Abstract
Triple A syndrome is a human autosomal recessive disorder characterized by an unusual array of tissue-specific defects. Triple A syndrome arises from mutations in a WD-repeat protein of unknown function called ALADIN (also termed Adracalin or AAAS). We showed previously that ALADIN localizes to nuclear pore complexes (NPCs), large multiprotein assemblies that are the sole sites of nucleocytoplasmic transport. Here, we present evidence indicating that NPC targeting is essential for the function of ALADIN. Characterization of mutant ALADIN proteins from triple A patients revealed a striking effect of these mutations on NPC targeting. A variety of disease-associated missense, nonsense, and frameshift mutations failed to localize to NPCs and were found predominantly in the cytoplasm. Microscopic analysis of cells from a triple A patient revealed no morphological abnormalities of the nuclei, nuclear envelopes, or NPCs. Importantly, these findings indicate that defects in NPC function, rather than structure, give rise to triple A syndrome. We propose that ALADIN plays a cell type-specific role in regulating nucleocytoplasmic transport and that this function is essential for the proper maintenance and/or development of certain tissues. Our findings provide a foundation for understanding the molecular basis of triple A syndrome and may lead to unique insights into the role of nucleocytoplasmic transport in adrenal function and neurodevelopment.
Triple A syndrome (also called Allgrove syndrome; Online Mendelian Inheritance in Man 231550) is a rare human autosomal recessive disorder (1, 2). Often, patients present with adrenocorticotropic hormone-resistant adrenal insufficiency, giving rise to potentially fatal hypoglycemic episodes. Triple A syndrome is distinguished from other adrenal insufficiencies, however, by the additional presence of achalasia of the lower esophageal sphincter leading to swallowing difficulties, alacrima (failure to produce tears), and neurological abnormalities. These four major symptoms, although characteristic of triple A syndrome, often are highly variable in severity and age of presentation and usually are progressive.
The gene mutated in triple A syndrome was identified recently (3, 4), and the protein product of this gene was named ALADIN (also called Adracalin or AAAS). ALADIN is a 60-kDa protein with a central ≈170-aa domain composed of four WD repeats. The flanking N and C termini, of ≈150 and 230 residues, respectively, contain no obvious structural domains. ALADIN is conserved in higher eukaryotes although there appears to be no direct homologue in lower eukaryotes such as Saccharomyces cerevisiae (refs. 3 and 4; unpublished data). WD-repeat domains are thought to be involved in the assembly of macromolecular complexes (5), suggesting that ALADIN also may act as a scaffold for multiprotein assemblies. However, because WD-repeat proteins play a role in a diverse range of cellular processes, the specific function of ALADIN is unknown.
Sequencing of the ALADIN gene from several triple A patients has identified a variety of mutations scattered throughout the gene (3, 4, 6–10). A large number of these mutations are nonsense, frameshift, or splice-site mutations that are predicted to truncate the C terminus of ALADIN, suggesting that this domain is important for the function of ALADIN. In addition, there are four point mutations, three of which (H160R, S263P, and V313A) are within the WD-repeat domain, whereas the fourth (Q15K) is close to the N terminus. There is no obvious correlation between different ALADIN mutations and disease phenotype. Patients with the same mutation, even patients within the same family, frequently display a high degree of variability in the clinical severity and age of onset of their symptoms (7, 11). This indicates that other factors play an important role in the pathogenesis of triple A syndrome.
In a proteomic analysis of mammalian nuclear pore complexes (NPCs), we identified ALADIN as a protein that biochemically fractionates with and localizes to NPCs (12). NPCs are large, multiprotein complexes that span the nuclear envelope (NE), forming a selective channel between the cytoplasm and nucleus (13). As the sole sites of nucleocytoplasmic transport, NPCs play a role in a variety of cellular processes, including cell-cycle progression, control of gene expression, and signal transduction. ALADIN could be involved in any of these aspects of NPC function by, for example, mediating the assembly of subdomains of the NPC or nucleating the formation of transport complexes.
The identification of ALADIN as a component of NPCs facilitates a more detailed analysis of the molecular basis of disease-causing mutations. Here, we show that mutations associated with triple A syndrome result in mislocalization of ALADIN from NPCs to the cytoplasm. Characterization of an ALADIN mutant cell line indicates that the absence of functional ALADIN does not result in morphological abnormalities in nuclei, NPCs, or NEs, indicating that the defect is functional rather than structural.
