Abstract
The Wingless (Wg) protein is a secreted glycoprotein involved in intercellular signaling. On activation of the Wg signaling pathway, Armadillo is stabilized, causing target genes to be activated by the transcription factor Pangolin (Pan). This study investigated the roles of Pan in the developing wing of Drosophila by clonal analysis. Three different aspects of wing development were examined: cell proliferation, wing margin specification, and wg self-refinement. Our results indicate that Pan function is critically required for all three of these processes. Consequently, lack of pan causes a severe reduction in the activity of the Wg target genes Distalless and vestigial within their normal domain of expression. Loss of pan function does not, however, lead to a derepression of these genes outside this domain. Thus, although Pan is positively required for the induction of Wg targets in the wing imaginal disk, it does not appear to play a default repressor function in the absence of Wg input. In contrast, lack of zygotic pan function causes a milder phenotype than that caused by the lack of wg function in the embryo. We show that this difference cannot be attributed to maternally provided pan product, indicating that a Pan repressor function usually prevents the expression of embryonic Wg targets. Together, our results suggest that for embryonic patterning the activator as well as repressor forms of Pan play important roles, while for wing development Pan operates primarily in the activator mode.
Wingless (Wg) plays important roles in Drosophila development. It is required for patterning of the embryonic epidermis (1, 2), for the proper establishment of the embryonic nervous system (3–6), and also for the specification, growth, and cell-fate assignment of adult appendages, such as the wing and the leg (7, 8). In the developing wing imaginal disk, wg is first involved in the definition of the wing versus notum primordium (9, 10). Later, Wg is secreted at the dorsoventral (D/V) compartment boundary of the wing disk, where it directs the formation of wing margin structures (11) and from where it acts as a morphogen to organize gene expression (12, 13). Wg also plays a role in restricting its own expression to cells immediately adjacent to the D/V boundary, a phenomenon referred to as wg self-refinement (14).
Wg exerts most if not all effects on cell-fate specification by regulating the transcription of target genes in responding cells. The key regulatory event in the Wg transduction pathway appears to be the posttranscriptional up-regulation of the β-catenin homolog Armadillo (Arm). Arm, in turn, confers transcriptional activator activity to the lymphoid-enhancing factor (LEF)/T cell factor (TCF) homolog Pangolin (Pan)/dTCF (15, 16). LEF/TCF proteins belong to the family of high-mobility-group transcription factors that bind to specific DNA sequences. Because the loss-of-pan-function phenotypes resemble those caused by loss of Wg signaling, it is likely that Pan acts as a transcriptional activator for Wg target genes. It was reported that in the absence of Wg input, Pan also functions as a transcriptional repressor in the embryo, possibly via the corepressor protein Groucho (17). Consistent with this repressor role of Pan, cells up-regulate the activity of a Ubx midgut enhancer if its Pan binding sites have been mutated (18). In addition, when Pan-binding sites were mutated in the even-skipped mesodermal enhancer, ectopic gene expression was observed in the dorsal mesoderm (19). Therefore, it is likely that the net balance of the Wg-dependent activator and Wg-independent repressor levels of Pan determines whether Wg targets are induced or repressed.
Here we wanted to address the function of Pan in imaginal-disk development. Is Pan critical for Wg signaling in imaginal cells? If so, does it also play a dual role in activating and repressing the transcription of Wg targets? To answer these questions, we set out to study the function of Pan by clonal analysis and removed pan function genetically in subsets of cells of the wing imaginal disk. Our results demonstrate that Pan is involved in all aspects of Wg signaling in the developing wing and functions primarily as an activator in this tissue, whereas it plays a dual role as an activator and repressor during embryogenesis.
Materials and Methods
Fly Stocks and Genetics.
For pan−/− clonal analysis, homozygous pan2/pan2 null mutant animals (16) were rescued with a P[tub-pan, w+] insertion on the left arm of the second chromosome. An arm-lacZ and the FRT40 transgenes were placed on the same chromosome arm by meiotic recombination. A y w f hsflp first and a P[f+] ck− FRT40 second chromosome were used to mark experimental clones with ck− and twin spots with f− in the adult wing (20). For sgg−/− pan−/− double-clonal analysis, a P[tub-pan, w+] insertion on the first chromosome was used. Larvae of the following genotype were generated for the induction of clones y w P[tub-pan, w+] FRT19/y sggD127 f36a FRT19; hsflp/+; pan2/pan2 or pan2/Dp(1;4)1021[y+]. For pan−/− germ-line clones, pan2/pan2 females were used that carried an ovoD1 transgene on the left arm of the third chromosome (21) as well as a P[tub-pan, w+] rescue construct on the same arm (recombined by x-ray-induced male recombination; see below).
