Abstract
Fig-pollinating wasps have provided model systems for developing and testing theories of the evolution of mutualism, sex allocation, and precision of adaptation. With few exceptions, previous studies have assumed one species of pollinator wasp per host fig species. Here we report genetic data demonstrating the coexistence of previously undetected cryptic fig wasp species in at least half of the host fig species surveyed. The substantial mitochondrial sequence differences (4.2–6.1%) imply old divergences (≈1.5–5.1 million years ago) among these species. Furthermore, some cryptic species pairs seem to be sister taxa, whereas others clearly are not, indicating both long-term coexistence on shared hosts and the colonization of novel fig species. These findings undermine the prevalent notion of strict one-to-one specificity between cospeciating figs and their pollinators, thereby challenging existing theory concerning the evolution and stability of mutualisms. Moreover, the incorporation of the genetic information significantly improves the fit of the observed sex ratios to predictions of local mate-competition theory, further strengthening support for sex allocation theory and the precision of adaptation.
Keywords: coevolution‖symbiosis‖fig wasp‖ficus‖local mate competition
Figs (Ficus spp. Moraceae) and their pollinating wasps (Agaonidae) constitute perhaps the most tightly integrated pollination mutualism known (1–5). The fig depends on the minute, pollen-bearing female wasps to pollinate the flowers and thereby initiate seed production. The mated female wasps, in turn, depend on the developing fig inflorescence for the production of their offspring, because each wasp offspring develops by consuming the contents of one would-be seed. The cycle begins when the mated female wasps locate a receptive tree and enter the enclosed fig inflorescences (syconia). As these foundress wasps pollinate the flowers, they also oviposit in some of them. Usually the foundresses die inside the syconium, and then both their offspring and the seeds begin to develop. Finally, after maturation, the offspring mate, and then the mated females collect pollen, leave their natal syconium, and fly off to find a receptive tree and begin the cycle anew.
The fig–wasp mutualism is both ancient and diverse, originating ≈90 million years ago (5) with >700 extant species of figs currently recognized (6). Both morphological (7, 8) and recent molecular studies (5, 9–11) broadly support the proposition of cocladogenesis and coadaptation between recognized genera of pollinating wasps and their respective sections of figs. These studies also suggest that finer-scale cospeciation of individual fig and wasp species is widespread. Furthermore, major fitness components in both the fig and the wasps are relatively easy to measure and interpret (4, 12–14). Combined, these attributes of figs and wasps provide a model system for both focal and comparative studies of the coevolution of costs and benefits involved in a mutualism (4, 12–15). Moreover, fig-pollinating wasps have been exploited extensively to both develop and test theories of sex allocation under conditions of local mate competition (LMC) (16–23). Here this theory predicts that, both within and among wasp species, as the number of foundresses that contribute to shared broods within syconia increases, the proportion of males (brood sex ratio) should increase from the extreme female bias expected with only one foundress. These studies have been generalized to investigate precision of adaptation and the situations promoting adaptive behavioral plasticity (20–23).
With few exceptions (2, 7, 8, 24–26), these previous studies have either suggested or assumed one species of pollinator wasp per host fig species. The degree to which this key assumption of host specificity is violated has profound implications for the understanding of fig-pollinator wasp interactions in particular as well as studies of adaptive sex allocation and of the coevolution of mutualisms in general. In this study we use recently developed microsatellite markers (27, 28) in combination with mitochondrial sequence analyses to show that the assumption of one species of pollinator wasp per host fig species is routinely violated. We discuss the implications of these findings with respect to our understanding of the fig–wasp mutualism. In addition we document their effects on the fit between observed sex ratios and the values predicted from LMC theory.
Materials and Methods
Collection and Microsatellite Analyses.