Materials and Methods
Molecular Biology.
Full-length and truncated ALADIN were PCR-amplified from IMAGE cDNA 3347423 (Incyte Pharmaceuticals, Palo Alto, CA) and cloned into the XhoI and EcoRI sites of pEGFP-C1. Point mutations were generated in GFP-ALADIN by QuikChange mutagenesis (Stratagene) according to the manufacturer's protocol.
Cell Culture and Transfection.
HeLa cells were maintained in DMEM supplemented with 10% FBS/1% penicillin-streptomycin/10 mM Hepes. GM12123 and GM03652 fibroblasts were obtained from Coriell Cell Repositories (Camden, NJ) and maintained in MEM supplemented with 15% FBS and 1× nonessential amino acids. Cells were transfected by using Lipofectamine Plus (Invitrogen) according to the manufacturer's recommendations.
Analysis of the ALADIN Gene in GM12123 Cells.
Genomic DNA was prepared from cultured cells by using standard procedures, and the ALADIN gene exons and intron–exon boundaries were PCR-amplified in triplicate. RNA was prepared by using the Absolutely RNA RT-PCR miniprep kit (Stratagene). The ALADIN transcript was amplified in triplicate by using the OneStep RT-PCR kit (Qiagen, Valencia, CA). Primers were chosen spanning the exon 12–exon 13 boundary and within exon 16. PCR and RT-PCR products were cloned into pCR2.1-TOPO for sequencing.
Immunofluorescence and Microscopy.
Cells were fixed in 2% formaldehyde in PBS and permeabilized in −20°C acetone for 2 min. Antibodies used were mAb414 (recognizes Nup358, Nup214, Nup153, and Nup62; Babco, Richmond, CA), 1:10; anti-Nup358 (14), 1:500; anti-Tpr (15), 1:500; anti-lamin B (16), 1:500; anti-importin-β (Affinity BioReagents, Neshanic Station, NJ), 1:100; and anti-transportin (Transduction Laboratories, Lexington, KY), 1:100. Secondary antibodies (Molecular Probes) conjugated to Alexa 594 or 488 were used at 1:50. Confocal microscopy was as described (12). Deconvolution microscopy was performed by using a Delta-vision microscope (Applied Precision, Issaquah, WA).
Electron Microscopy.
Cells were fixed, resin-embedded, sectioned by using standard methods, and viewed with a Philips CM120 transmission electron microscope.
Results
ALADIN Localizes near Nup358 on the Cytoplasmic Face of NPCs.
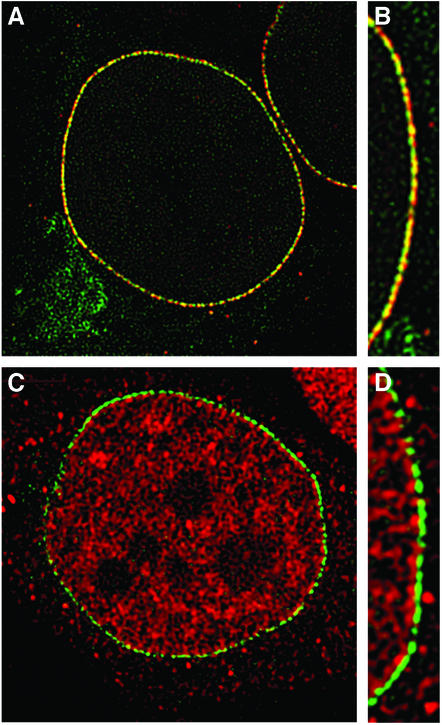
We recently demonstrated that ALADIN biochemically fractionates with and localizes to NPCs (12). To characterize further the association of ALADIN with NPCs, we analyzed its localization with respect to known nucleoporins. HeLa cells were transfected with GFP-ALADIN and then fixed and labeled with antibodies to Nup358 and Tpr [nucleoporins that localize to the cytoplasmic and nucleoplasmic filaments of NPCs, respectively (14, 17)]. Using deconvolution microscopy, we observed a close overlap between the localization of GFP-ALADIN and Nup358, with Nup358 appearing slightly more distal to the midplane of NPCs (Fig. 1 A and B). There was, however, no overlap between GFP-ALADIN and Tpr (Fig. 1 C and D). These results verify that ALADIN is a nucleoporin and indicate further that it localizes close to Nup358, probably at the cytoplasmic face of NPCs.