X-Ray-Induced Male Recombination.
Third instar larvae were irradiated with 1,500 rad (Philips MG 160; 150 kV, 14 mA for 3 min with a 25-cm focus distance and a 2-mm Plexiglas filter).
Induction of Clones.
pan−/− and sgg−/− pan−/− clones were induced by heat shock at 34°C for 30 min 1–4 days after egg deposition (by collecting eggs for 3 days, followed by a 1-day incubation before induction). Imaginal disks of third instar larvae were isolated for immunohistochemistry 1–3 days after flp induction. Germ-line clones were induced by applying heat shocks at late third instar or pupal stages.
Immunohistochemistry.
Imaginal disks were stained as described (22). Antibodies against Dll (provided by S. Cohen, EMBL, Heidelberg), Vg (provided by S. Carroll, University of Wisconsin, Madison), and Wg (provided by S. Cohen) were diluted 1:500. Rabbit anti-β-Gal polyclonal antibody (Cappel) was used at 1:2,000 and mouse anti-β-Gal monoclonal antibody (Promega Z378A) was used at 1:1,000 to mark pan−/− clones.
Wing Mounting.
Dissected wings were stored in a glycerin:ethanol 1:3 solution, then washed twice in 100% ethanol and mounted in Euparal.
Cuticle Preparation.
Cuticle preparations for germ-line clones were performed by picking individual embryos onto slides, washing them with bleach, water, and methanol, and adding Hoyer's solution. After placing cover slips, the slides were heated at 65°C for 3–5 h.
Results
Pan Is Required for Proliferation and Survival of Wing Cells.
The pan gene is located on the small fourth chromosome, which does not usually undergo meiotic or mitotic recombination. Therefore, it is technically difficult to induce pan mutant cell clones. Here we used a pan transgene to circumvent this problem. The expression levels of pan appear to be critical for Drosophila development. High experimental Pan levels cause lethality, whereas low Pan levels fail to rescue pan mutant animals (not shown). Eventually, three insertions of a pan-rescuing transgene were recovered, in which the transcription of a full-length pan cDNA is driven by the ubiquitously active promoter of the tubulin alpha1 gene. The insertion on chromosome arm 2L, which exhibits highest rescuing activity (100% larval, 70% adult), was used to generate an arm-lacZ tub-pan[2L] FRT40 chromosome. By the use of an hsp70-flp transgene, marked clones were induced in a pan2 null mutant background. Clones that lack arm-lacZ activity also lack pan activity; by contrast, sibling twin-spot clones inherit two copies of either transgene.
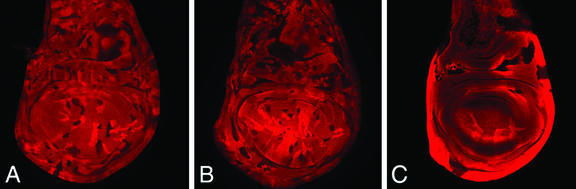
Induction of recombination during second larval instar resulted in pan mutant clones and twin spots of similar size throughout the wing imaginal disk (Fig. 1A). Earlier induction, during first larval instar, caused a reduced size of pan clones compared with their twin spots in the wing-blade primordium (Fig. 1B). When recombination was induced during late embryogenesis, only twin spots but no pan mutant clones were observed in the wing pouch (Fig. 1C). We interpret these observations as an indication that pan null mutant wing cells cannot proliferate normally and are lost from the wing epithelium once transient perdurance of Pan protein ceases in mutant cells. This behavior is reminiscent of that observed for clones mutant for other components of the Wg signaling transduction pathway, such as arm, Dfrizzled/Dfrizzled2, legless, or pygopus (13, 23, 24).
Figure 1.
Loss of Pan function leads to cell proliferation defects in the developing wing blade. pan−/− clones are marked by the absence of lacZ activity (shown in red), and their twin spots are marked by increased activity (bright red). The clones were induced at the second instar stage (A), at the first instar stage (B), or during late embryogenesis (C).
Pan Acts as a Positive Regulator of Wg Target Genes.
