Abstract
Membrane lipids were once thought to be homogenously distributed in the 2D surface of a membrane, but the lipid raft theory suggests that cholesterol and sphingolipids partition away from other membrane lipids. Lipid raft theory further implicates these cholesterol-rich domains in many processes such as signaling and vesicle traffic. However, direct characterization of rafts has been difficult, because they cannot be isolated in pure form. In the first functional proteomic analysis of rafts, we use quantitative high-resolution MS to specifically detect proteins depleted from rafts by cholesterol-disrupting drugs, resulting in a set of 241 authentic lipid raft components. We detect a large proportion of signaling molecules, highly enriched versus total membranes and detergent-resistant fractions, which thus far biochemically defined rafts. Our results provide the first large-scale and unbiased evidence, to our knowledge, for the connection of rafts with signaling and place limits on the fraction of plasma membrane composed by rafts.
Membrane lipid rafts (1–3), despite years of study, remain elusive and controversial entities. Many biophysical, biochemical, and microscopy studies suggest that lipid rafts are real and not artifacts of detergent extractions (4–7). The general lipid composition of rafts is understood to such a degree that rafts can be regenerated in synthetic vesicles containing only lipids (8), but the protein composition of rafts remains contentious. Lipid rafts are functionally defined by their dependence on cholesterol, but this definition cannot be translated to a generally agreed on purification scheme. Two common methods have been used to isolate rafts, resistance to either high pH or nonionic detergents, and in both methods rafts are separated from other proteins by density gradient centrifugation. Detergent resistance is the much more widely used of the two but, as with any biochemical fractionation, both methods are plagued by issues of contamination from nonraft proteins. Here we overcome this problem by applying a recently described method in quantitative proteomics [stable isotope-labeling with amino acids in cell culture (SILAC) (9)] to directly determine the subset of cholesterol-dependent proteins in the biochemical preparation.
Methods
SILAC.
Two populations of HeLa cells were grown in leucine-deficient DMEM/10% dialyzed FBS. One population was supplemented with normal isotopic abundance l-leucine (Leu, 105 mg/liter, Sigma) and the other with 99% isotopic abundance l-leucine-3,3,3-D3 (LeuD3, 107.4 mg/liter, Aldrich), as described (9). Each population was grown for at least three passages encompassing a minimum of seven population doublings. Four 14-cm plates each of Leu and LeuD3 cells were serum-starved for 18 h before treatment with cholesterol-disrupting agents and preparation of detergent- and pH/carbonate-resistant fractions.
Detergent- and pH/Carbonate-Resistant Fractions.
After serum starvation, one population of cells was treated with a cholesterol-disrupting drug [nystatin, 60 min at 50 μg/ml; filipin, 30 min at 25 μg/ml; or methyl-β-cyclodextrin (MβCD), 60 min at 5 mM, all at 37°C], and the other was treated with an appropriate carrier control. In most cases, the Leu population of cells received the drug to more easily differentiate contaminating proteins from authentic proteins being altered by the treatment (see Results and Discussion). When reverse labeling was performed in some experiments, similar results were observed. A low-density detergent-resistant fraction was prepared as described (10). Basically Leu- and LeuD3-encoded cells were lysed for 1 h at 4°C in 1% Triton X-100 (in 25 mM Mes, pH 6.5), clarified, and the protein content of each lysate measured by Coomassie (Pierce). Equal amounts of protein from the Leu (treated) and LeuD3 (untreated) lysates were combined and mixed with an equal volume of 90% sucrose in Mes-buffered saline (MBS) (150 mM NaCl/25 mM Mes, pH 6.5) for a final sucrose concentration of 45%. This solution was then placed in the bottom of an ultracentrifuge tube as the base of a discontinuous sucrose gradient. Additional layers consisting of 35% and 5% sucrose in MBS were gently placed on top, and the whole gradient was centrifuged at 166,000 × g for 18 h at 4°C. The resulting low-density light-scattering band (≈18% sucrose) was extracted, diluted 4× in MBS, and centrifuged an additional 2 h (166,000 × g, 4°C) to pellet the detergent-resistant material. The pH/carbonate-resistant fraction was prepared in a similar way, with the exception that the cells were lysed in 100 mM Na2CO3, pH 11.0, and mechanically disrupted by 10 strokes in a Dounce homogenizer and three 20-s bursts of a probe sonicator, as described (11).