Figure 1.
ALADIN localizes to the cytoplasmic face of NPCs. (A and B) Colocalization of GFP-ALADIN (green) and Nup358 (red). (C and D) Colocalization of GFP-ALADIN (green) and Tpr (red).
The C-Terminal Domain of ALADIN Is Essential for NPC Targeting.
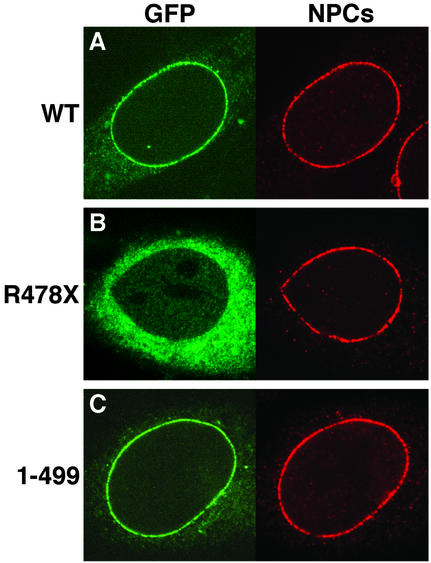
Several of the mutations identified in triple A patients result in truncation of the C terminus of ALADIN, suggesting that the C-terminal domain is important for the function of ALADIN (3, 4, 7–10). We analyzed the subcellular localization of the shortest pathogenic C-terminal truncation of ALADIN, the nonsense mutation R478X, which lacks the C-terminal 69 aa and terminates after glycine 477. Transfection of GFP-ALADINR478X revealed that this mutant no longer localizes to NPCs but is found predominantly in the cytoplasm (Fig. 2B; compare with wild-type localization in Fig. 2A). Identical results were obtained with several other pathogenic ALADIN mutations that disrupt larger stretches of the C terminus (data not shown). A shorter, non-disease-associated deletion of the C terminus (GFP-ALADIN1–499) retains wild-type NPC localization (Fig. 1C), indicating that residues between amino acids 478 and 499 are essential for targeting ALADIN to NPCs. All known nonsense, frameshift, and splice-site mutations encode for an ALADIN protein lacking this domain and, therefore, will not localize to NPCs. These results indicate that the C-terminal domain of ALADIN is essential for its correct targeting to NPCs. The observation that this domain is deleted in a large number of triple A patients further indicates that NPC targeting is essential for normal ALADIN function.
Figure 2.
The C-terminal domain of ALADIN is essential for NPC targeting. The localization of GFP-tagged ALADIN constructs is shown on the left of each image (green). (A) Wild-type ALADIN. (B) ALADINR478X. (C) ALADIN1–499. The localization of NPCs is shown on the right of each image (red) by immunostaining with mAb414 (recognizes Nup358, Nup214, Nup153, and Nup62).
The N-Terminal Domain of ALADIN Is also Essential for NPC Targeting.