Fig wasps were collected in the vicinity of the Panama Canal, Republic of Panama between January 1997 and December 1999. Three sets of samples were collected. First, for each of eight host fig species, wasps collected from 10–65 different syconia (distinct familial lineages) were characterized with recently developed microsatellite loci (27, 28). These analyses revealed previously undetected cryptic species (see below). Subsequently, the mitochondrial cytochrome oxidase subunit I (COI) gene was sequenced from one to eight individuals per species in this sample to confirm the species status. Further, these data were combined with sequences from earlier studies for phylogenetic analyses (5). Where undescribed cryptic wasp species were confirmed, we used the name of the described species (29) in association with that host tree followed by sp. A, sp. B, etc. Second, 121 fruits (46 with multiple foundresses) of Ficus obtusifolia from four crops were collected for sex-ratio analyses and genotyping of brood to quantify actual contributions by each foundress to multifoundress fruits. Levels of inbreeding were estimated from genotypes of female wasps from 453 fruits (22, 23, 27, 28). Third, single-foundress fruits from two subsequent crops on the same individual F. obtusifolia tree were collected for estimating variables that characterize the fitness outcomes for each partner of the fig–wasp interaction. Specifically, in fruits with only single foundresses, the wasp offspring were reared, sexed, and counted. The seeds were counted from the entire fruit, and undeveloped flowers from a section of approximately one quarter of each fruit. We then compared the outcomes of the interactions of each of the coexisting pollinator species and the host tree.
Mitochondrial Sequence Analyses.
Sequences from the 3′ end of the COI gene (816 bp) were obtained by using standard molecular methods and insect primers. Phylogenetic analyses were conducted with PAUP* 4.0b1 (30). Maximum-likelihood methods were used to reconstruct the phylogenies. The general reversible model with rate heterogeneity (REV Γ) was used, and the parameters of the model were estimated from the data. The tree topologies were estimated by using a heuristic algorithm with branch swapping (tree bisection–reconnection). Time divergences were estimated by using published rates for other insects that were also estimated by using COI sequences and independent calibration points (31–34). Those rates have been estimated to be 1.2% per million years (31), 1.5% per million years (32), 2.3% per million years (33), and 2.2–2.8% per million years (34). We present the range of the suggested dates of divergence based on the two extreme calibrations (1.2–2.8% per million years) and using net divergences (35). That partially corrects for the potential bias generated by not taking into account the sequence divergence already present in the ancestral population at the time of its divergence into evolutionarily distinct lineages (35).
The phylogenetic relationships among cryptic species were tested statistically by using one-tailed Kishino–Hasegawa tests (36, 37). The alternative topologies were defined by forcing haplotypes of each cryptic pair of species associated to the same fig species to be monophyletic or paraphyletic. The alternative trees then were reconstructed by performing a heuristic search constrained on the unresolved topology and forcing the monophyly or paraphyly of each cryptic pair of species. Those tests were conducted with a larger data set that included additional sequence from the 3′ end (916 bp) of COI and the complete (672 bp) cytochrome oxidase subunit II (COII) gene from 22 individual wasps representing 15 species of Pegoscapus (including all cryptic species in this genus described here) and 3 species of Tetrapus. P values in the text refer to results of the tests of monophyly by using the COI–COII data set except for the two haplotype groups of Tetrapus americanus, which were only sampled for COI.
Sex Ratio Analyses.
Rigorous testing of LMC theory with haplodiploid organisms such as fig-pollinating wasps requires knowing the number of foundresses that contribute to a focal brood and estimating the average level of inbreeding in the study population. Previous tests have used the dead bodies of female wasps in focal syconia to estimate foundress number and a weighted average across many syconia to estimate inbreeding levels. These estimates involve a series of assumptions (refs. 18, 19, 22, and 23 and see below). The use of the microsatellites allows precise identification of the parentage of offspring in focal broods and a direct estimate of the level of inbreeding. Here the number of foundresses that actually contributed to broods was estimated by genotyping all sons (maximum 50) and 10–50 daughters from each of 46 fruits of F. obtusifolia that had contained more than one dead foundress body. The inbreeding coefficient (Fis) used to calculate the expected sex ratio (18) is estimated directly from heterozygosities observed in the microsatellites (27, 28). Using the estimated Fis, we generated the expected sex ratios for broods with more than one foundress. In the case of single-foundress broods, we used the average sex ratio of 75 fruits with only a single foundress as the expected value. Then we calculated the fit between predicted and observed brood sex ratios using two different methods for assigning foundress numbers to the broods. First, as in previous studies, we used the number of bodies found within the syconia. Second, we used the genetic evidence. Finally, we compared the two sets of calculations.
