foc levels, leaving unassembled Fob. The Foc is a small IMP with two TMs, which assembles into a ring structure comprising 10 subunits (34). Purified Foc subunits self-assemble into a ring-like complex when present in detergent solution (35). However, membrane insertion and assembly of the functional Fo subcomplex in vivo could be more complicated and require assistance, for instance, to ensure the proper timing of subsequent assembly events. YidC could be involved in the insertion of Fo components, assembly of the Fo subcomplex, or both. In support of a combined function of YidC, Oxa1p seems to be involved in both insertion and assembly of the F1Fo ATPase. In the mitochondria of an OXA1 deletion strain, the amounts of several Fo sector subunits were dramatically reduced; subunit c (Atp9) was decreased by ≈40%. Interestingly, it was shown that when the intermembrane space AAA protease YME1 was deleted in combination with OXA1, F1Fo ATPase subunits were properly localized even in the absence of Oxa1p, and 80% of the oligomycin-sensitive ATPase activity could be restored (10). Therefore, it seems that in mitochondria Oxa1p assists and protects the assembling ATPase from proteases without being absolutely required for membrane insertion of the Fo subunits. Irrespective of the precise molecular mechanism, the involvement of the YidC/Oxa1p protein family in the biogenesis of energy-transducing membrane protein complexes seems to be evolutionarily conserved, as already suggested by Dujardin and coworkers (36).
A reduced integration/assembly of the terminal oxidase is mostly likely the primary cause of the loss of PMF upon depletion of YidC in vivo. At present, we cannot exclude that other effects of YidC depletion injure the integrity of the inner membrane and hence cause a dissipation of the PMF. However, because the translocation of presecretory proteins is only marginally sensitive to YidC depletion (7) and the levels of key Sec components are not reduced upon YidC depletion, a jammed or otherwise compromised Sec translocase seems an unlikely explanation for the observed effects.
It has been shown before that the PMF influences the membrane integration and assembly of a variety of membrane proteins. The PMF stimulates the insertion and translocation of periplasmic domains of membrane proteins (37, 38), the oligomerization of protomers into complexes (39), and the transmembrane topology of membrane proteins (40). In this respect, the relatively mild effects of YidC depletion on the integration of the Sec-dependent IMPs Lep and FtsQ could be due to the reduced PMF rather than the lack of YidC (7, 13). Also, the putative YidC substrate M13 procoat was shown to require a PMF for proper membrane integration (38). However, it seems unlikely that the M13 procoat insertion defect under YidC-depleting conditions is caused solely by PMF reduction, because the insertion of a mutated, PMF-independent M13 procoat was also shown to be impaired upon depletion of YidC (8), although the effect was less pronounced.
Our study identifies two E. coli IMPs that strongly depend on YidC for proper biogenesis: CyoA and Foc. These proteins have in common that they are rather small integral membrane proteins with two closely spaced TMs. They both participate in multisubunit oligomeric complexes. These could be natural substrates of the YidC-dependent membrane protein insertion pathway in E. coli that is also used by certain phage coat proteins. Future experiments should focus on the mechanism of integration and assembly of CyoA and Foc.
Acknowledgments
We thank G. van Eikenhorst and Dr. K. Krab for advice and assistance with the TPP+ measurements. This work was supported by the Earth and Life Science Foundation and the Council for Chemical Sciences of the Netherlands Organization for Scientific Research, which are subsidized by the Netherlands Organization for Scientific Research.