High-Performance Liquid Chromatography/Tandem MS (LC/MS/MS), Database Searching, and Quantitation.
In most experiments, the pellet from the second centrifugation was solubilized in a small volume of 6 M urea/2 M thiourea (in 10 mM Hepes, pH 8.0), from which the proteins were precipitated by diluting 5× with absolute ethanol and adjusted to 50 mM NaCH3COO with a 2.5 M stock solution, pH 5.0. After the solution was allowed to stand for 2 h, the precipitated proteins were pelleted by centrifugation for 10 min at 12,000 × g at room temperature (r.t.). The pellet from this step was then resolubilized in urea/thiourea, reduced (1 μg of DTT per 50-μg estimated sample protein for 30 min at r.t.) (12), alkylated (5 μg of iodoacetamide per 50-μg sample protein for 30 min at r.t.), digested with LysC (1 μg/50 μg sample protein for 3 h at r.t.), diluted 4× with 50 mM NH4HCO3, and digested with trypsin (1 μg/50 μg sample protein for 12 h at r.t.). For MβCD-treated detergent-resistant fractions, samples were prepared as above or by first partially resolving the proteins by SDS/PAGE and then in-gel digesting (12) the entire lane after cutting it into six fractions. Peptide mixtures were then desalted by using stop and go extraction (STAGE) tips (13) and bomb-loaded onto reversed-phase analytical columns for LC (14). Peptides were eluted from the analytical columns by three-step linear 2.5-h gradients running from 5.6% to 64% acetonitrile and sprayed directly into the orifice of a QSTAR-Pulsar quadrupole time-of-flight (TOF) hybrid MS (PE-Sciex, Thornhill, Ontario, Canada) (14). Proteins were identified by LC/MS/MS (15) by information-dependent acquisition of fragmentation spectra for multiply charged peptides that were then searched against the Human International Protein Index database (ftp://ftp.ebi.ac.uk/pub/databases/IPI/old/HUMAN) by using mascot (Matrix Science, London). The following search parameters were used in all mascot searches: maximum of one missed trypsin cleavage, cysteine carbamidomethylation, methionine oxidation, leucine with three extra deuterium atoms, and a maximum 0.2-Da error tolerance in both the MS and MS/MS data. Protein hits with one ions score >45 or two >30 were considered identified with no manual inspection. All other hits were manually verified by using accepted rules for peptide fragmentation in a quadrupole–TOF hybrid MS. Possible redundancy arising from combining datasets was minimized by blasting (ftp://ftp.ncbi.nih.gov/blast/executables), a FASTA library of all of the amino acid sequences against itself. Any two sequences with 100% overlap of one sequence with the other were considered redundant. mascot outputs were parsed to obtain peptide-pair lists, and extracted ion chromatograms (XICs) were generated from survey scan data by using SPINX (SILAC Peptide INtegration by XIC), an in-house script. Where one peptide in a pair had not been selected for sequencing, its mass was calculated, and its elution time window was predicted.
Measurement of Cholesterol Content.
Serum-starved HeLa cells were treated with MβCD, nystatin, or filipin as described, washed three times with PBS, and scraped into PBS. Total cellular lipid content was extracted from 10-μg (total protein, determined by the Coomassie method) aliquots of each treatment as described (16), scaling all volumes down by 103. Cholesterol content of each sample was measured by using the Amplex red kit (Molecular Probes), according to the manufacturer's directions.
Preparation of Total Membranes.
Serum-starved HeLa cells were scraped into homogenization buffer [20 mM Hepes, pH 7.4/255 mM sucrose/1 mM EDTA with Complete Protease Inhibitors (Roche Diagnostics)] and homogenized by 15 strokes of a Dounce homogenizer (17). Nuclei, mitochondria, and large cellular debris were pelleted by centrifugation at 10,000 × g for 10 min. The postnuclear supernatant was transferred to an ultracentrifuge tube and centrifuged for 2 h at 245,000 × g to pellet remaining membranes. These proteins were then prepared for LC/MS/MS as described.