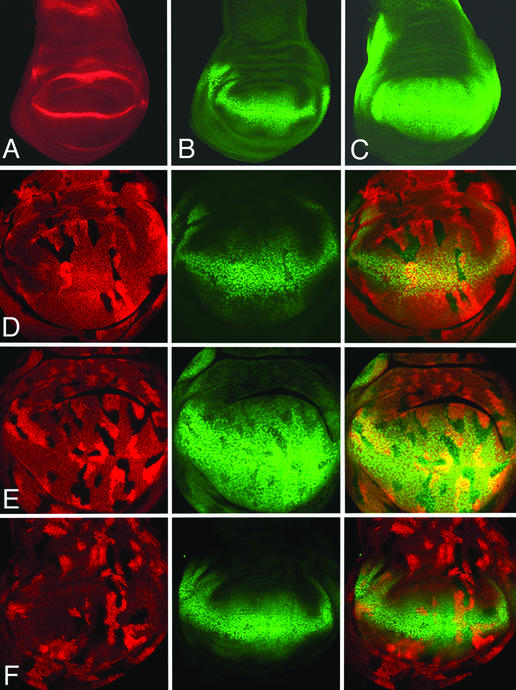
Wg is expressed in the wing pouch of late third instar disks in a thin stripe of cells destined to form the presumptive wing margin (Fig. 2A). Wg emanating from this stripe acts at long range to activate the expression of a number of genes, such as Distalless (Dll) (Fig. 2B) and vestigial (vg) (Fig. 2C), which control wing development (12, 13, 25, 26). Although it has not been demonstrated that Dll and vg are direct target genes of Wg signaling, their expression levels serve as valuable readout for Wg transduction.
Figure 2.
Expression of Wg target genes in pan−/− and groucho−/− clones. In wild-type disks, wg is expressed (red) at the D/V compartment boundary. Dll (B) and vg (C) expression (green) straddles the stripe of wg expression in the wing pouch. Reduced expression of Dll (D, green) and vg (E, green) is observed in pan−/− clones [marked by the absence of lacZ activity (red in D and E)]. No ectopic expression is observed in pan−/− clones outside the normal Dll or vg expression domain. groucho mutant cells express normal levels of Dll expression (F). groucho−/− clones are marked by the absence of CD2 staining (red), whereas Dll expression is detected by antibody staining (green).
We generated pan mutant clones during the second larval instar and analyzed the expression of Dll and vg. pan mutant cells situated within the Dll and vg expression domain exhibit reduced expression levels of these genes (Fig. 2 D and E). pan mutant cells outside or straddling the outer limits of Dll and vg expression do not show a transcriptional up-regulation of these genes. These results indicate that Pan functions as a positive regulator for Wg signaling. Moreover, loss of Pan activity does not cause a derepression of target genes in cells receiving low or no Wg input.
It has previously been reported that Pan and Groucho function together to repress embryonic Wg targets in the absence of Wg signaling (17). To confirm our conclusion that Pan does not function as a transcriptional repressor in wing cells, we investigated the role of groucho in regulating Wg responsive genes. Loss of groucho function did not affect Dll expression in grouchoE48 homozygous cells (Fig. 2F).
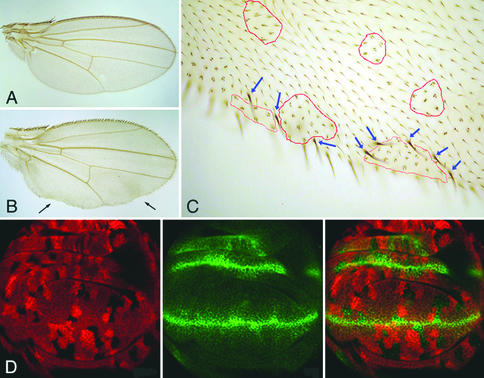
By using the trichome and bristle marker crinkled we analyzed the adult fate of pan mutant cells in wing patterning. Peripheral pan−/− clones lead to the loss of wing margin structures, whereas clones in the central part of the wing blade exhibit no obvious defects (Fig. 3B). At higher magnification, we found that clones at the wing margin can cause the formation of ectopic wing margin bristles in adjacent wild-type territory (Fig. 3C), reminiscent of the behavior of dishevelled mutant clones (see below). Again, therefore, the phenotypes from loss of pan, which we have observed in pan mutant clones of developing and adult wings, resemble those from loss of functions of other positive regulators of the Wg signaling pathway. No obvious planar polarity defects were observed in pan mutant clones on adult wings. In sum, we interpret our results as an indication that Pan functions as a positive mediator of Wg outputs.