Abbreviations
- IMP
inner membrane protein
- TM
transmembrane segment
- PMF
proton-motive force
- ACMA
9-amino-6-chloro-2-methoxyacridine
- TPP+
tetraphenylphosphonium
- IMV
inverted inner membrane vesicle
- DCIP
2,6-dichlorindophenol
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Herskovits A A, Bochkareva E S, Bibi E. Mol Microbiol. 2000;38:927–939. doi: 10.1046/j.1365-2958.2000.02198.x. [DOI] [PubMed] [Google Scholar]
- 2.Manting E H, Driessen A J M. Mol Microbiol. 2000;37:226–238. doi: 10.1046/j.1365-2958.2000.01980.x. [DOI] [PubMed] [Google Scholar]
- 3.Scotti P A, Urbanus M L, Brunner J, de Gier J W, von Heijne G, van der Does C, Driessen A J M, Oudega B, Luirink J. EMBO J. 2000;19:542–549. doi: 10.1093/emboj/19.4.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nouwen N, Driessen A J M. Mol Microbiol. 2002;44:1397–1405. doi: 10.1046/j.1365-2958.2002.02972.x. [DOI] [PubMed] [Google Scholar]
- 5.Kuhn A. FEMS Microbiol Rev. 1995;17:185–190. doi: 10.1111/j.1574-6976.1995.tb00201.x. [DOI] [PubMed] [Google Scholar]
- 6.de Gier J W, Scotti P A, Sääf A, Valent Q A, Kuhn A, Luirink J, von Heijne G. Proc Natl Acad Sci USA. 1998;95:14646–14651. doi: 10.1073/pnas.95.25.14646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samuelson J C, Chen M Y, Jiang F L, Möller I, Wiedmann M, Kuhn A, Phillips G J, Dalbey R E. Nature. 2000;406:637–641. doi: 10.1038/35020586. [DOI] [PubMed] [Google Scholar]
- 8.Samuelson J C, Jiang F, Yi L, Chen M, de Gier J W, Kuhn A, Dalbey R E. J Biol Chem. 2001;276:34847–34852. doi: 10.1074/jbc.M105793200. [DOI] [PubMed] [Google Scholar]
- 9.Luirink J, Samuelsson T, de Gier J W. FEBS Lett. 2001;501:1–5. doi: 10.1016/s0014-5793(01)02616-3. [DOI] [PubMed] [Google Scholar]
- 10.Lemaire C, Hamel P, Velours J, Dujardin G. J Biol Chem. 2000;275:23471–23475. doi: 10.1074/jbc.M002045200. [DOI] [PubMed] [Google Scholar]
- 11.Hell K, Neupert W, Stuart R A. EMBO J. 2001;20:1281–1288. doi: 10.1093/emboj/20.6.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stuart R. Biochim Biophys Acta. 2002;1592:79–87. doi: 10.1016/s0167-4889(02)00266-5. [DOI] [PubMed] [Google Scholar]
- 13.Urbanus M L, Scotti P A, Fröderberg L, Sääf A, de Gier J W L, Brunner J, Samuelson J C, Dalbey R E, Oudega B, Luirink J. EMBO Rep. 2001;2:524–529. doi: 10.1093/embo-reports/kve108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Laan M, Houben E N G, Nouwen N, Luirink J, Driessen A J M. EMBO Rep. 2001;2:519–523. doi: 10.1093/embo-reports/kve106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houben E N G, Scotti P A, Valent Q A, Brunner J, de Gier J W, Oudega B, Luirink J. FEBS Lett. 2000;476:229–233. doi: 10.1016/s0014-5793(00)01735-x. [DOI] [PubMed] [Google Scholar]
- 16.Houben E N G, Urbanus M L, van der Laan M, ten Hagen-Jongman C M, Driessen A J M, Brunner J, Oudega B, Luirink J. J Biol Chem. 2002;277:35880–35886. doi: 10.1074/jbc.M205556200. [DOI] [PubMed] [Google Scholar]
- 17.Beck K, Eisner G, Trescher D, Dalbey R E, Brunner J, Müller M. EMBO Rep. 2001;2:709–714. doi: 10.1093/embo-reports/kve154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Urbanus M L, Fröderberg L, Drew D, Björk P, de Gier J W, Brunner J, Oudega B, Luirink J. J Biol Chem. 2002;277:12718–12723. doi: 10.1074/jbc.M200311200. [DOI] [PubMed] [Google Scholar]