Results and Discussion
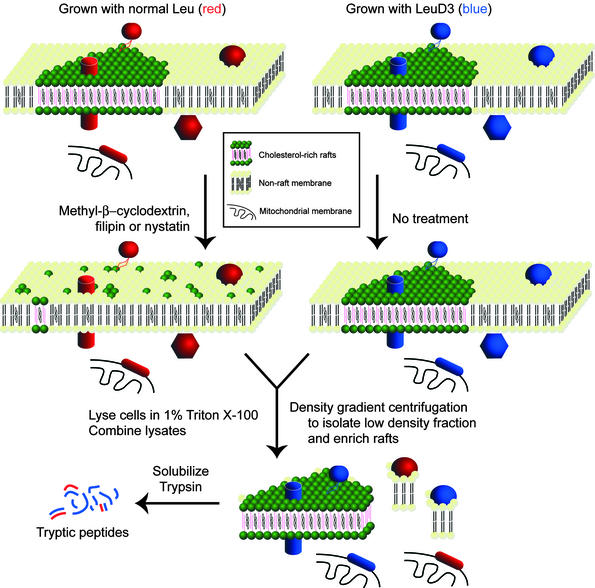
We encode all of the proteins in one of two cell populations by metabolically labeling with deuterium-substituted leucine (9). One of these populations is treated with a cholesterol-disrupting drug to break up lipid rafts in those cells. The treated and untreated cells are then combined and fractionated biochemically. No rafts originating from the treated cells will be isolated, because the drug treatment will have significantly disrupted their structural integrity (Fig. 1). The prior isotope-encoding will then reveal specific quantitative changes of only the cholesterol-dependent proteins in the resistant fraction (Figs. 1 and 2).
Figure 1.
Isolation scheme. Leu-labeled HeLa cells (depicted by red proteins/peptides) were treated with a cholesterol-disrupting agent, lysed, combined with lysates of LeuD3-labeled untreated HeLa cells (depicted by blue proteins/peptides), and used to prepare a detergent-resistant fraction. Because rafts in the drug-treated cells have lost their structural integrity they no longer are purified in the detergent-resistant fraction, whereas nonraft contaminants originating from treated and untreated samples will copurify. Tryptic peptides were then prepared from isolated detergent- or pH/carbonate-resistant fractions, as described in Methods.
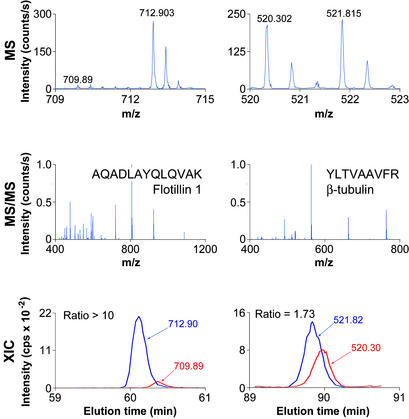
Figure 2.
MS and chromatograms. Representative MS, MS/MS, and extracted ion chromatograms (XIC) for peptides from flotillin 1 and β-tubulin, two proteins identified in this study. Multiply charged peptides observed in MS mode were selected for fragmentation (MS/MS). Ion chromatograms (intensity vs. time) of Leu- and LeuD3-containing peptides identified by MS/MS were extracted from the series of MS scans and integrated by using SPINX (see Methods).
We used two raft isolation schemes (detergent and pH/carbonate resistance) and three cholesterol-disrupting drugs (nystatin, filipin, and MβCD) to generate six treated/untreated pairs for quantitative proteomic analysis. Equal amounts of lysate protein from the treated (normal leucine, Leu) and untreated (triply deuterated leucine, LeuD3) cells were combined and used to prepare a single detergent-resistant fraction. Combining the lysates before isolation of the resistant fractions eliminates any potential variability from differences in individual preparations. Subsequent quantitative analysis by MS revealed cholesterol-depleted proteins through the lower peak heights of Leu versus LeuD3 peptides (Fig. 2). The complete list of identified proteins, including measured ratios and accession numbers, is published as supporting information on the PNAS web site, www.pnas.org.
