Abstract
Acquisition of a cardiac fate by embryonic mesodermal cells is a fundamental step in heart formation. Heart development in frogs and avians requires positive signals from adjacent endoderm, including bone morphogenic proteins, and is antagonized by a second secreted signal, Wnt proteins, from neural tube. By contrast, mechanisms of mesodermal commitment to create heart muscle in mammals are largely unknown. In addition, Wnt-dependent signals can involve either a canonical β-catenin pathway or other, alternative mediators. Here, we tested the involvement of Wnts and β-catenin in mammalian cardiac myogenesis by using a pluripotent mouse cell line (P19CL6) that recapitulates early steps for cardiac specification. In this system, early and late cardiac genes are up-regulated by 1% DMSO, and spontaneous beating occurs. Notably, Wnt3A and Wnt8A were induced days before even the earliest cardiogenic transcription factors. DMSO induced biochemical mediators of Wnt signaling (decreased phosphorylation and increased levels of β-catenin), which were suppressed by Frizzled-8/Fc, a soluble Wnt antagonist. DMSO provoked T cell factor-dependent transcriptional activity; thus, induction of Wnt proteins by DMSO was functionally coupled. Frizzled-8/Fc inhibited the induction of cardiogenic transcription factors, cardiogenic growth factors, and sarcomeric myosin heavy chains. Likewise, differentiation was blocked by constitutively active glycogen synthase kinase 3β, an intracellular inhibitor of the Wnt/β-catenin pathway. Conversely, lithium chloride, which inhibits glycogen synthase kinase 3β, and Wnt3A-conditioned medium up-regulated early cardiac markers and the proportion of differentiated cells. Thus, Wnt/β-catenin signaling is activated at the inception of mammalian cardiac myogenesis and is indispensable for cardiac differentiation, at least in this pluripotent model system.
The earliest event in heart formation is commitment of mesodermal cells to a cardiogenic “fate” and their migration into anterolateral regions of the embryo during late gastrulation (1, 2). In this process, morphogenic movements and cardiac fate determination are believed to be regulated by multiple extracellular cues such as bone morphogenetic proteins (BMPs), transforming growth factor-β, and fibroblast growth factors (FGFs) secreted from underlying endodermal cells (3). This general model is based on a series of elegant findings in amphibians (Xenopus laevis) and avians, as early heart formation is most easily observed in embryos that develop outside the mother, becoming accessible to explant experiments and mRNA or protein injection (4–8). Contrasting evidence suggests that precardial tissue from newly gastrulated quail embryos can differentiate into beating muscle cells in the absence of endoderm (9, 10), implying that several discrete steps initiate heart development: a specification step before gastrulation, which leads to the appearance of myocardial precursor cells, and a subsequent step during gastrulation, in which endoderm serves to enhance the rate of myocyte differentiation and degree of heart tube morphogenesis. Therefore, it is important to elucidate when, where, and how mesodermal cells are instructed to assume the cardiac fate for understanding the entire body of mechanisms that operate later in heart development. However, for mammalian cardiac muscle, these initial instructive events are largely unproven.
Wnt/Wg genes, related to wingless in Drosophila, encode a number of secreted proteins that play critical roles in the development of many organisms, especially in cell fate and patterning (11–13). Notably, the prototype wingless itself collaborates with the BMP homologue, decapentaplegic, to specify the rudimentary heart tube in flies (14–17). In the absence of Wnt proteins, cells undertake active measures to maintain low levels of the Wnt signaling protein, β-catenin. Under baseline circumstances, β-catenin is phosphorylated at its N terminus by glycogen synthase kinase 3β (GSK-3β), targeting β-catenin for destruction by the ubiquitin–proteosome pathway (18). Wnt binding to the Frizzled (Fz) family of serpentine receptors activates an associated downstream component, Dishevelled, which leads to inactivation of GSK-3β, thereby stabilizing β-catenin. Accumulation of β-catenin in the cytosol results in its translocation to the nucleus (no specific control is known at this step), its interaction with T cell factor (TCF)/lymphoid enhancer factor transcription factors, and transcription of Wnt-responsive genes (19). Thus, beyond the role of membrane-associated β-catenin in adherens junctions, soluble β-catenin has a pivotal function in the canonical Wnt signal transduction pathway. In addition, Wnt proteins also activate an alternative cascade, involving protein kinase C and Jun N-terminal kinase (18, 20, 21).
Recent provocative studies using Xenopus and chick embryos indicate that Wnt proteins are potent negative regulators of heart muscle specification in those species (6–8, 22). Apart from the a priori concern that such findings are not necessarily predictive of mammalian biology, isoform and pathway differences both likely exist. For instance, in Xenopus, Wnt3A and -8 were inhibitors acting via GSK-3 (6), whereas Wnt11 was an inducer of heart formation through the β-catenin-independent pathway (23).