Figure 3.
pan−/− clones at the wing margin are associated with loss-of-Wg-signaling phenotypes. (A) Wild-type wing. (B) Small pan−/− clones show no abnormality within the wing blade, and clones at the wing margin (see arrows) lead to the loss of wing margin structures. pan−/− clones are marked with crinkled. The clones can be recognized under a high magnification (see red outlines). Ectopic bristles are observed surrounding pan−/− clones at the wing margin (see blue arrows). No effect was observed for pan−/− clones at the wing blade (red outlines). In wing imaginal disks, Wg is ectopically expressed in pan−/− clones near the D/V boundary (D). pan−/− clones are marked by the absence of lacZ activity (red). Wg expression is detected by anti-Wg antibody staining (green).
Pan Is Required for wg Self-Refinement.
It was reported that Wg refines its own expression domain at the D/V boundary of third instar wing disks (14). The mechanism of this self-refinement, however, has not been definitely established. The original observation was made with dishevelled mutant cells, which ectopically express Wg if situated close to the D/V boundary. In a complementary study, Dishevelled was shown to interact molecularly with the intracellular domain of Notch (27). Because Notch signaling appears to function as prime inducer of wg transcription (28), these observations raised the possibility that an intersection of the Wg and Notch signaling pathways at the level of Dishevelled caused the attenuation of Notch signaling, and hence an autoregulatory reduction of wg transcription (14, 27). This conclusion implies that the distal components of the Wg pathway, such as Arm and Pan, would be dispensible for wg self-refinement.
Here we sought to test this hypothesis by analyzing the requirement for Pan in wg self-refinement. pan−/− clones in the wing pouch were analyzed for Wg expression. No ectopic Wg expression was detected in clones distant from the D/V boundary. However, pan mutant clones close to this boundary exhibit ectopic Wg expression (Fig. 3D), indicating that Wg refines its expression through a mechanism involving Pan, and hence the distal portion of the Wg signal transduction cascade. Consistent with this result we found that such clones can cause the formation of ectopic wing margin bristles in adjacent wild type cells (Fig. 3C). Because these structures are normally induced by high levels of Wg signaling, we interpret their ectopic presence to be a consequence of ectopic Wg production in the adjacent pan mutant cells. However, because we used an antibody to monitor Wg expression in this study, we cannot rule out an alternative possibility to explain the excess Wg protein observed in pan mutant cells; loss of Pan function presumably leads to the up-regulation of Dfrizzled2, which in turn may cause a stabilization of Wg protein at the cell surface (29).
Evidence against Pan-Independent Outputs of Arm Signaling.
Having established a system to eliminate all pan function in the developing wing cells of Drosophila, we wanted to address the question of whether the Wg pathway is distally branched, or, in other words, whether the regulation of Pan activity is the sole output of Arm signaling. To activate Arm maximally without overexpression, we sought to remove the function of Shaggy (Sgg), an upstream negative regulator of the Wg signaling pathway. If at the same time Pan is also removed, we can ask what, if any, aspect of Arm signaling can bypass Pan.
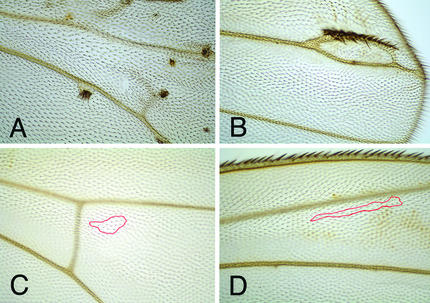
Larvae of the following genotype were generated: y w P[tub-pan] FRT19/y sggD127 f36a FRT19; hsp70-flp/+; pan2/pan2 and their siblings which carry a wild-type copy of the pan gene y w P[tub-pan] FRT19/y sggD127 f36a FRT19; hsp70-flp/+; pan2/Dp[y+]. Clones double mutant for sgg and pan (sgg−/−; pan−/−) and sgg single mutant control clones (sgg−/−; pan+/−) were induced by a heat shock during first or second instar stage of these larvae and analyzed in the adult wing by means of their forked mutant phenotype. As reported previously (30), sgg−/− single mutant clones gave rise to tufts of ectopic bristles on the adult wing (Fig. 4 A and B). This attempt to form ectopic wing margin structures is typical for wing cells subject to a gain of Wg signaling. No ectopic bristles were observed when clones were double mutant for sgg and pan (Fig. 4 C and D). At the wing margin clones with this double mutant genotype exhibit phenotypes similar to those observed for pan single mutant clones (data not shown). These results indicate that the loss of Pan function causes a block in Wg signal transduction and argue against a Pan-independent branch downstream of Sgg. However, from these results we cannot exclude the possibility that Wg can signal independently of Pan during other developmental processes.