- 19.Model P, Jovanovic G, Dworkin J. Mol Microbiol. 1997;24:255–261. doi: 10.1046/j.1365-2958.1997.3481712.x. [DOI] [PubMed] [Google Scholar]
- 20.van der Does C, Manting E H, Kaufmann A, Lutz M, Driessen A J M. Biochemistry. 1998;37:201–210. doi: 10.1021/bi972105t. [DOI] [PubMed] [Google Scholar]
- 21.de Vrije T, Tommassen J, de Kruijff B. Biochim Biophys Acta. 1987;900:63–72. doi: 10.1016/0005-2736(87)90278-1. [DOI] [PubMed] [Google Scholar]
- 22.Elferink M G L, Hellingwerf K J, Belkum M J, Poolman B, Konings W N. FEMS Microbiol Lett. 1984;21:293–298. [Google Scholar]
- 23.Nouwen N, van der Laan M, Driessen A J M. FEBS Lett. 2001;508:103–106. doi: 10.1016/s0014-5793(01)03033-2. [DOI] [PubMed] [Google Scholar]
- 24.Lanzetta P A, Alvarez L J, Reinach P S, Candia O A. Anal Biochem. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- 25.Matsushita K, Patel L, Kaback H R. Biochemistry. 1984;23:4703–4714. doi: 10.1021/bi00315a028. [DOI] [PubMed] [Google Scholar]
- 26.Lindquist S, Craig E A. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 27.Oliver D B, Beckwith J. Cell. 1982;30:311–319. doi: 10.1016/0092-8674(82)90037-x. [DOI] [PubMed] [Google Scholar]
- 28.Raivio T L, Silhavy T J. J Bacteriol. 1997;179:7724–7733. doi: 10.1128/jb.179.24.7724-7733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleerebezem M, Tommassen J. Mol Microbiol. 1993;7:947–956. doi: 10.1111/j.1365-2958.1993.tb01186.x. [DOI] [PubMed] [Google Scholar]
- 30.Kleerebezem M, Crielaard W, Tommassen J. EMBO J. 1996;15:162–171. [PMC free article] [PubMed] [Google Scholar]
- 31.Park S J, Tseng C P, Gunsalus R P. Mol Microbiol. 1995;15:473–482. doi: 10.1111/j.1365-2958.1995.tb02261.x. [DOI] [PubMed] [Google Scholar]
- 32.Gennis R B, Stewart V. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 217–261. [Google Scholar]
- 33.Pedersen P L, Ko Y H, Hong S. J Bioenerg Biomembr. 2000;32:325–332. doi: 10.1023/a:1005594800983. [DOI] [PubMed] [Google Scholar]
- 34.Jiang W, Hermolin J, Fillingame R H. Proc Natl Acad Sci USA. 2001;98:4966–4971. doi: 10.1073/pnas.081424898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arechaga I, Butler P J, Walker J E. FEBS Lett. 2002;515:189–193. doi: 10.1016/s0014-5793(02)02447-x. [DOI] [PubMed] [Google Scholar]
- 36.Bonnefoy N, Chalvet F, Hamel P, Slonimski P P, Dujardin G. J Mol Biol. 1994;239:201–212. doi: 10.1006/jmbi.1994.1363. [DOI] [PubMed] [Google Scholar]
- 37.Whitley P, Zander T, Ehrmann M, Haardt M, Bremer E, von Heijne G. EMBO J. 1994;13:4653–4661. doi: 10.1002/j.1460-2075.1994.tb06788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao G, Kuhn A, Dalbey R E. EMBO J. 1995;14:866–875. doi: 10.1002/j.1460-2075.1995.tb07068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Dalen A, Schrempf H, Killian J A, de Kruijff B. EMBO Rep. 2000;1:340–346. doi: 10.1093/embo-reports/kvd067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kiefer D, Kuhn A. EMBO J. 1999;18:6299–6306. doi: 10.1093/emboj/18.22.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]