A genetic analysis of the Wnt family in mammals, even confined to its potential involvement in cardiac myogenesis, is far from straightforward, given extraordinary diversity and overlapping expression of the ligands and receptors. Besides Fz proteins, lipoprotein-receptor-related proteins also couple Wnts to β-catenin (24). In addition, no known promoter has sufficient early, potent, and specific expression to undertake a conditional deletion of Wnt receptors or conditional expression of Wnt inhibitors in the presumptive heart-forming region. For these reasons, multipotential cells that can be made to adopt the cardiac fate in culture provide valuable models, necessary at least as an interim step, for understanding the earliest mechanisms of cardiac determination. By using one such system, mouse P19CL6 cells (25–31), we demonstrate an essential role for the Wnt/β-catenin pathway in mammalian cardiac myogenesis.
Materials and Methods
Materials and Reagents.
P19CL6 mouse embryonic carcinoma cells were kindly provided by I. Komuro (Chiba University, Chiba, Japan). Wnt3A, control conditioned media (CM), and the corresponding expression plasmids were provided by S. Takada (Kyoto University, Kyoto; ref. 32). The constitutively active GSK-3β vector (pcDNA3-GSK-βA9), with Ser-9 replaced by alanine, was provided by M. Parsons (University of Toronto, Toronto; ref. 33). TOPFLASH (a firefly luciferase reporter plasmid, driven by two sets of three copies of the TCF binding site and herpes simplex virus thymidine kinase minimal promoter) and FOPFLASH (identical except for inactivating mutations of the TCF sites) were purchased from Upstate Biotechnology (Lake Placid, NY). pRL-cytomegalovirus (CMV; a constitutive, CMV-driven control, encoding Renilla luciferase) was from Promega, DMSO and lithium chloride (LiCl) were from Sigma, and Fz-4 and -8/Fc chimeric proteins were from R & D Systems.
Cell Culture, Differentiation, and Transfection.
Cells were grown on 10-cm dishes in α-MEM (Invitrogen) supplemented with 10% FBS (HyClone), penicillin, and streptomycin. To induce differentiation, cells were seeded at a 1:40 dilution with α-MEM/10% FBS/1% DMSO. For each experiment, cardiomyocyte differentiation was apparent in the control cultures as spontaneous beating, starting at days 9–10. To obtain stable transformants incorporating GSK-3βA9-hemagglutinin (HA) vs. the vector control, cells were transfected by using Lipofectamine 2000 (Invitrogen) and maintained in medium containing 500 μg/ml geniticin (Invitrogen). After 10–14 days, 60 colonies were picked and screened by RT-PCR. To detect exogenous GSK-3β selectively, the 5′ primer corresponded to the N terminus of GSK-3β and the 3′ primer to the Haemophilus influenzae HA epitope tag.
RT-PCR.
RNA was isolated by using TRIzol (Invitrogen). Sequences of primers and probes corresponding to each mRNA are available on request. RNA (0.1 μg) was subjected to quantitative RT-PCR by using the TaqMan One-Step RT-PCR Master Mix reagent (Applied Biosystems). The RT and PCR were run sequentially by using a 7700 Sequence Detector System (Applied Biosystems). The copy number for each transcript is expressed relative to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), used as a constitutive control. RT-PCR for FGF8 and β-actin was done as described (34).
Immunocytochemistry.
Cells were seeded on glass cover slips and cultured with or without DMSO for 3 days. Cells were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100 for 5 min. To detect phosphorylated β-catenin, cells were incubated overnight with rabbit Ab to phospho-β-catenin (Ser-33/Ser-37/Thr-41; Cell Signaling Technology, Beverly, MA) in Tris-buffered saline/3% BSA and then for 1 h with goat Ab to rabbit IgG conjugated with Alexa Fluor 488 (Molecular Probes). Immunostaining for sarcomeric myosin heavy chains (MHCs) was performed by using FITC-conjugated MF20 Ab (35). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI).
Western Blot Analysis.