Figure 4.
Loss of pan function can revert the phenotype of sgg−/− clones. sgg−/− clones invariably form ectopic bristles (A and B), whereas pan−/−sgg−/− clones never show ectopic bristles (C and D). Clones of pan−/−sgg−/− double mutants are marked by forked bristles within the red outline.
Activator and Repressor Function of Pan in the Embryo.
The strong loss-of-Wg-signaling phenotype observed with pan null mutant cells is in apparent contradiction to the mild segment polarity defect of pan null mutant embryos. This difference could be accounted for by three explanations. First, it could be due to maternal pan product partially perduring to embryonic stages during which Wg signaling defines the cuticular pattern. Second, the loss of pan could lead to a partial derepression of Wg target genes if Pan exerts a repressor function in the absence of Wg signaling. Finally, part of the Wg signal could bypass Pan. The third possibility is unlikely given our findings in the wing disk, although we cannot formally rule out the existence of an embryonic pathway that is able to influence Wg target gene transcription in a Pan-independent manner. To discriminate between the more likely possibilities one and two, we sought to remove any potential maternal component of pan.
Female germ-line clones were generated by using the FLP recombination target (FRT)-FLP technique in combination with an FRT80 (3L) chromosome carrying both a P[ovoD] and a P[tub-pan] transgene. This chromosome was obtained by x-ray-induced recombination in males. We had to use a P[tub-pan] transgene insertion with lower rescuing activity than the one on 2L because, in contrast to the P[ovoD] on chromosome arm 3L, any single P[ovoD] on 2L fails to cause total sterility of females (21). Germ-line clones were induced in rescued pan−/− animals by means of a heat shock-driven hsp70-flp transgene. Embryos derived from resulting mosaic females and fertilized with pan mutant sperm lack both maternal and zygotic pan function.
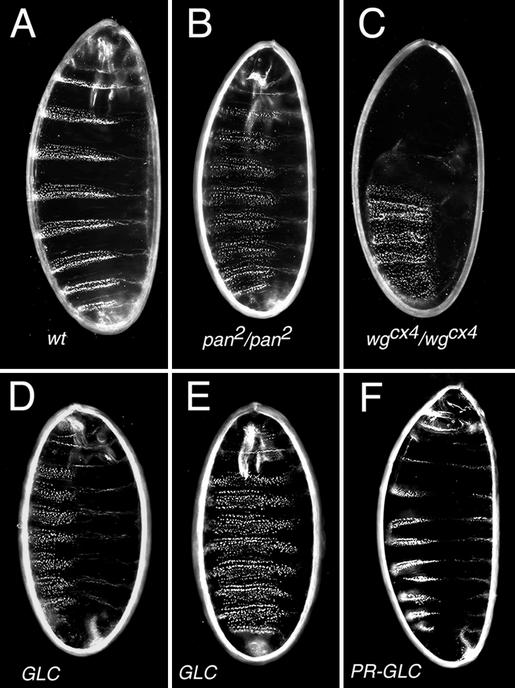
Unfortunately, rescued mosaic females were rare, weak, and subfertile. To obtain 40 fertilized eggs that developed cuticle, 600 such females were needed. To our surprise, pan−/− mutants devoid of maternal contribution did not show phenotypes more severe than those that received maternal pan function (Fig. 5 D–F). To further examine the possibility that removing maternal contribution leads to embryonic death before the formation of cuticle, the ovaries of these females were examined by 4′,6-diamidino-2-phenylindole and phalloidin staining. All of the stages examined during oogenesis appeared normal (data not shown). Hence, the most likely explanation for this weak segment polarity phenotype is a role for pan in repressing Wg-target genes in the absence of Wg activity. The analysis of embryos lacking wg function as well as maternal and zygotic pan function would provide an unequivocal answer. However, for technical reasons (see above) we failed to obtain such embryos. Instead we analyzed cuticles from embryos that only lacked zygotic pan and wg functions, and those resemble pan rather than wg mutants (data not shown). This observation is consistent with previous results from Cavallo et al. (17) and suggests a dual role for Pan during embryogenesis. Embryos devoid of maternal pan+ product can be rescued paternally (pan+ chromosome marked with y+). Although the cuticular phenotype is restored to that of wild-type embryos, not all paternally rescued animals survived to adult stage (Fig. 5F). Together, our results indicate that, in contrast to the wing imaginal disk, the maternal and zygotic pan products function both as transcriptional activator and repressor of Wg targets.