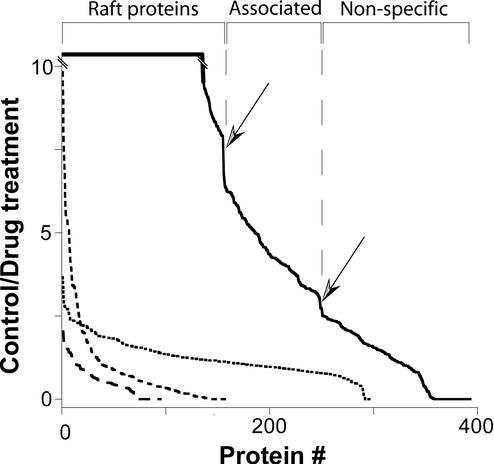
In pH/carbonate-resistant rafts, all three drugs worked as expected, decreasing the levels of a large group of proteins in the preparation, including several known raft proteins. However, although changes were seen with each drug, the magnitude of the effects was unremarkable, typically reaching no more than 2.5-fold. Furthermore, when plotted in order of decreasing ratio, no distinct group of values was discernible that might represent raft-specific proteins (Fig. 3). Even though the drug effects in this preparation were largely specific for known or plausible raft proteins (i.e., higher depletion ratios seen for integral-membrane proteins, hydrophobically modified proteins, etc.), the size of the effects made it difficult to determine a threshold over which proteins could confidently be considered as true raft components. The pH/carbonate preparation also contained a higher than expected number of proteins from cellular locations such as the mitochondria that, while membranous, are unlikely to contain rafts (1).
Figure 3.
Control/drug-treated ratios. Ratios for detergent-resistant proteins (solid line, control/MβCD; medium dashes, control/filipin; long dashes, control/nystatin) and pH/carbonate-resistant proteins (short dashes, average of ratios for nystatin, filipin, and MβCD) plotted from largest to smallest. Discontinuities in the detergent-resistant plot are marked by arrows and correspond roughly to ratios of 7.5 and 3.
Nystatin and filipin were even less efficient at depleting proteins in detergent-resistant fractions, resulting in only very small differences between untreated- and drug-treated samples (Fig. 3). We attribute this weak effect to the propensity of these drugs to sequester cholesterol but not fully remove it from the membrane, an action that may not be disruptive enough to deplete raft proteins from the detergent-resistant fraction.
In contrast to nystatin and filipin, the cholesterol-chelating drug MβCD had a striking effect on a specific subset of proteins in the detergent-resistant raft preparation. In fact, many proteins were so effectively excluded by MβCD from the biochemically isolated rafts that often Leu-containing peptides from those proteins were undetectable, whereas the corresponding LeuD3-labeled peptide was easily detected (assigned ratios' >10′). Sharp drops in the solid curve in Fig. 3 (indicated by arrows) suggest that the proteins can be classified into three distinct categories: those with a ratio >7.5, between 7.5 and 3, and those <3.
There were 145 proteins with ratios <3, most of which were cytoplasmic proteins, nuclear factors, or mitochondrial enzymes (Table 1, bottom). The ratios for these proteins indicate they are not drastically affected by cholesterol depletion, and so the presence of most of them in the detergent-resistant fraction is a result of their high cellular expression levels, making them inherently difficult to completely separate from the proteins of interest (Fig. 1). Contaminating proteins, mostly epidermal keratins and serum proteins, were detected only in their unlabeled form and therefore have ratios close to zero.
Table 1.