Results and Discussion
Cryptic Pollinating Wasp Species and Implications for the Mutualism.
In four of the eight host fig species surveyed, genetic data revealed distinct, cryptic species within what were thought to be single wasp species (29). Specifically, a distinct subset of the wasps that were associated with each host fig species does not share alleles in 77–89% of the microsatellite loci sampled. At these loci, either the length ranges of the alleles do not overlap between the distinct cryptic wasp species or the locus only amplifies in one of the species (Table 1) (observed ranges of allele lengths for the microsatellite loci and sample sizes for each cryptic species are available at the MEN web site, http://snook.bio.indiana.edu/MENotes/easy_search.html). Therefore, these loci define distinct genetic groupings of wasps. Furthermore, in all cases studied the distinct microsatellite groupings correspond to distinct monophyletic groupings of mitochondrial COI haplotypes (Fig. 1), therefore confirming the status of these species. Moreover, the genetic distances between the COI genes characterizing different cryptic species pairs are large (4.2–6.6%; Table 1), suggesting that these species are old and have diverged from each other at times ranging between 1.5 and 5.1 million years ago (5, 31–35). In one species pair, we observed a low frequency of F1 hybrids between two cryptic species (Table 1) but no evidence of back crosses or genetic introgression. The complete lack of introgression suggests that hybrids have negligible fitness.
Table 1.
Summary of the microsatellite and sequence data used to distinguish cryptic species associated with nine Panamanian species of host fig
| Ficus species | Pollinator | No. of broods (crops) | Diagnostic loci (total) | COI sequence divergence, average/net | No. of fixed differences (amino acid) |
|---|---|---|---|---|---|
| F. obtusifolia | P. hoffmeyeri sp. A | 338 (14) | 10 (13) | 4.34/4.20 | 32 (1) |
| P. hoffmeyeri sp. B | 115 (12) | ||||
| P. hoffmeyeri A × B | 4 (4) | ||||
| F. popenoei | P. gemellus sp. A | 28 (4) | 7 (9) | 6.62/6.14 | 43 (7) |
| P. gemellus sp. B | 228 (8) | ||||
| F. bullenei | P. gemellus sp. A | 4 (1) | 8 (9) | 4.70/4.64 | 29 (0) |
| P. gemellus sp. C | 6 (1) | ||||
| F. perforata | P. insularis sp. A | 12 (3) | 10 (11) | 6.11/5.65 | 43 (3) |
| P. insularis sp. B | 13 (2) | ||||
| F. citrifolia | P. tonduzi | 14 (2) | 0 (5) | — | — |
| F. nymphaefolia | P. piceipes | 10 (2) | 0 (8) | — | — |
| F. near trigonata | P. lopesi | 10 (2) | 0 (3) | — | — |
| F. pertusa | P. silvestrii | 10 (1) | 0 (2) | — | — |
| F. maxima | T. americanus | — | — | 10.76/9.25 | 61 (8) |
Female wasps from each of n broods (separate fruits) were sampled from different crops (in parentheses) and identified to species by using the diagnostic loci available (see Results and Discussion). Average sequence divergences between cryptic Pegoscapus species are given as the percentage of differences at all COI sites (816 bp) from one to eight individuals sequenced per species. Divergence among the cryptic Tetrapus pollinators is given from a COI sample of 730 bp. The number of fixed nucleotide differences between cryptic species observed in the COI region is shown (and the number leading to an amino acid change). P. hoffmeyeri A × B hybrids all showed one allele from each of the parental species for each of the analyzed loci.