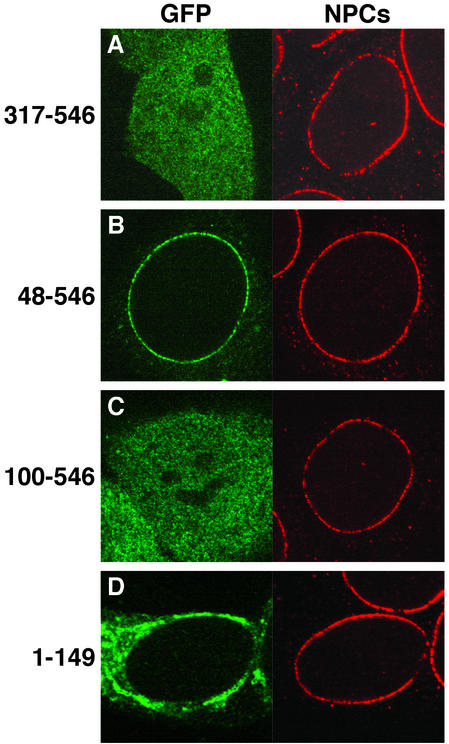
Interestingly, the C-terminal domain of ALADIN, although necessary for NPC localization, is not sufficient, because this domain alone cannot target a GFP fusion protein to NPCs (GFP-ALADIN317–546; Fig. 3A). To further define domains of ALADIN that are required for NPC targeting, we constructed a series of truncations from the N terminus of ALADIN. GFP-ALADIN48–546 retains wild-type NPC localization (Fig. 3B) whereas a larger truncation, GFP-ALADIN100–546, no longer localizes to NPCs (Fig. 3C). This indicates that residues in the N-terminal domain are also important for targeting ALADIN to NPCs. However, our results suggest that the N and C termini of ALADIN play different roles in NPC targeting because proteins with C-terminal truncations accumulate in the cytoplasm, whereas proteins with N-terminal truncations are found throughout the cell (compare Figs. 2B and 3C). We reasoned that the N-terminal domain of ALADIN might function as a cytoplasmic-targeting/retention signal, and to test this we fused this domain alone (amino acids 1–149) to GFP. We found that, although GFP-ALADIN1–149 is predominantly cytoplasmic, it appears to form aggregates (Fig. 3D), and thus it is uncertain whether cytoplasmic localization is a specific function of ALADIN′s N-terminal domain. Nonetheless, it is clear that both N- and C-terminal domains of ALADIN are required for correct targeting to NPCs. These domains may interact with different factors to cooperatively target ALADIN to NPCs or may be required for correct protein folding.
Figure 3.
The N-terminal domain of ALADIN is essential for NPC targeting. The localization of GFP-tagged ALADIN constructs is shown on the left of each image (green). (A) ALADIN317–546. (B) ALADIN48–546. (C) ALADIN100–546. (D) ALADIN1–149. The localization of NPCs is shown on the right of each image (red) by immunostaining with mAb414.
Mutations in the WD-Repeat Domain of ALADIN Affect NPC Targeting.
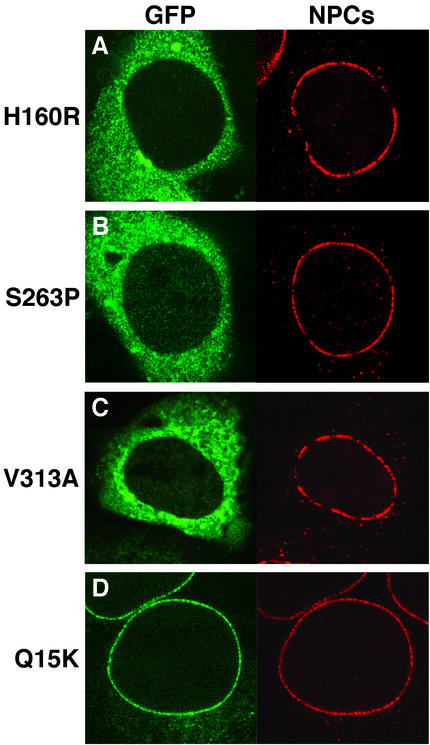
We examined the subcellular localization of the point mutations identified in triple A patients (4, 6, 9). Like the C-terminal truncations, GFP-ALADINH160R, GFP-ALADINS263P, and GFP-ALADINV313A are mislocalized to the cytoplasm (Fig. 4 A–C). Because these mutations are in the WD-repeat domain (repeats 1, 3, and 4, respectively), they may disrupt assembly of the WD-repeat propeller structure, resulting in a misfolded protein. Intriguingly, GFP-ALADINQ15K still localized to NPCs (Fig. 4D), suggesting that this residue may be critical for the interaction of ALADIN with a protein(s) essential for the function of ALADIN but not involved in NPC localization. It may be of significance that the Q15K mutation is associated with milder symptoms (9) or a later onset of disease (7).
Figure 4.
Mutations in the WD-repeat domain of ALADIN affect NPC targeting. The localization of GFP-tagged ALADIN constructs is shown on the left of each image (green). (A) ALADINH160R. (B) ALADINS263P. (C) ALADINV313A. (D) ALADINQ15K. The localization of NPCs is shown on the right of each image (red) by immunostaining with mAb414.
ALADIN Mutation Results in Functional Rather than Structural Defects.