Examples of identified proteins
| Identification no. | Name | Ratio |
|---|---|---|
| Raft proteins (>7.5) | ||
| P08174 | CD55 | >10 |
| P07948 | Lyn tyrosine kinase | >10 |
| Q969J8 | Flotillin 1 | >10 |
| P15328 | Folate receptor | >10 |
| P05186 | Alkaline phosphatase | >10 |
| O00161 | SNAP-23 | >10 |
| P27105 | Stomatin | >10 |
| P21589 | 5′-nucleotidase | >10 |
| Gsα1, Gqα, Goα2, Giα1, Giα2, Giα3, Gα11, Gα12, Gα13, Gβ1, Gβ2, Gβ4, Gγ12 | >10 | |
| P07947 | YES tyrosine kinase | >10 |
| Q9NV17 | Hypothetical protein FLJ10709 | >10 |
| Q14699 | KIAA0084 | >10 |
| P18206 | Vinculin | 9.4 ± 0.1 |
| Q8WWV8 | Scribble | >10 |
| P14923 | Plakoglobin 1 | >10 |
| Q9Z0J8 | Kilon | >10 |
| Q9BZM4 | ULBP3 | 8.2 ± 0.7 |
| Raft-associated proteins (7.5 < × < 3.0) | ||
| Q03135 | Caveolin-1 | 3.3 ± 0.4 |
| P38606 | V-ATPase A | 5.3 ± 0.8 |
| P21281 | V-ATPase B | 4.3 ± 0.4 |
| P21283 | V-ATPase C | 5.2 ± 0.7 |
| Q9Y5K8 | V-ATPase D | 4.4* |
| O75348 | V-ATPase G | 6.3 ± 0.3 |
| Q9U112 | V-ATPase H | 4.3 ± 0.7 |
| Q15904 | V-ATPase S1 | 3.4 ± 0.4 |
| P29312 | 14-3-3ζ/δ | 5.5* |
| P11233 | RalA | 4.6 ± 0.9 |
| P082389 | HSP-90β | 4.7 ± 0.5 |
| O94905 | Chromosome 8 ORF 2 | 4.9 ± 1.2 |
| Q9UHA4 | MEK binding partner 1 | 4.4 ± 0.5 |
| Q9Y3U1 | Hypothetical 66.9 kDa protein | 6.2 ± 0.4 |
| Q9Y490 | Talin | 5.0 ± 1.3 |
| Q14642 | Inositol-1,4,5-P3 5-phosphatase | 5.4* |
| Nonspecific proteins (<3.0) | ||
| Q16891 | Mitofilin, mitochondria | 2.1* |
| P02786 | Transferrin receptor | 2.7 ± 0.2 |
| P21796 | VDAC1, mitochondria | 1.3 ± 0.1 |
| P45880 | VDAC2, mitochondria | 1.5 ± 0.4 |
| O00410 | Nuclear importin-3 | 1.5 ± 0.3 |
| P12236 | Mitochondrial adenine translocator | 1.6 ± 0.3 |
| O95613 | Pericentrin 2, nuclear | 1.1* |
| P04843 | Ribophorin I | 1.4 ± 0.1 |
| Q92896 | E-selectin | 1.6 ± 0.2 |
SWISS-PROT/TrEMBL identifiers, protein names, and control/MβCD ratios (±SE) for selected proteins.
Ratio based on one peptide.
MβCD was much more effective at disrupting cholesterol than either nystatin or filipin, because treatment with it depletes the cells of virtually all cholesterol relative to untreated cells (Table 2). The heavy dependence of raft integrity on cholesterol (2, 3, 18) means that such efficient removal of cholesterol from cells should profoundly alter raft structure and lead to very large ratios in the SILAC assay (Figs. 1 and 2). Therefore, the two groups with high ratios in Fig. 3 (241 proteins in total) should represent authentic raft components. If this is true, known raft markers should fall into either of these categories, which is indeed what we observed. Remarkably, every generally accepted raft protein expected to be expressed in epithelium-derived HeLa cells (2, 3) and identified in this study had ratios placing them in either the medium or high ratio groups (Table 1).
Table 2.