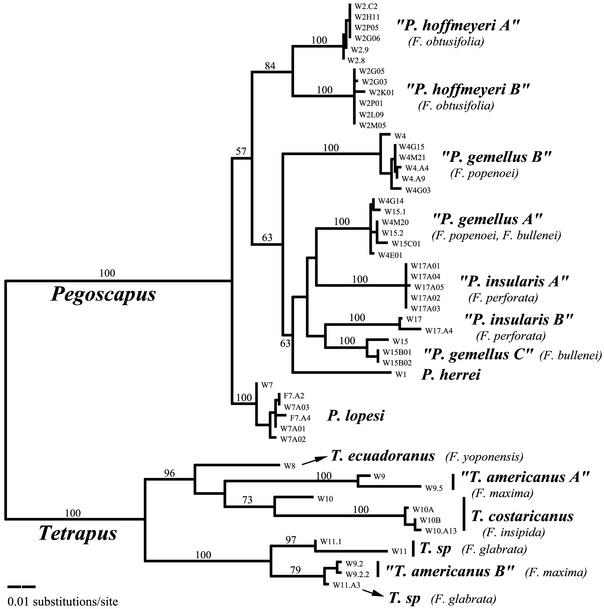
Figure 1.
The maximum-likelihood tree [−ln(L) = 4044.21] of 53 COI haplotypes from Pegoscapus spp. pollinators of six species of neotropical strangler figs and Tetrapus spp. pollinators of four species of free-standing figs. Host names of the cryptic species are shown in parentheses. The numbers above branches are bootstrap values (>50%, 500 replications) for the corresponding nodes of the neighbor-joining tree. In some cases cryptic species pollinating the same host are sister taxa (F. obtusifolia), in other cases they are not (F. popenoei and F. maxima), and in other cases the relationships are not resolvable with the available data (see Results and Discussion).
Two lines of evidence from the present study suggest that the existence of cryptic pollinator wasp species is likely to be a pervasive pattern across fig-wasp taxa worldwide. First, we have both microsatellite and mtDNA sequence data from only a relatively small number of individual wasps in the four cases where cryptic species were not detected (Table 1). Thus, our estimate of the prevalence of cryptic species in this genus (50%) is likely to be conservative. Second, we have mitochondrial evidence of additional cryptic species in T. americanus, the pollinator of one species of Panamanian free-standing fig (Ficus maxima, sect. Pharmacosycea Pharmacosycea) (Fig. 1). The genus Tetrapus represents the basal genus for all other genera of fig-pollinating wasps and diverged from them ≈80 million years ago (5). Therefore, the existence of cryptic species in both the most basal and one of the more derived genera suggests that multiple species of pollinators per fig is a recurring theme throughout the long history of the fig–wasp mutualism. In addition, previous studies have reported multiple morphologically distinct wasp species associated with single host fig species belonging to other, phylogenetically interposed genera (2, 8, 11, 24–26, 38).
Phylogenetic analyses demonstrate that although some cryptic species pairs sharing the same host seem to be sister taxa, others almost certainly are not (Fig. 1). This observation is relevant to the more general question of cospeciation, because the presence of nonsister cryptic species on the same host reveals that wasp species must have shifted from one host fig to another. For example, in Ficus popenoei, the two wasp species are not sister taxa (monophyly rejected, P < 0.02), indicating that at least one of these species has colonized this host after the mutualism with the other had already been established. Similarly, the two cryptic species pollinating F. maxima are not sister taxa (P < 0.0001), indicating a colonization of this host as well. Conversely, in the host fig, F. obtusifolia, the two wasp species seem to be sister taxa (paraphyly rejected, P < 0.01). In Ficus perforata and Ficus bullenei, the relationships among the associated cryptic wasp species are ambiguous (neither paraphyly nor monophyly can be rejected). Finally, we found that one group of genetically indistinguishable pollinators (P. gemellus sp. A) is shared between the hosts F. popenoei and F. bullenei (29) (Table 1 and Fig. 1).
Together, these findings hold at least three potentially important implications for our understanding of fig–wasp mutualisms. First, our findings undermine the idea of strict one-to-one specificity. Existing cases in which multiple wasp species per host are reported frequently involve different wasp species that associate with a host fig in different parts of its range (24–26). In contrast to these allopatric cases, there are few studies demonstrating that multiple pollinators routinely and successfully associate with a given host in sympatry (7, 24–26, 38, 39). However, even in these studies in which different wasp species have been found to sympatrically inhabit the same host, at the vast majority of sites the different wasp species do not overlap (24, 25). Thus, although cases to the contrary are known, one-to-one specificity is thought generally to characterize the relationship between figs and wasps (2, 7, 24–26, 40). Our data combined with these other reports (2, 7, 11, 24–26, 38–40) strongly suggest that this overly simple view requires revision.