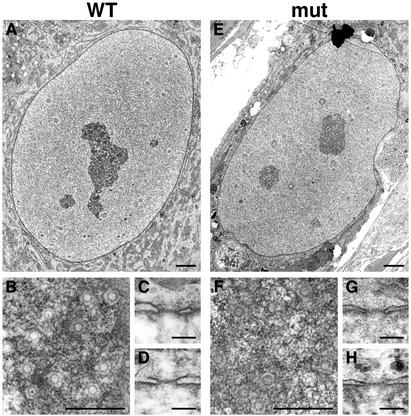
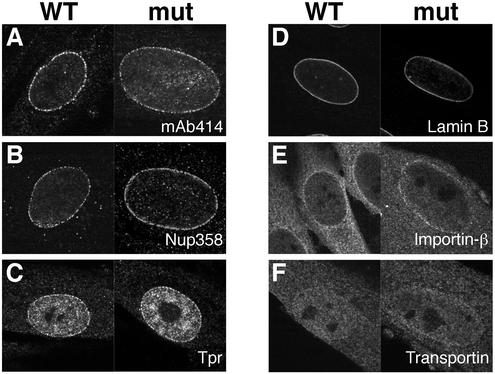
Deletion of nucleoporins in both yeast (18) and vertebrates (19) can result in improper NE and/or NPC assembly as well as defects in nuclear transport. To characterize the effects of ALADIN mutations on NPC structure, we identified a human fibroblast cell line in the Coriell Cell Repositories (GM12123) derived from a patient with the symptoms of triple A syndrome. Sequence analysis of the ALADIN gene from this cell line identified a splice-site mutation in the donor site of intron 14 (IVS14 + 1 → C; data not shown). RT-PCR analysis of RNA from the GM12123 cell line identified two different, aberrantly spliced ALADIN transcripts, the predicted protein products of which lack the C-terminal, NPC-targeting domain (data not shown). Therefore, if expressed, these protein products would be nonfunctional. To determine whether the absence of functional ALADIN results in any morphological defects, we examined the GM12123 cells by electron and immunofluorescence microscopy. Using electron microscopy, we observed no obvious morphological defects in the nuclei, NEs, or NPCs of GM12123 cells (Fig. 5 E–H) compared with a normal fibroblast cell line (GM03652; Coriell Cell Repositories) (Fig. 5 A–D). NPC structure in tangential sections (Fig. 5 B and F) and cross-sections (Fig. 5 C, D, G, and H) was identical in both wild-type and mutant cells. To analyze further nuclear structure in ALADIN mutant cells, we performed immunofluorescence by using antibodies that recognize nucleoporins [mAb414 (recognizes Nup358, Nup214, Nup153, and Nup62), Nup358, and Tpr], the nuclear lamina (lamin B), and nucleocytoplasmic transport factors (importin-β and transportin). We observed no differences in the localization of any of these proteins in mutant cells (Fig. 6 A–F Right) compared with wild-type cells (Fig. 6 A–F Left). Thus, a defect in the targeting of ALADIN to NPCs does not appear to result in disruption of NPC/NE structure.
Figure 5.
ALADIN mutant cells are morphologically normal. Shown are electron microscopy images of wild-type (A–D) and ALADIN mutant (E–H) cells showing normal nuclei (A and E) and normal NPCs in tangential section (B and F) and cross section (C, D, G, and H). (Bars: A and E, 2 μm; B and F, 500 nm; C, D, G, and H, 100 nm.)
Figure 6.
ALADIN mutant cells are morphologically normal. Immunostaining of nuclear structures in wild-type (Left) and ALADIN mutant (Right) cells is shown. Cells were immunostained with mAb414 (A), anti-Nup358 (B), anti-Tpr (C), anti-lamin B (D), anti-importin-β (E), and anti-transportin (F).
Discussion
Although ALADIN is known to be the gene mutated in triple A syndrome, until now, the specific function of this protein and the consequence of its mutation have not been investigated. To begin to understand the function of ALADIN, we analyzed further its localization to NPCs by deconvolution microscopy and found that ALADIN localizes close to Nup358. Although these results suggest that ALADIN localizes to the cytoplasmic face of NPCs, this technique is limited in resolution and a more definitive localization to distinct substructures of the NPC will require higher-resolution immunoelectron microscopy studies. We also characterized the localization of mutant ALADIN proteins associated with triple A syndrome. Intriguingly, we found a strong correlation between mutations that cause triple A syndrome and the disruption of ALADIN's ability to localize to NPCs. This association between mislocalization of ALADIN and disease indicates that targeting of ALADIN to NPCs is essential for its normal function.