Cholesterol remaining after drug treatment
| Treatment | Cholesterol remaining, % ± SE |
|---|---|
| Control | 100 ± 6.6 |
| MβCD | 4.3 ± 2.0 |
| Nystatin | 81.1 ± 6.7 |
| Filipin | 66.3 ± 6.7 |
In addition to being separable by ratio, these proteins also group similarly based on their known relationships with membranes. The group with the highest ratios consists mostly of proteins with at least one transmembrane domain or with a hydrophobic modification such as a GPI-anchor, a double acylation, or a palmitoyl group (Table 1) that are sufficient for targeting proteins to rafts (19). The group with medium ratios consists mostly of proteins that are not themselves embedded in the membrane but that are known or thought to bind to such proteins (Table 1). We have termed these groups “raft proteins” and “raft-associated proteins,” respectively. That these proteins can be grouped similarly based on two properties, their susceptibility to MβCD treatment and their association with membranes, is additional strong evidence that the groupings are real and not a statistical artifact. One oddity in this classification is the raft marker caveolin. The measured ratio for caveolin places it in the “raft-associated” category when it would be expected to be in the “raft” category, suggesting that there may be unknown structural properties of caveolin that affect its interactions with cholesterol and thus its susceptibility to disruption by MβCD.
The importance of using a functional property (such as response to cholesterol disruption) to increase the specificity of proteomic characterization of biochemical fractions is apparent from examination of our own data and comparison with those of others. In the MβCD experiment described above, 352 noncontaminant proteins were quantified from detergent-resistant fractions, but almost one-third of these turned out to be nonspecific copurifying proteins. Conversely, a small number of proteins that would otherwise be expected to be nonspecific appear in the raft and raft-associated protein groups. Although we cannot be certain, the appearance of intermediate filament proteins such as vimentin and keratins proteins may be a result of secondary interactions with true raft proteins. When raft proteins are depleted by MβCD, the proteins bound to them are also depleted (20). We have no explanation for the presence of nucleolin and certain ribosomal proteins.
There are few proteomics studies of raft proteins, and none of them is quantitative. Bini et al. (21) reported the identification of 19 proteins in a detergent-resistant fraction from Jurkat T cells, but less than half of these fall into the raft classifications described here (Fig. 3 and supporting information on the PNAS web site). von Haller et al. (22) identified ≈70 proteins in a detergent-resistant fraction also from Jurkat T cells Of these, only two-thirds are found to be authentic raft proteins (Fig. 3 and supporting information on the PNAS web site), and our data suggest that the remaining one-third are false positives. Without the functional specificity of cholesterol depletion, as used in the experiment reported here, these proportions are exactly what would be expected from biochemical purification alone. In addition, our data also indicate that the pH/carbonate-resistant preparation is less specific in isolating raft proteins and more difficult to interpret than the detergent-resistant method.
In total, 703 proteins were identified in detergent-resistant fractions and 585 in pH/carbonate-resistant fractions. Of the 703 detergent-resistant proteins, 392 were quantifiable and revealed 241 authentic raft proteins. Identification of a set of true raft proteins directly illuminates questions of interest in the raft and signaling fields. Lipid rafts are often linked to signal transduction pathways, in particular as coordinators of the initial events in the cascades (3). Most of the signaling pathways studied in the context of rafts are tyrosine kinase cascades, and the data presented here provide unbiased support for this association (Table 1). Interestingly, among the raft and raft-associated proteins identified here are a significant number of serine/threonine kinases/phosphatases as well as numerous heterotrimeric G protein subunits (Table 1), suggesting that rafts may be more general signaling coordinators. Certain signaling molecules were previously known to be enriched in rafts, but this is the first demonstration, to our knowledge, of such an enrichment on a large scale.