Figure 5.
Embryos maternally and zygotically mutant for pan display segment polarity phenotypes resembling those of zygotic pan mutant embryos. The alternation of naked cuticle and denticle belts in wild-type embryos (A) is replaced by a continuous lawn of denticles in wg−/− mutant embryos (C). The zygotic pan mutant phenotype is milder than that of wg mutants (B). Embryos derived from pan mutant germ-line clones (GLCs; D and E) do not show a more severe phenotype than the zygotic mutants. Paternal contribution can rescue the phenotype of pan−/− GLC-derived animals (PR-GLC; F).
Discussion
Pan is the transcriptional mediator of the Wg signal transduction pathway. It is encoded by a gene located on the fourth chromosome. This location does not permit a functional analysis on the basis of mitotic recombination. Here we used a pan rescue construct to circumvent this problem. However, our transgene is driven by a foreign promoter and thus only partially effective. Rescued animals were not healthy and exhibited reduced fertility. Ideally, a genomic fragment with the endogenous regulatory regions should be used as rescue construct, as was done, for example, in the cubitus interruptus (ci) gene (31) which encodes the transcriptional mediator of the Hedgehog (Hh) signal transduction pathway. However, the pan gene is unusually large (>40 kb) and hence refractory to this approach. Below we discuss our findings and also indicate caveats and limitations of each conclusion.
No Apparent Role of a Pan Repressor in Wing Disk Cells.
The genetic loss of pan function in wing imaginal disks causes a cell-autonomous reduction of Dll and vg expression. Similar observations were made with dsh and arm mutant clones (12). The pouch expression of the Dll and vg genes depends critically on Wg input (13); the apparent residual activity of these genes in pan mutant cells may therefore reflect perdurance, of either pan function or of their own products. It is unlikely that the low levels of Dll and vg products reflect a transcriptional derepression of their genes due to the removal of a Pan repression function, because pan−/− clones outside the normal realms of Dll and vg do not up-regulate these genes. This observation is surprising in the view of the dual roles that have been proposed for Pan in the embryo. Reduction of embryonic pan activity partially suppressed wg and arm mutant phenotypes, and led to the derepression of Wg-responsive genes (17). The apparent absence of a Pan repressor function in the wing primordium also contrasts with the situation in the Hh pathway. Genetic removal of ci results in a derepression of the Hh target gene dpp in wing disks (31, 32), indicating that Ci represses dpp transcription in the absence of Hh signaling. Thus, it appears that the contributions of Pan repressor and activator functions vary in different tissues and/or developmental stages.
Our analysis does not exclude, however, the following two possibilities in which a Pan repressor function may nevertheless play a role in wing development. First, in analogy to the Hh pathway where not all target genes are subject to repression by Ci (e.g., ptc; ref. 31), it is possible that some Wg targets other than Dll and vg are indeed derepressed in pan mutant cells. However, this is not very likely because pan mutant clones in adult wings only display loss-of-wg phenotypes. Second, it is possible that even low levels of pan expression may suffice to provide effective repressor function. As we argued above, wing pouch cells do not survive in complete absence of pan transcription. Thus, at the moment of analysis, pan−/− cells probably still contain at least some Pan protein. Our observation that groucho mutant cells do not up-regulate Dll expression renders this second possibility unlikely.
After completion of this study, Chan and Struhl (33) reported that membrane-tethered Arm can effectively transduce the Wg signal in an arm mutant background, and suggested that Arm may usually function by raising the nuclear ratio of activator to repressor forms of Pan in response to Wg, either by selectively exporting a Pan repressor form from the nucleus or by generating a Pan activator form in the cytoplasm. Although our inability to observe a derepression of Wg targets argues against the existence of a constitutive repressor function of Pan during wing development (see above), our experiments do not rule out a scenario in which Wg signaling regulates the balance of putative activator and repressor forms of Pan, mainly because the genetic removal of the pan gene would concomitantly affect the levels of both forms.
An Absolute Requirement for Pan in Wg/Arm Signaling.