Second, our findings undermine the idea of tight cospeciation and suggest that neither the number of wasp species associated with a particular host fig nor the evolutionary relationships among them are fixed. The case of genetically indistinguishable pollinators associated with F. popenoei and F. bullenei likely presents an early stage of a wasp species colonizing a novel host. Such colonization has the potential to lead to hybridization and genetic introgression between the host species (5). Thus colonization events such as the one we have detected here (also see refs. 9 and 11) can potentially provide a mechanism for apparent cases of incongruence that have been observed between fig and wasp phylogenies (2, 5, 10, 11, 41).
Third, some theories suggest that the presence of multiple symbionts produces an unstable situation for a mutualism, and that colonization events provide a likely scenario for the transition from mutualism to parasitism (4, 42–47). For example, in several cases reported in the moths that either pollinate or parasitize Yucca flowers, a colonization of a novel host has been followed by a transition from mutualism to parasitism (46, 48). Similarly, the African fig Ficus sycomorus (2, 4, 8, 49) has two associated wasp species, Ceratosolen arabicus and Ceratosolen galili, yet the latter has ceased pollinating and is effectively a parasite. C. galili is only related distantly to the pollinating species C. arabicus, suggesting an analogous evolutionary pattern (2, 4, 5, 8–11). However, in all Panamanian cases, both of the cryptic species successfully reproduce, pollinate, and induce seed production in the host fig. In the most thoroughly sampled host species, any differences between the two cryptic species in the outcome of their interaction with the host seem to be minimal (Table 2), and there is little evidence for a shift from mutualism to parasitism.
Table 2.
Comparisons of the interactions of different cryptic wasp species with F. obtusifolia
| n | Brood size, mean ± SD | Female wasps, mean ± SD | Seeds, mean ± SD | Estimated flowers, mean ± SD | Proportion of flowers developed, median | Proportion seeds, median | |
|---|---|---|---|---|---|---|---|
| 1998 crop | |||||||
| P. hoffmeyeri A | 32 | 200.5 ± 39.1 | 183.7 ± 36.9 | 183.1 ± 36.1 | 470.1 ± 64.8 | 0.88 | 0.47 |
| P. hoffmeyeri B | 9 | 223.3 ± 52.4 | 208.4 ± 51.2 | 210.2 ± 37.0 | 502.8 ± 92.8 | 0.92 | 0.47 |
| Test value | t = −1.435 | t = −1.630 | t = −1.981 | t = −1.211 | U = 78.50 | U = 137.00 | |
| P | 0.159 | 0.111 | 0.055 | 0.233 | 0.039 | 0.826 | |
| 1999 crop | |||||||
| P. hoffmeyeri A | 15 | 163.8 ± 26.3 | 152.9 ± 24.7 | 171.3 ± 40.3 | 364.7 ± 55.8 | 0.94 | 0.52 |
| P. hoffmeyeri B | 37 | 158.0 ± 33.9 | 146.4 ± 33.0 | 193.9 ± 27.6 | 362.2 ± 69.7 | 0.94 | 0.56 |
| Test value | t = 0.590 | t = 0.687 | t = −2.335 | t = 0.123 | U = 272.50 | U = 180.00 | |
| P | 0.558 | 0.496 | 0.024 | 0.903 | 0.920 | 0.049 |
We collected data on the reproductive success of the pollinators and an individual F. obtusifolia over two successive reproductive events. Only single-foundress fruits were compared. We determined the species of the foundress by genotyping a single daughter wasp per fruit. We then compared the reproductive success of the different wasp species as well as the production of major components of fig reproductive success associated with them (10). Counted and estimated numbers were tested with a t test, and proportions were tested with the Mann–Whitney U test. After using the Dunn–Sidak sequential Bonferroni corrections, no test shows a significant difference.