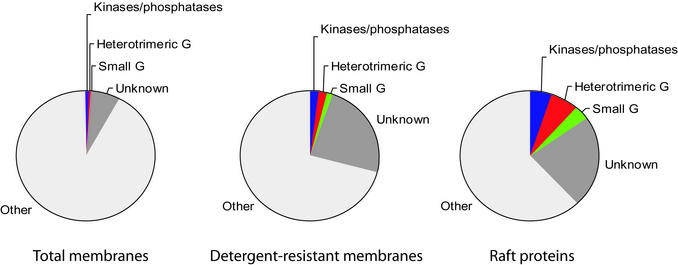
Original models suggested rafts are relatively rare in membranes [“rafts in an ocean” (23, 24)], but based on molar lipid abundances some estimates now predict rafts may constitute up to 30% or more of plasma membrane area in certain cells (25). If rafts did cover one-third of the cell surface, their purification versus the whole membrane could result in only a 3-fold enrichment as an upper limit. To directly address the question of raft abundance, we analyzed a postnuclear/postmitochondria total membrane fraction by LC/MS, identifying 162 of the most abundant proteins. We then used these and the other data discussed above to compare the number of protein kinases/phosphatases, small GTP-binding proteins (excluding those involved in membrane transport), and heterotrimeric G proteins in each fraction. Fig. 4 shows the proportion of these three classes of signaling molecules in total membrane fractions, detergent-resistant fractions, and the set of cholesterol-dependent raft proteins identified here. The proportion increases to much more than 10-fold from total membranes to cholesterol-dependent raft proteins. Fig. 4 also indicates there is a large percentage of unknown proteins in rafts that are also enriched, suggesting that the proportion of signaling molecules may be even higher, and that many interesting raft proteins remain to be characterized. Even allowing for a small diluting effect by ER membranes in the total membrane fraction, the >10-fold enrichment of many signaling protein families in rafts versus total membranes means that the percentage of the membrane occupied by rafts has to be much lower than 10%. However, this does not imply that rafts are of a uniform or static size and composition.
Figure 4.
Enrichment of signaling molecules. Protein kinases/phosphatases, heterotrimeric G protein subunits/effectors, small molecular weight G proteins/exchange factors, and unknown proteins identified as raft proteins as a fraction of the total proteins in total membranes, detergent-resistant membranes, and raft proteins.
On the basis of previous biochemical isolations, rafts are also proposed to reside in membrane compartments other than the plasma membrane (10, 26). Clearly, both raft lipids and raft proteins are synthesized in the endoplasmic reticulum/Golgi before transport to the plasma membrane and, indeed, many protein synthesis proteins and vesicle trafficking proteins are among the proteins identified here. However, this does not demonstrate that rafts per se exist in those compartments, only that those proteins are in a detergent-resistant, cholesterol-dependent state while residing there. Mitochondria too have been proposed to contain rafts (21), but the data presented here strongly argue against this possibility. On the other hand, we do find that most of the subunits of the lysosomal proton pump as well as other degradative pathway components are sensitive to MβCD, supporting previous studies that found raft markers in these organelles (27). The data further demonstrate the considerable differences between detergent- and pH/carbonate-resistant fractions. Typically, at least one-third of the proteins in detergent-resistant fractions are nonraft proteins (see above and refs. 21 and 22), but pH/carbonate-resistant fractions likely contain at least 75% nonraft proteins. Due to the very high levels of proteins unreceptive to cholesterol disruption in the pH/carbonate-resistant fraction, we conclude that this preparation is a poor model for studying lipid rafts.
Clearly, biochemical fractionation alone is not sufficiently specific to unequivocally assign localizations, but use of stable isotope encoding to add a functional dimension greatly increases specificity. By requiring simultaneous matching of the two most widely accepted criteria for raft proteins, the study presented here provides, to our knowledge, the first large-scale characterization of raft protein components.
Supplementary Material
Acknowledgments
L.J.F. was supported by a European Molecular Biology Organization long-term fellowship, and C.L.d.H. was supported by a Canadian Institutes of Health Research postdoctoral fellowship. We thank Sebastian Schuck for help with the cholesterol assay and members of the Center for Experimental BioInformatics for constructive comments and discussion. The Center for Experimental BioInformatics is supported by the Danish National Research Foundation.