Cells were seeded on six-well dishes (1.67 × 106 cells per well) and cultivated with or without 1% DMSO and 500 ng/ml Fz/Fc chimeric protein. After 3 days, cells were harvested in PBS at 4°C, centrifuged at 2,000 × g for 5 min, and resuspended in 20 mM Tris⋅HCl (pH 7.5), 25 mM sodium fluoride, and 1 mM EDTA, containing a protease inhibitor mixture (Roche Molecular Biochemicals). Cells were incubated on ice for 20 min, followed by 30 strokes in a Dounce homogenizer, and centrifuged at 100,000 × g for 30 min. The supernatant was collected as the soluble, cytoplasmic fraction and subjected to electrophoresis in 10% SDS-polyacrylamide gels. Proteins were transferred to poly(vinylidene difluoride) membranes, which were incubated sequentially in Tris-buffered saline, 2% BSA, 0.05% Tween 20, and then mouse Ab to β-catenin (BD Biosciences) or goat Ab to total actin (C-11; Santa Cruz Biotechnology) overnight at 4°C. Bound Ab was visualized by using horseradish peroxidase (HRP)-conjugated goat Ab to mouse IgG (Zymed) or donkey Ab to goat IgG (Santa Cruz Biotechnology) and enhanced chemiluminescence reagents (Amersham Biosciences). To detect phosphorylated β-catenin, cells were lysed with RIPA buffer (0.15 mM NaCl/0.05 mM Tris⋅HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS) with 10 nM calyculin A, 10 nM okadaic acid, and a protease inhibitor mixture. Western blotting was done as described previously by using rabbit Ab to phospho-β-catenin and HRP-conjugated goat Ab to rabbit IgG (Santa Cruz Biotechnology).
Luciferase Assays.
Cells seeded and cultured as described previously were transfected 1 day after plating by using Lipofectamine 2000 in serum-free α-MEM for 6 h. Transfections contained 0.5 μg of TOPFLASH or FOPFLASH plus 0.1 μg of pRL-CMV as the cotransfected control. Medium containing 10% FBS with or without DMSO was changed 6 and 48 h after transfection. Cells were lysed 4 days after DMSO treatment, and luciferase was assayed by using the Dual-Luciferase system (Promega). Firefly luciferase activity, indicating TCF-dependent transcription, was normalized to the Renilla luciferase activity of each extract. TOPFLASH activity induced by Wnt3A CM vs. control CM was measured after 18 h. Activity of the GSK-3βA9 expression vector was corroborated by transient transfection in 293T cells. Cells were passaged at a 1:2.5 dilution into 24-well dishes, and TOPFLASH, pRL-CMV, and PGK-Wnt3A were cotransfected by calcium-phosphate precipitation along with pcDNA3-GSK-3βA9-HA vs. the empty vector control (36). Twenty-four hours after transfection, the medium was changed to DMEM supplemented with 0.2% BSA. Forty-eight hours after transfection, cells were lysed and luciferase activities were assayed as described previously.
Statistical Analysis.
Results, shown as the mean ± SE, were compared by ANOVA followed by Scheffé's test, with P < 0.05 considered significant.
Results
Wnt3A and -8 Are Early Responses in Differentiating P19CL6 Cells.
We first used quantitative RT-PCR (QRT-PCR) analyses to map the temporal changes of cardiac-specific genes in differentiating P19CL6 cells, in comparison to their potential regulators (Fig. 1A). Without DMSO, the cells did not express any of the cardiac-specific genes assayed. After 1% DMSO treatment, the cells expressed the cardiac transcription factors Nkx2.5, GATA4, MEF2C, and Tbx5 as early stage markers within 3–8 days. Spontaneous beating was visible at days 9–10, and continued for 4–8 days. Among these markers, GATA4 and Nkx2.5 were expressed earliest (as early as 3 days after DMSO), and GATA4 protein was detected at 8 days (Fig. 1B). α-MHC was seen as early as day 8 and increased until day 12, corresponding to the onset of spontaneous beating. Concordant with the presence of α-MHC mRNA (Fig. 1A) and protein (Fig. 1B), MF20-positive differentiated myocytes were observed on day 12 (Fig. 1C).
Figure 1.
Wnt3A and Wnt8 are early responses in differentiating P19CL6 cells. (A) Gene expression was determined by QRT-PCR, normalized to GAPDH, and represented as relative copy number. (B) Western blot, showing induction of GATA4 and sarcomeric MHCs vs. tubulin as the control. (C) Immunocytochemistry, showing induction of sarcomeric MHCs. Cells were cultivated for 12 days with (Right) or without (Left) 1% DMSO, and stained with MF20 (red) and 4′,6-diamidino-2-phenylindole (blue). (Bar = 50 μm.) (D) FGF8 expression by RT-PCR.
Neither BMP2 nor BMP4 was expressed in the absence of DMSO. Both were induced to low levels within 4–5 days, with expression highest on day 8 and persisting through day 12 (Fig. 1A). Studies have implicated endogenous BMPs as essential for cardiogenesis in these cells (26, 27). By contrast to the delayed and sustained expression of BMP2 and BMP4, Wnt3A and Wnt8A were each induced as early as 2 days after DMSO treatment, with expression greatest on day 3 (Fig. 1A). Thereafter, both Wnt3A and -8A mRNA levels were quickly down-regulated on day 4, with low or undetectable expression at all later times. Like Wnt3A and -8A, FGF8 was strongly induced at 2–4 days and decreased progressively thereafter (Fig. 1D). Wnt11, which is required for Xenopus cardiogenesis (23), was not expressed (data not shown).