Truncation of the N or C terminus, as well as mutations in the WD-repeat domain, can lead to mislocalization of ALADIN, which suggests that NPC targeting is complex and may depend on interactions with multiple factors and/or proper protein folding. As a member of the WD-repeat family of proteins (5), ALADIN could have a variety of functions at NPCs, such as NPC assembly, nucleation of transport complexes, or formation of other complexes. Aberrant localization of ALADIN could disrupt these essential functions in a variety of ways. The recessive nature of triple A disease makes it unlikely that ALADIN mislocalization titrates essential factors from NPCs in a dominant-negative manner. Instead, failure of ALADIN to target to NPCs may result in aberrant localization or stabilization of other factors or may render ALADIN itself unstable, resulting in a functional null. Although the specific consequences of ALADIN's mislocalization are unknown, it does not result in obvious morphological defects. Our data show that nuclei, NPCs, and NEs are morphologically normal in ALADIN mutant cells. Indeed, the disease itself is not lethal and most tissues are unaffected, indicating that essential NPC functions are retained. In agreement with this, we found that several nucleoporins and transport factors are localized normally in ALADIN mutant cells. However, there are many proteins that we were unable to test and it is possible that mutations in ALADIN result in specific mislocalization of other nucleoporins or transport factors. Identification of proteins that interact with ALADIN will enable us to study how their localization and/or function is compromised in triple A syndrome and may allow us to understand the pathogenesis of triple A syndrome more clearly.
The tissue specificity of triple A syndrome is particularly interesting, because it provides evidence that an individual nucleoporin can have tissue-specific functions. The basis for this tissue specificity currently is unclear. ALADIN is ubiquitously, although not equally, expressed in all tissues tested (refs. 3, 4, and 11; unpublished results). It appears, therefore, that ALADIN has essential functions specific to the affected tissues or that there is a threshold, below which loss of functional ALADIN cannot be tolerated in certain tissues. We favor a model in which ALADIN mutations result in a specific defect in NPC function. For example, ALADIN may be involved in the nucleocytoplasmic transport of a specific protein(s) that plays an important role in the development and/or maintenance of adrenal cortex, lachrymal gland, esophageal sphincter, and neurons. This model predicts that mislocalization of ALADIN disrupts the subcellular localization of other protein(s), which, in turn, results in defects in these specific tissues. Of particular interest in this respect is the point mutation Q15K, which does not disrupt NPC targeting but, instead, may disrupt the interaction of ALADIN with such a protein. ALADIN may be stably associated with NPCs and function as a docking site for putative transport cargoes. Alternatively, ALADIN may shuttle on and off NPCs, as described for other nucleoporins (20, 21), and play a more dynamic role in nucleocytoplasmic transport.
An interesting aspect of triple A syndrome is the high degree of variability in severity and age of onset, seen even between patients with the same mutation and similar genetic backgrounds (7, 11). Clearly, other factors also are involved in triple A syndrome, and recent reports suggest that there may be another causative gene (11). These additional factors may be involved in nucleocytoplasmic transport or in signaling pathways that act through ALADIN at NPCs. The identification of proteins that play a role in triple A syndrome may help us to understand how essential processes in adrenal function and neurodevelopment are linked to NPC function. As the first nucleoporin to be linked to a heritable human disease, ALADIN offers a unique opportunity to study the function of a specific nucleoporin and NPCs in general. Our observations provide a foundation for understanding the molecular basis of triple A syndrome. Future work may clarify the role of ALADIN at NPCs, allowing us to understand further how ALADIN mutations lead to triple A syndrome and to potentially develop therapeutic strategies.
Acknowledgments
We thank the Johns Hopkins University School of Medicine microscope facility for assistance and S. J. Collis for help and advice. This work was funded by the American Cancer Society.