Abbreviations
- SILAC
stable isotope labeling with amino acids in cell culture
- MβCD
methyl β-cyclodextrin
- LeuD3
l-leucine-5,5,5-d3
- LC/MS
high-performance liquid chromatography/MS
- MS/MS
tandem MS
- r.t.
room temperature
References
- 1.van Meer G, Lisman Q. J Biol Chem. 2002;277:25855–25858. doi: 10.1074/jbc.R200010200. [DOI] [PubMed] [Google Scholar]
- 2.Galbiati F, Razani B, Lisanti M P. Cell. 2001;106:403–411. doi: 10.1016/s0092-8674(01)00472-x. [DOI] [PubMed] [Google Scholar]
- 3.Simons K, Toomre D. Nat Rev Mol Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 4.Friedrichson T, Kurzchalia T V. Nature. 1998;394:802–805. doi: 10.1038/29570. [DOI] [PubMed] [Google Scholar]
- 5.Varma R, Mayor S. Nature. 1998;394:798–801. doi: 10.1038/29563. [DOI] [PubMed] [Google Scholar]
- 6.Pralle A, Keller P, Florin E L, Simons K, Horber J K. J Cell Biol. 2000;148:997–1008. doi: 10.1083/jcb.148.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harder T, Scheiffele P, Verkade P, Simons K. J Cell Biol. 1998;141:929–942. doi: 10.1083/jcb.141.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dietrich C, Bagatolli L A, Volovyk Z N, Thompson N L, Levi M, Jacobson K, Gratton E. Biophys J. 2001;80:1417–1428. doi: 10.1016/S0006-3495(01)76114-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ong S E, Blagoev B, Kratchmarova I, Kristensen D B, Steen H, Pandey A, Mann M. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 10.Brown D A, Rose J K. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- 11.Smart E J, Ying Y S, Mineo C, Anderson R G. Proc Natl Acad Sci USA. 1995;92:10104–10108. doi: 10.1073/pnas.92.22.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shevchenko A, Wilm M, Vorm O, Mann M. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 13.Rappsilber J, Ishihama Y, Mann M. Anal Chem. 2003;I75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 14.Rappsilber J, Ryder U, Lamond A I, Mann M. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mann M, Hendrickson R C, Pandey A. Annu Rev Biochem. 2001;70:437–473. doi: 10.1146/annurev.biochem.70.1.437. [DOI] [PubMed] [Google Scholar]
- 16.Bligh F C, Dyer W J. Can J Biochem Physiol. 1959;37:911–937. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 17.Foster L J, Yaworsky K, Trimble W S, Klip A. Am J Physiol. 1999;276:C1108–C1114. doi: 10.1152/ajpcell.1999.276.5.C1108. [DOI] [PubMed] [Google Scholar]
- 18.Simons K, Ikonen E. Science. 2000;290:1721–1726. doi: 10.1126/science.290.5497.1721. [DOI] [PubMed] [Google Scholar]
- 19.Zacharias D A, Violin J D, Newton A C, Tsien R Y. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 20.Blagoev B, Kratchmarova I, Ong S E, Nielsen M, Foster L J, Mann M. Nat Biotechnol. 2003;21:315–318. doi: 10.1038/nbt790. [DOI] [PubMed] [Google Scholar]
- 21.Bini L, Pacini S, Liberatore S, Valensin S, Pellegrini M, Raggiaschi R, Pallini V, Baldari C T. Biochem J. 2002;369:301–309. doi: 10.1042/BJ20020503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Haller P D, Donohoe S, Goodlett D R, Aebersold R, Watts J D. Proteomics. 2001;1:1010–1021. doi: 10.1002/1615-9861(200108)1:8<1010::AID-PROT1010>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 23.Simons K, Ikonen E. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 24.Simons K, van Meer G. Biochemistry. 1988;27:6197–6202. doi: 10.1021/bi00417a001. [DOI] [PubMed] [Google Scholar]
- 25.Renkonen O, Kaarainen L, Simons K, Gahmberg C G. Virology. 1971;46:318–326. doi: 10.1016/0042-6822(71)90033-x. [DOI] [PubMed] [Google Scholar]
- 26.Simons K, Wandinger-Ness A. Cell. 1990;62:207–210. doi: 10.1016/0092-8674(90)90357-k. [DOI] [PubMed] [Google Scholar]
- 27.Dermine J F, Duclos S, Garin J, St-Louis F, Rea S, Parton R G, Desjardins M. J Biol Chem. 2001;276:18507–18512. doi: 10.1074/jbc.M101113200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.