The Wnt/β-Catenin Pathway Is Activated at the Early Stage of Differentiation.
To assess whether functional Wnt signaling is activated at the time of Wnt induction, we examined cytosolic β-catenin, the crux of the canonical Wnt signal transmission pathway (13, 37). Cells treated with or without DMSO were harvested and lysed on day 3, cytosolic protein was fractionated, and β-catenin protein levels were examined by Western blot (Fig. 2A). DMSO caused the accumulation of soluble β-catenin; accumulation of β-catenin was also observed in the total cell lysates (Fig. 2B). Simultaneous treatment with 500 ng/ml Fz-8/Fc chimeric protein, an antagonist for Wnt8A and potentially for other Wnts, decreased β-catenin to the basal level (Fig. 2 A and B), indicating that its accumulation is regulated by an autocrine or paracrine circuit in this system, involving endogenous Wnts. Conversely, as expected, phosphorylated β-catenin (the form targeted for degradation) was decreased by DMSO treatment for 3 days, determined by immunoblotting and immunocytochemistry with Ab to the phosphorylated epitope (Fig. 2 B and C). Furthermore, phosphorylated β-catenin was rescued by treatment with Fz-8/Fc (Fig. 2B). As a third criterion to confirm the activation of the canonical Wnt pathway, TCF-dependent transcriptional activity was measured (Fig. 2D). DMSO treatment for 4 days provoked a 9-fold increase in luciferase activity. No activation was seen by using the corresponding control reporter with mutated TCF binding sites. Thus, Wnt induction by DMSO was functionally coupled to activation of the β-catenin pathway.
Figure 2.
The Wnt/β-catenin-signaling pathway is activated at the onset of cardiac myogenesis. (A) Increased soluble β-catenin. Cells were treated with (+) or without (−) 1% DMSO for 3 days. The cytosolic fraction was subjected to Western blot analysis. β-Catenin was specifically decreased by Fz-8/Fc, an extracellular antagonist of Wnt signaling. (B) Decreased phosphorylated β-catenin and increased total β-catenin in whole-cell lysates. Cells were cultured as in A and Western blotting was done by using Abs to phospho-β-catenin (Top) and total β-catenin (Middle). (C) Decreased phosphorylated β-catenin (green), shown by immunostaining. (Left) Without DMSO. (Right) With DMSO. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (blue). (Bar = 50 μm.) (D) TCF/lymphoid enhancer factor-dependent transcription. Cells were cultured ± DMSO and transfected with TOPFLASH or FOPFLASH (inactive, mutant TCF sites) along with pRL-CMV. Luciferase activity was determined after 4 days of treatment.
The Wnt/β-Catenin-Signaling Pathway Was Required for Cardiac Differentiation.
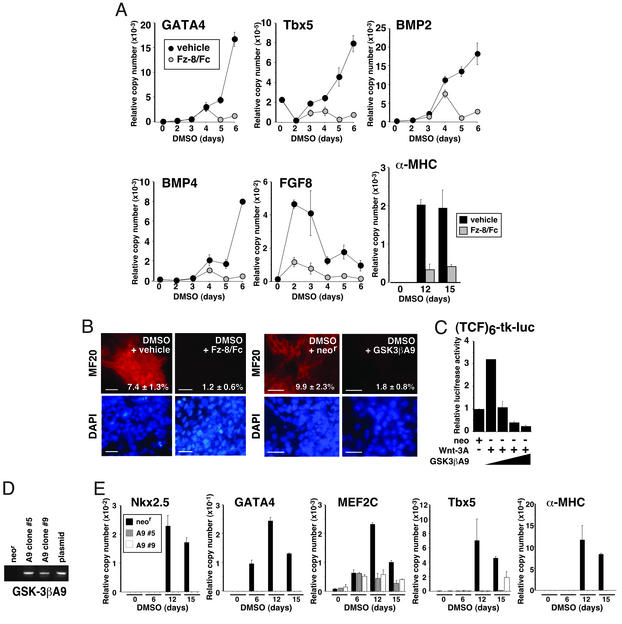
To explore the possible role of Wnt-mediated signaling in early cardiac myogenesis, we first monitored differentiation induced by DMSO, with and without 200 ng/ml Fz-8/Fc. Treatment with the soluble Wnt inhibitor prevented GATA4 and Tbx5 induction by DMSO, at least through day 6 (Fig. 3A). Likewise, Fz-8/Fc inhibited the expression of BMP2, BMP4, and FGF8. Thus, the Wnt pathway lies upstream to the induction of these three cardiac differentiation factors (Fig. 3A). Similar results were obtained by using Fz-4/Fc. Correspondingly, Fz-8/Fc decreased the proportion of MF20-positive cells 6- to 7-fold (day 12; Fig. 3B Left), and suppression continued for at least 15 days (Fig. 3A).