Abbreviations
- NE
nuclear envelope
- NPC
nuclear pore complex
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Allgrove J, Clayden G S, Grant D B, Macaulay J C. Lancet. 1978;1:1284–1286. doi: 10.1016/s0140-6736(78)91268-0. [DOI] [PubMed] [Google Scholar]
- 2.Clark A J, Weber A. Endocr Rev. 1998;19:828–843. doi: 10.1210/edrv.19.6.0351. [DOI] [PubMed] [Google Scholar]
- 3.Tullio-Pelet A, Salomon R, Hadj-Rabia S, Mugnier C, de Laet M H, Chaouachi B, Bakiri F, Brottier P, Cattolico L, Penet C, et al. Nat Genet. 2000;26:332–335. doi: 10.1038/81642. [DOI] [PubMed] [Google Scholar]
- 4.Handschug K, Sperling S, Yoon S J, Hennig S, Clark A J, Huebner A. Hum Mol Genet. 2001;10:283–290. doi: 10.1093/hmg/10.3.283. [DOI] [PubMed] [Google Scholar]
- 5.Smith T F, Gaitatzes C, Saxena K, Neer E J. Trends Biochem Sci. 1999;24:181–185. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- 6.Persic M, Prpic I, Huebner A, Severinski S. J Pediatr Gastroenterol Nutr. 2001;33:503–504. doi: 10.1097/00005176-200110000-00017. [DOI] [PubMed] [Google Scholar]
- 7.Sandrini F, Farmakidis C, Kirschner L S, Wu S M, Tullio-Pelet A, Lyonnet S, Metzger D L, Bourdony C J, Tiosano D, Chan W Y, et al. J Clin Endocrinol Metab. 2001;86:5433–5437. doi: 10.1210/jcem.86.11.8037. [DOI] [PubMed] [Google Scholar]
- 8.Schmittmann-Ohters K, Huebner A, Richter-Unruh A, Hauffa B P. Horm Res. 2001;56:67–72. doi: 10.1159/000048093. [DOI] [PubMed] [Google Scholar]
- 9.Houlden H, Smith S, De Carvalho M, Blake J, Mathias C, Wood N W, Reilly M M. Brain. 2002;125:2681–2690. doi: 10.1093/brain/awf270. [DOI] [PubMed] [Google Scholar]
- 10.Goizet C, Catargi B, Tison F, Tullio-Pelet A, Hadj-Rabia S, Pujol F, Lagueny A, Lyonnet S, Lacombe D. Neurology. 2002;58:962–965. doi: 10.1212/wnl.58.6.962. [DOI] [PubMed] [Google Scholar]
- 11.Huebner A, Kaindl A M, Braun R, Handschug K. Endocr Res. 2002;28:733–739. doi: 10.1081/erc-120016998. [DOI] [PubMed] [Google Scholar]
- 12.Cronshaw J M, Krutchinsky A N, Zhang W, Chait B T, Matunis M J. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allen T D, Cronshaw J M, Bagley S, Kiseleva E, Goldberg M W. J Cell Sci. 2000;113:1651–1659. doi: 10.1242/jcs.113.10.1651. [DOI] [PubMed] [Google Scholar]
- 14.Wu J, Matunis M J, Kraemer D, Blobel G, Coutavas E. J Biol Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- 15.Fontoura B M, Dales S, Blobel G, Zhong H. Proc Natl Acad Sci USA. 2001;98:3208–3213. doi: 10.1073/pnas.061014698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hytiroglou P, Choi S W, Theise N D, Chaudhary N, Worman H J, Thung S N. Hum Pathol. 1993;24:169–172. doi: 10.1016/0046-8177(93)90296-s. [DOI] [PubMed] [Google Scholar]
- 17.Frosst P, Guan T, Subauste C, Hahn K, Gerace L. J Cell Biol. 2002;156:617–630. doi: 10.1083/jcb.200106046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doye V, Hurt E. Curr Opin Cell Biol. 1997;9:401–411. doi: 10.1016/s0955-0674(97)80014-2. [DOI] [PubMed] [Google Scholar]
- 19.Wu X, Kasper L H, Mantcheva R T, Mantchev G T, Springett M J, van Deursen J M. Proc Natl Acad Sci USA. 2001;98:3191–3196. doi: 10.1073/pnas.051631598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindsay M E, Plafker K, Smith A E, Clurman B E, Macara I G. Cell. 2002;110:349–360. doi: 10.1016/s0092-8674(02)00836-x. [DOI] [PubMed] [Google Scholar]
- 21.Daigle N, Beaudouin J, Hartnell L, Imreh G, Hallberg E, Lippincott-Schwartz J, Ellenberg J. J Cell Biol. 2001;154:71–84. doi: 10.1083/jcb.200101089. [DOI] [PMC free article] [PubMed] [Google Scholar]