Several bifurcations appear to occur in the Wnt/Wg pathway downstream of Frizzled and Dishevelled, fuelling at least three pathways with signaling activity, such as the canonical Wnt/β-catenin pathway, which activates target genes in the nucleus most likely through TCF/Pan; the planar cell polarity pathway, which involves JNK and cytoskeletal rearrangements; and the Wnt/Ca2+ pathway, which activates phospholipase C and PKC (34). Here we wanted to address the linearity of the canonical Wnt/β-catenin pathway and asked whether all β-catenin/Arm signaling involves output by means of TCF/Pan proteins. Maximal stimulation of Arm signaling was obtained by removal of Sgg, the kinase that normally marks Arm for degradation. On the basis of our observations that sgg pan double-mutant clones behave like pan single-mutant clones, we concluded that Arm signaling cannot bypass Pan. In a similar but more comprehensive study, we have recently addressed the equivalent question for the Hh pathway (35): is there a branch of Hh signaling that bypasses Ci? The answer was unambiguously negative, despite many previous reports of apparent Ci/Gli-independent Hh outputs. For both pathways, the debate about this issue is nurtured in part by the different phenotypes of ci vs. hh and pan vs. wg mutant embryos, respectively, owing to a default repressor function of the nuclear mediators in these pathways.
It is important to point out, however, that, because of technical difficulties, we have not assayed the effect of the sgg pan double-mutant genotype in multiple developmental settings or with sensitive readouts. The sgg pan approach suffers from another deficit compared with the ptc ci analysis; whereas ptc appears to be a dedicated negative component of the Hh pathway, the kinase encoded by sgg plays multiple roles and its genetic removal may affect cells in ways beyond the constitutive activation of the Arm signaling pathway. On the other hand, our main finding in the adult wing is corroborated by our previous study in the embryo, where loss of Pan function can totally block the phenotype caused by constitutively activated Arm (15). Together, these results leave little room for the possibility that Arm can partially bypass Pan to regulate Wg targets in Drosophila development.
Acknowledgments
We thank G. Struhl, P. Simpson, M. Peifer, and N. Perrimon for fly stocks, S. Cohen and S. Carroll for antibodies, and H. E. Varmus for comments on the manuscript. This work was supported by the Swiss National Science Foundation and the Kanton of Zurich.
Abbreviations
- Wg
Wingless
- D/V
dorsoventral
- Arm
Armadillo
- Pan
Pangolin
- Dll
Distalless
- vg
vestigial
- Sgg
Shaggy
- Ci
Cubitus interruptus
- Hh
Hedgehog
- TCF
T cell factor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Bejsovec A, Martinez Arias A. Development (Cambridge, UK) 1991;113:471–485. doi: 10.1242/dev.113.2.471. [DOI] [PubMed] [Google Scholar]
- 2.Nusslein-Volhard C, Wieschaus E. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 3.Bhat K M. Development (Cambridge, UK) 1996;122:2921–2932. doi: 10.1242/dev.122.9.2921. [DOI] [PubMed] [Google Scholar]
- 4.Chu-LaGraff Q, Doe C Q. Science. 1993;261:1594–1597. doi: 10.1126/science.8372355. [DOI] [PubMed] [Google Scholar]
- 5.Patel N H, Schafer B, Goodman C S, Holmgren R. Genes Dev. 1989;3:890–904. doi: 10.1101/gad.3.6.890. [DOI] [PubMed] [Google Scholar]
- 6.Phillips R G, Whittle J R. Development (Cambridge, UK) 1993;118:427–438. doi: 10.1242/dev.118.2.427. [DOI] [PubMed] [Google Scholar]
- 7.Cadigan K M, Nusse R. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 8.Wodarz A, Nusse R. Annu Rev Cell Dev Biol. 1998;14:59–88. doi: 10.1146/annurev.cellbio.14.1.59. [DOI] [PubMed] [Google Scholar]