Figure 3.
The Wnt/β-catenin-signaling pathway was required for cardiac differentiation. (A) Fz-8/Fc suppresses the induction of GATA4, TBX5, BMP2, BMP4, FGF8, and α-MHC, determined by QRT-PCR. Cells were treated with DMSO plus diluent or 200 ng/ml Fz-8/Fc. Equivalent results were obtained by using Fz-4/Fc. (B Left) Fz-8/Fc suppresses the induction of sarcomeric MHC protein. (Right) Constitutively active GSK-3β (GSK3βA9) suppresses the induction of sarcomeric MHC protein. (Bar = 25 μm.) (C) GSK3βA9 inhibits Wnt3A-induced TCF transcriptional activity. 293T cells were cotransfected with PGK-neo, PGK-Wnt3A, and pcDNA3-GSK-3βA9-HA as shown, plus the TOPFLASH and pRL-CMV reporter genes. (D) GSK-3βA9-HA expression in stably transformed P19CL6 cells, by RT-PCR. Cells were stably transfected with the pcDNA3 expression vectors shown. Plasmid, 5 ng of pcDNA3-GSK-3βA9-HA as template. (E) GSK-3βA9 suppresses DMSO-induced cardiac gene expression.
To determine specifically whether the β-catenin pathway for Wnt signaling was responsible for Wnt-dependent cardiac myogenesis, we used a constitutively active form of GSK-3β (pcDNA3-GSK-3βA9-HA). In pilot studies by transient cotransfection of 293T cells (Fig. 3C), we confirmed that this vector inhibited Wnt3A-induced transcription of the TCF reporter gene. We then obtained stable transformants of P19CL6 cells harboring GSK-3βA9 vs. the control vector, neor (Fig. 3D). GSK-3βA9 suppressed the induction of cardiac transcription factors by DMSO through at least 15 days, with identical results in independent lines (Fig. 3E and data not shown). By contrast, cardiac differentiation was impaired in none of the clonal isolates bearing the selectable marker neor alone, assayed at equivalent passage number. Likewise, GSK-3β suppressed the prevalence of sarcomeric MHC staining, whereas transfection with neor had no effect (Fig. 3B Right). Together, these findings with soluble Fz proteins and activated GSK-3β implicate signaling by endogenous Wnts, via β-catenin, as required for early cardiac determination in this setting.
The Wnt/β-Catenin-Signaling Pathway Enhances Cardiac Myogenesis.
We next explored the reciprocal possibility of promoting cardiac differentiation in this system by supplying exogenous Wnt or potentiating β-catenin. We first confirmed that Wnt3A-conditioned medium could activate TCF-dependent transcription in P19CL6 cells (Fig. 4A Left). Although control CM had no effect (from L cells stably transfected with just the neomycin-resistance gene), Wnt3A-conditioned medium markedly enhanced the inductive effect of DMSO on Nkx2.5, GATA4, MEF2C, and Tbx5 (Fig. 4A Right). α-MHC, otherwise expressed no sooner than day 8, was detected on day 6 in cultures receiving Wnt3A CM. Interestingly, Wnt3A CM also enhanced the expression of both BMP2 and BMP4. In accordance with the enhanced expression of cardiogenic factors, the prevalence of MF20-positive cells was increased nearly 3-fold on day 12 by Wnt3A CM (Fig. 4B).
Figure 4.
The Wnt/β-catenin-signaling pathway enhances cardiac myogenesis. (A) Wnt3A CM increases TCF-dependent transcription. P19CL6 cells cotransfected with TOPFLASH and pRL-CMV were assayed after 18 h in Wnt3A or control (neo) CM. (B) Wnt3A CM induces cardiac-specific markers (0–15 days) and BMPs (6 days), shown by QRT-PCR. Wnt3A CM induces sarcomeric MHC, shown by immunostaining. (Bar = 50 μm.) (C) LiCl enhances cardiomyogenesis. Induction of cardiac-specific markers was analyzed by QRT-PCR (day 5). NaCl was added as the control.