Finally, our discovery of cryptic species of fig-pollinating wasps mirrors similar findings in several other mutualisms where molecular techniques have revealed a much greater cryptic diversity of the participants than suspected previously (47, 48, 50, 51). Methodologically, the demonstration of the widespread existence of cryptic diversity in several very different mutualistic systems suggests that phylogenetic studies (in figs or other host–symbiont systems) that use genetic information from only one or a few individuals per species to represent the evolutionary history of one side of a mutualism or parasitism (e.g., refs. 9–11 and 52) are likely to grossly underestimate actual ecological and coevolutionary complexity. Conceptually, this underappreciated diversity challenges much of the existing theory concerning the evolution and stability of mutualisms (4, 42–47, 53). Existing theory has often been formulated on the basis of overly simplistic representations of real systems. Future progress in the study of mutualisms almost certainly will depend on the development and testing of models that more closely reflect the actual natural histories of these remarkable systems.
Cryptic Pollinating Wasp Species and Implications for Sex-Allocation Studies.
The existence of cryptic species also has profound implications for studies of sex allocation and precision of adaptation in fig-pollinating wasps. Previous tests of LMC theory have used the number of dead foundresses present in a fig fruit to estimate the number of mothers contributing eggs to the broods. Specifically, it is usually assumed that each foundress contributes offspring equally and at similar sex ratios to common broods (17–23, 54, 55). However, genotyping broods from 46 F. obtusifolia fruits with multiple dead foundresses showed that neither of these assumptions are met. In fact, in only 10 (22%) of these broods did more than one foundress actually contribute offspring, and half of these 10 broods consisted of two different cryptic species, which means that both the inbreeding and LMC levels are higher than thought previously in both coexisting species. By using the numbers of foundresses that actually contributed to broods rather than the number of dead foundresses, we significantly improved the overall fit of observed sex ratio to theory (for the two species combined: t = 3.94, df = 45, P < 0.001; Fig. 2). The improvement of fit was similar in both cryptic species when they were considered individually.
Figure 2.
Brood sex ratio vs. the number of potentially contributing foundresses (number of dead foundress bodies) (A) and the actual number of genetically distinguishable broods (B). Genetic data significantly improve the fit of observed sex ratios with predictions of LMC theory (t = 3.94, df = 45, P < 0.001). Predicted sex ratio (line) for multifoundress broods uses an inbreeding coefficient of 0.87 estimated from five microsatellite loci. The mean sex ratio of single-foundress broods was 0.083 (square). Triangles represent two data points outside the range of the graph. The numbers of foundresses that actually contributed to broods was estimated by genotyping all sons (maximum 50) and 10–50 daughters from each of 46 fruits that had contained more than one dead foundress body from four crops of F. obtusifolia.
Furthermore, in agreement with theoretical predictions (21–23), the sex ratios of single-foundress broods in the rarer species (sp. B; ≈20% of all wasps) were significantly more female-biased than the more common species (sp. A; Mann–Whitney U = 81.0, n = 41, P < 0.05). This result is relevant to the testing of theory in two important ways. First, this finding provides an additional, unanticipated line of support for the general predictions of LMC theory. Second, the present study suggests that the sex-ratio responses of distinct wasp species were lumped in the past, which likely includes studies of the variance in brood sex ratios of these wasp populations (21–23). Here, theory predicts very precise sex ratios and that the variance in single-foundress broods should be less than binomial. Because the mean brood sex ratios of these species are different, such lumping will tend to falsely inflate the estimates of variance. The recognition of cryptic species is therefore likely to resolve the previous reports that variances of brood sex ratio in these species were higher than expected. Thus, the recognition of cryptic species seems to improve the fit of both mean and variance in fig-wasp sex-ratio responses to theoretical predictions.
Finally, Hamilton (56) suggested that sex allocation in general and LMC in particular was “the section of evolutionary theory that best proves the power and accuracy of the Neodarwinian paradigm as a whole.” The improved fit of empirical data to the predicted values of the model as revealed by the microsatellite markers suggests that this part of evolutionary theory has even more power and accuracy than thought previously.