- 9.Sharma R P, Chopra V L. Dev Biol. 1976;48:461–465. doi: 10.1016/0012-1606(76)90108-1. [DOI] [PubMed] [Google Scholar]
- 10.Williams J A, Paddock S W, Carroll S B. Development (Cambridge, UK) 1993;117:571–584. doi: 10.1242/dev.117.2.571. [DOI] [PubMed] [Google Scholar]
- 11.Couso J P, Bishop S A, Martinez Arias A. Development (Cambridge, UK) 1994;120:621–636. doi: 10.1242/dev.120.3.621. [DOI] [PubMed] [Google Scholar]
- 12.Neumann C J, Cohen S M. Development (Cambridge, UK) 1997;124:871–880. doi: 10.1242/dev.124.4.871. [DOI] [PubMed] [Google Scholar]
- 13.Zecca M, Basler K, Struhl G. Cell. 1996;87:833–844. doi: 10.1016/s0092-8674(00)81991-1. [DOI] [PubMed] [Google Scholar]
- 14.Rulifson E J, Micchelli C A, Axelrod J D, Perrimon N, Blair S S. Nature. 1996;384:72–74. doi: 10.1038/384072a0. [DOI] [PubMed] [Google Scholar]
- 15.Brunner E, Peter O, Schweizer L, Basler K. Nature. 1997;385:829–833. doi: 10.1038/385829a0. [DOI] [PubMed] [Google Scholar]
- 16.van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, et al. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 17.Cavallo R A, Cox R T, Moline M M, Roose J, Polevoy G A, Clevers H, Peifer M, Bejsovec A. Nature. 1998;395:604–668. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- 18.Riese J, Yu X, Munnerlyn A, Eresh S, Hsu S C, Grosschedl R, Bienz M. Cell. 1997;88:777–787. doi: 10.1016/s0092-8674(00)81924-8. [DOI] [PubMed] [Google Scholar]
- 19.Knirr S, Frasch M. Dev Biol. 2001;238:13–26. doi: 10.1006/dbio.2001.0397. [DOI] [PubMed] [Google Scholar]
- 20.Burke R, Basler K. Development (Cambridge, UK) 1996;122:2261–2269. doi: 10.1242/dev.122.7.2261. [DOI] [PubMed] [Google Scholar]
- 21.Chou T B, Perrimon N. Genetics. 1996;144:1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Struhl G, Basler K. Cell. 1993;72:527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]
- 23.Chen C M, Struhl G. Development (Cambridge, UK) 1999;126:5441–5452. doi: 10.1242/dev.126.23.5441. [DOI] [PubMed] [Google Scholar]
- 24.Kramps T, Peter O, Brunner E, Nellen D, Froesch B, Chatterjee S, Murone M, Zullig S, Basler K. Cell. 2002;109:47–60. doi: 10.1016/s0092-8674(02)00679-7. [DOI] [PubMed] [Google Scholar]
- 25.Kim J, Sebring A, Esch J J, Kraus M E, Vorwerk K, Magee J, Carroll S B. Nature. 1996;382:133–138. doi: 10.1038/382133a0. [DOI] [PubMed] [Google Scholar]
- 26.Williams J A, Paddock S W, Vorwerk K, Carroll S B. Nature. 1994;368:299–305. doi: 10.1038/368299a0. [DOI] [PubMed] [Google Scholar]
- 27.Axelrod J D, Matsuno K, Artavanis-Tsakonas S, Perrimon N. Science. 1996;271:1826–1832. doi: 10.1126/science.271.5257.1826. [DOI] [PubMed] [Google Scholar]
- 28.Diaz-Benjumea F J, Cohen S M. Development (Cambridge, UK) 1995;121:4215–4225. doi: 10.1242/dev.121.12.4215. [DOI] [PubMed] [Google Scholar]
- 29.Cadigan K M, Fish M P, Rulifson E J, Nusse R. Cell. 1998;93:767–777. doi: 10.1016/s0092-8674(00)81438-5. [DOI] [PubMed] [Google Scholar]
- 30.Blair S S. Dev Biol. 1992;152:263–278. doi: 10.1016/0012-1606(92)90134-3. [DOI] [PubMed] [Google Scholar]
- 31.Methot N, Basler K. Cell. 1999;96:819–831. doi: 10.1016/s0092-8674(00)80592-9. [DOI] [PubMed] [Google Scholar]
- 32.Dominguez M, Brunner M, Hafen E, Basler K. Science. 1996;272:1621–1625. doi: 10.1126/science.272.5268.1621. [DOI] [PubMed] [Google Scholar]
- 33.Chan S K, Struhl G. Cell. 2002;111:265–280. doi: 10.1016/s0092-8674(02)01037-1. [DOI] [PubMed] [Google Scholar]
- 34.Huelsken J, Birchmeier W. Curr Opin Genet Dev. 2001;11:547–553. doi: 10.1016/s0959-437x(00)00231-8. [DOI] [PubMed] [Google Scholar]
- 35.Methot N, Basler K. Development (Cambridge, UK) 2001;128:733–742. doi: 10.1242/dev.128.5.733. [DOI] [PubMed] [Google Scholar]