We next used LiCl, which binds and inhibits GSK-3β and, hence, activates Wnt signaling selectively via the β-catenin/TCF pathway (13, 38). At 10 μM, LiCl significantly increased the expression of Nkx2.5, GATA4, Tbx5, BMP2, and BMP4 at day 5 and MF20-positive cells at day 12 (Fig. 4C). LiCl itself had an inductive effect on each of the early cardiac markers and on sarcomeric MHCs even in the absence of DMSO. However, cells treated with LiCl or Wnt3A CM alone did not show spontaneous beating (data not shown), suggesting that additional signals conferred by DMSO were necessary for terminal differentiation, beyond just induction of the factors shown here.
Discussion
In summary, activation of the Wnt/β-catenin-signaling cascade was an early event in the cardiogenic differentiation of pluripotent P19CL6 cells, as measured by Wnt3A and Wnt8A induction, hypophosphorylation of β-catenin, accumulation of β-catenin, and TCF/lymphoid enhancer factor-dependent transcription. Blocking Wnt–receptor interactions with soluble Fz proteins and inhibition of β-catenin with GSK-3β evoked equivalent effects, largely or completely blocking the cardiogenic pathway, including induction of Tbx5 and GATA4, the earliest marker of differentiation here and in microarray profiles of this system (30). Intriguingly, lymphoid enhancer factor 1 itself was an early response to DMSO in the study cited (30). Thus, endogenous Wnts mediate cardiogenesis in P19CL6 cells and do so via the canonical β-catenin pathway. By contrast, endogenous BMPs are later responses of the cells to DMSO (BMP2 and BMP4, Fig. 1; BMP5, ref. 30), required but insufficient to trigger the cardiac fate (26, 27). Although other endogenous signals might coexist, our data support the conclusion that Wnts, coupled to β-catenin, are the long-sought essential early targets of DMSO for cardiac specification. We emphasize that, in Drosophila, the cardiogenic signal from Wg itself depends on armadillo, the β-catenin homologue (15).
In agreement with this interpretation, cardiac differentiation was augmented by extracellular and intracellular interventions that stimulate this pathway, Wnt3A CM and LiCl, respectively. Our conclusions differ, obviously, from inhibitory roles found for Wnt3A and -8 in Xenopus and chicks (6–8). Differences from mammalian cardiac specification exist in both species, e.g., in the apparent involvement of activins (23, 39, 40). Apart from potential phylogenetic dissimilarities, the studies also differ inherently (cultured cells vs. explants and embryos) in the stage of maturation, perturbations, and mix of cell types present. Indirect inductive interactions are possible even in this clonal line (30, 41). Like BMPs, FGF8 was induced by DMSO and suppressed by the Wnt inhibitor (Figs. 1D and 3A). Although no direct extrapolation to mammals is yet available from the inductive function of this gene in avian cardiogenesis (42, 43), we speculate that Wnt3A and -8 act at least in part indirectly by controlling the creation of endoderm or endoderm-like cells as the source of these cardiogenic factors.
Our results do not contradict the stimulation of cardiogenesis by Wnt11 via the noncanonical pathway in Xenopus and in P19 cells (the parent line for the clone we used). Only sufficiency, not a requirement for Wnt11, was tested (23), and perhaps both classes of Wnt signal operate in mammals. Germ-line deletion of β-catenin is lethal at gastrulation (44), obscuring any role of the gene in cardiac progenitors, and the formation of multiple hearts in mutant mice after conditional deletion of β-catenin (45) is discordant with our findings only at first glance. In this case, β-catenin was deleted exclusively from embryonic endoderm, redirecting endodermal cell fate as a cell-autonomous event. By design, the experiment does not address the function of β-catenin in normal cardiac progenitors themselves.
Although the function of endogenous Wnts emerges more clearly from inhibitor studies than from stimulation of the pathway, that Wnt CM and LiCl promote the formation of heart muscle cells has auspicious translational implications, in addition to posing questions for stem cell biology and mammalian cardiac development more broadly. Endogenous stem cells from bone marrow and perhaps other sites are recruited to damaged heart muscle and there commit to a cardiac fate, albeit in numbers too small to be effective in repair (46). Nothing whatsoever is known of the inductive signals for cardiac myocyte formation in adults.
Acknowledgments
We thank R. J. Schwartz, T. Miura, H. Oh, Y. Hamamori, M. Xie, and D. Zhang for helpful suggestions. This work was supported in part by National Institutes of Health grants (to M.D.S.) and the M. D. Anderson Foundation Professorship (to M.D.S.).
Abbreviations
- BMP
bone morphogenetic protein
- GSK-3β
glycogen synthase kinase 3β
- MHC
myosin heavy chain
- QRT-PCR
quantitative RT-PCR
- TCF
T cell factor
- CM
conditioned media
- CMV
cytomegalovirus
- HA
hemagglutinin
- LiCl
lithium chloride
- FGF
fibroblast growth factor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Garcia-Martinez V, Schoenwolf G C. Dev Biol. 1993;159:706–719. doi: 10.1006/dbio.1993.1276. [DOI] [PubMed] [Google Scholar]
- 2.Harvey R P, Rosenthal N, editors. Heart Development. San Diego: Academic; 1998. [Google Scholar]
- 3.Beddington R S, Robertson E J. Cell. 1999;96:195–209. doi: 10.1016/s0092-8674(00)80560-7. [DOI] [PubMed] [Google Scholar]
- 4.Schultheiss T M, Burch J B, Lassar A B. Genes Dev. 1997;11:451–462. doi: 10.1101/gad.11.4.451. [DOI] [PubMed] [Google Scholar]
- 5.Eisenberg C A, Eisenberg L M. Dev Dyn. 1999;216:45–58. doi: 10.1002/(SICI)1097-0177(199909)216:1<45::AID-DVDY7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 6.Schneider V A, Mercola M. Genes Dev. 2001;15:304–315. doi: 10.1101/gad.855601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marvin M J, Di Rocco G, Gardiner A, Bush S M, Lassar A B. Genes Dev. 2001;15:316–327. doi: 10.1101/gad.855501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tzahor E, Lassar A B. Genes Dev. 2001;15:255–260. doi: 10.1101/gad.871501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalez-Sanchez A, Bader D. Dev Biol. 1990;139:197–209. doi: 10.1016/0012-1606(90)90288-t. [DOI] [PubMed] [Google Scholar]
- 10.Antin P B, Taylor R G, Yatskievych T. Dev Dyn. 1994;200:144–154. doi: 10.1002/aja.1002000206. [DOI] [PubMed] [Google Scholar]
- 11.Arias A M, Brown A M C, Brennan K. Curr Opin Genet Dev. 1999;9:447–454. doi: 10.1016/s0959-437x(99)80068-9. [DOI] [PubMed] [Google Scholar]
- 12.Bejsovec A. Curr Biol. 1999;9:R684–R687. doi: 10.1016/s0960-9822(99)80439-4. [DOI] [PubMed] [Google Scholar]
- 13.Moon R T, Bowerman B, Boutros M, Perrimon N. Science. 2002;296:1644–1646. doi: 10.1126/science.1071549. [DOI] [PubMed] [Google Scholar]
- 14.Wu X, Golden K, Bodmer R. Dev Biol. 1995;169:619–628. doi: 10.1006/dbio.1995.1174. [DOI] [PubMed] [Google Scholar]
- 15.Park M Y, Wu X S, Golden K, Axelrod J D, Bodmer R. Dev Biol. 1996;177:104–116. doi: 10.1006/dbio.1996.0149. [DOI] [PubMed] [Google Scholar]
- 16.Frasch M. Nature. 1995;374:464–467. doi: 10.1038/374464a0. [DOI] [PubMed] [Google Scholar]
- 17.Lee H H, Frasch M. Development (Cambridge, UK) 2000;127:5497–5508. doi: 10.1242/dev.127.24.5497. [DOI] [PubMed] [Google Scholar]
- 18.Liu C, Li Y, Semenov M, Han C, Baeg G H, Tan Y, Zhang Z, Lin X, He X. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- 19.Cadigan K M, Nusse R. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 20.Kuhl M. Semin Cell Dev Biol. 2002;13:243–249. doi: 10.1016/s1084-9521(02)00050-2. [DOI] [PubMed] [Google Scholar]
- 21.Weston C R, Davis R J. Curr Opin Genet Dev. 2002;12:14–21. doi: 10.1016/s0959-437x(01)00258-1. [DOI] [PubMed] [Google Scholar]
- 22.Olson E N. Science. 2001;291:2327–2328. doi: 10.1126/science.1060063. [DOI] [PubMed] [Google Scholar]
- 23.Pandur P, Lasche M, Eisenberg L M, Kuhl M. Nature. 2002;418:636–641. doi: 10.1038/nature00921. [DOI] [PubMed] [Google Scholar]
- 24.Mao J, Wang J, Liu B, Pan W, Farr G H, 3rd, Flynn C, Yuan H, Takada S, Kimelman D, Li L, Wu D. Mol Cell. 2001;7:801–809. doi: 10.1016/s1097-2765(01)00224-6. [DOI] [PubMed] [Google Scholar]
- 25.Skerjanc I S. Trends Cardiovasc Med. 1999;9:139–143. doi: 10.1016/s1050-1738(99)00017-1. [DOI] [PubMed] [Google Scholar]
- 26.Monzen K, Shiojima I, Hiroi Y, Kudoh S, Oka T, Takimoto E, Hayashi D, Hosoda T, Habara-Ohkubo A, Nakaoka T, et al. Mol Cell Biol. 1999;19:7096–7105. doi: 10.1128/mcb.19.10.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monzen K, Hiroi Y, Kudoh S, Akazawa H, Oka T, Takimoto E, Hayashi D, Hosoda T, Kawabata M, Miyazono K, et al. J Cell Biol. 2001;153:687–698. doi: 10.1083/jcb.153.4.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anisimov S, Tarasov K, Riordon D, Wobus A, Boheler K. Mech Dev. 2002;117:25–74. doi: 10.1016/s0925-4773(02)00177-6. [DOI] [PubMed] [Google Scholar]
- 29.Paquin J, Danalache B A, Jankowski M, McCann S M, Gutkowska J. Proc Natl Acad Sci USA. 2002;99:9550–9555. doi: 10.1073/pnas.152302499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng C F, Wei Y, Levsky J M, McDonald T V, Childs G, Kitsis R N. Physiol Genomics. 2002;9:145–155. doi: 10.1152/physiolgenomics.00027.2002. [DOI] [PubMed] [Google Scholar]
- 31.Hiroi Y, Kudoh S, Monzen K, Ikeda Y, Yazaki Y, Nagai R, Komuro I. Nat Genet. 2001;28:276–280. doi: 10.1038/90123. [DOI] [PubMed] [Google Scholar]
- 32.Muroyama Y, Fujihara M, Ikeya M, Kondoh H, Takada S. Genes Dev. 2002;16:548–553. doi: 10.1101/gad.937102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohteki T, Parsons M, Zakarian A, Jones R G, Nguyen L T, Woodgett J R, Ohashi P S. J Exp Med. 2000;192:99–104. doi: 10.1084/jem.192.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee S H, Lumelsky N, Studer L, Auerbach J M, McKay R D. Nat Biotechnol. 2000;18:675–679. doi: 10.1038/76536. [DOI] [PubMed] [Google Scholar]
- 35.Oh H, Taffet G E, Youker K A, Entman M L, Overbeek P A, Michael L H, Schneider M D. Proc Natl Acad Sci USA. 2001;98:10308–10313. doi: 10.1073/pnas.191169098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jordan M, Schallhorn A, Wurm F M. Nucleic Acids Res. 1996;24:596–601. doi: 10.1093/nar/24.4.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papkoff J R, Schryver B, Polakis P. Mol Cell Biol. 1996;16:2128–2134. doi: 10.1128/mcb.16.5.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen R H, Ding W V, McCormick F. J Biol Chem. 2000;275:17894–17899. doi: 10.1074/jbc.M905336199. [DOI] [PubMed] [Google Scholar]
- 39.Logan M, Mohun T. Development (Cambridge, UK) 1993;118:865–875. doi: 10.1242/dev.118.3.865. [DOI] [PubMed] [Google Scholar]
- 40.Matzuk M M, Kumar T R, Bradley A. Nature. 1995;374:356–360. doi: 10.1038/374356a0. [DOI] [PubMed] [Google Scholar]
- 41.Smith S C, Reuhl K R, Craig J, McBurney M W. J Cell Physiol. 1987;131:74–84. doi: 10.1002/jcp.1041310112. [DOI] [PubMed] [Google Scholar]
- 42.Alsan B H, Schultheiss T M. Development (Cambridge, UK) 2002;129:1935–1943. doi: 10.1242/dev.129.8.1935. [DOI] [PubMed] [Google Scholar]
- 43.Frank D U, Fotheringham L K, Brewer J A, Muglia L J, Tristani-Firouzi M, Capecchi M R, Moon A M. Development (Cambridge, UK) 2002;129:4591–4603. doi: 10.1242/dev.129.19.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haegel H, Larue L, Ohsugi M, Fedorov L, Herrenknecht K, Kemler R. Development (Cambridge, UK) 1995;121:3529–3537. doi: 10.1242/dev.121.11.3529. [DOI] [PubMed] [Google Scholar]
- 45.Lickert H, Kutsch S, Kanzler B, Tamai Y, Taketo M M, Kemler R. Dev Cell. 2002;3:171–181. doi: 10.1016/s1534-5807(02)00206-x. [DOI] [PubMed] [Google Scholar]
- 46.Jackson K A, Majka S M, Wang H, Pocius J, Hartley C J, Majesky M W, Entman M L, Michael L H, Hirschi K K, Goodell M A. J Clin Invest. 2001;107:1395–1402. doi: 10.1172/JCI12150. [DOI] [PMC free article] [PubMed] [Google Scholar]