Acknowledgments
We gratefully acknowledge useful suggestions and comments from Egbert Leigh, Jr., Sunshine Van Bael, Ian Sanders, Michel Chapuisat, Emmanuelle Jousselin, Finn Kjellberg, Jannie Nielsen, Hans Christian Petersen, and Jeanette Bot. Adalberto Gomez and Maritza Lopez helped with field work. This work was supported by the Smithsonian Tropical Research Institution and the Swiss Science Foundation.
Abbreviations
- COI
cytochrome oxidase subunit I
- LMC
local mate competition
Footnotes
References
- 1.Corner E J H. Wayside Trees of Malaya. Singapore: Government Printer Office; 1952. [Google Scholar]
- 2.Wiebes J T. Annu Rev Ecol Syst. 1979;10:1–12. [Google Scholar]
- 3.Janzen D H. Annu Rev Ecol Syst. 1979;10:13–51. [Google Scholar]
- 4.Herre E A. In: Levels of Selection in Evolution. Keller L, editor. Princeton: Princeton Univ. Press; 1999. pp. 209–237. [Google Scholar]
- 5.Machado C A, Jousselin E, Kjellberg F, Compton S G, Herre E A. Proc R Soc London Ser B. 2001;268:685–694. doi: 10.1098/rspb.2000.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berg C C. Experientia. 1989;45:605–611. [Google Scholar]
- 7.Ramirez W. Ann M Bot Gard. 1974;61:770–780. [Google Scholar]
- 8.Berg C C, Wiebes J T. African Fig Trees and Fig Wasps. Amsterdam: North–Holland; 1992. [Google Scholar]
- 9.Herre E A, Machado C A, Bermingham E, Nason J D, Windsor D M, McCafferty S S, Van Houten W, Bachman K. J Biogeogr. 1996;23:521–530. [Google Scholar]
- 10.Weiblen G D. Syst Biol. 2001;50:243–267. [PubMed] [Google Scholar]
- 11.Kerdelhue C, Le Clainche I, Rasplus J Y. Mol Phylogenet Evol. 1999;11:401–414. doi: 10.1006/mpev.1998.0590. [DOI] [PubMed] [Google Scholar]
- 12.Herre E A. Experientia. 1989;45:637–647. [Google Scholar]
- 13.Anstett M C, Bronstein J L, Hossaert-McKey M. J Evol Biol. 1996;9:417–428. [Google Scholar]
- 14.Herre E A, West S A. Proc R Soc London Ser B. 1997;264:1501–1507. [Google Scholar]
- 15.Bronstein J L, McKey D. Experientia. 1989;45:601–604. [Google Scholar]
- 16.Hamilton W D. Science. 1967;156:477–488. doi: 10.1126/science.156.3774.477. [DOI] [PubMed] [Google Scholar]
- 17.Hamilton W D. In: Sexual Selection and Reproductive Competition in Insects. Blum M S, Blum N A, editors. New York: Academic; 1979. pp. 167–220. [Google Scholar]
- 18.Frank S A. Evolution (Lawrence, Kans) 1985;39:949–964. doi: 10.1111/j.1558-5646.1985.tb00440.x. [DOI] [PubMed] [Google Scholar]
- 19.Herre E A. Science. 1985;228:896–898. doi: 10.1126/science.228.4701.896. [DOI] [PubMed] [Google Scholar]
- 20.Herre E A. Nature. 1987;329:627–629. [Google Scholar]
- 21.West S A, Herre E A. Evolution (Lawrence, Kans) 1998;52:475–485. doi: 10.1111/j.1558-5646.1998.tb01647.x. [DOI] [PubMed] [Google Scholar]
- 22.Herre A E, West S A, Cook J M, Compton S G, Kjellberg F. In: Social Competition and Cooperation in Insects and Arachnids I: The Evolution of Mating Systems. Choe J C, Crespi B, editors. Princeton: Princeton Univ. Press; 1997. pp. 226–239. [Google Scholar]
- 23.Herre E A, Machado C A, West S A. In: Adaptationism and Optimality. Orzack S H, Sober E, editors. New York: Cambridge Univ. Press; 2001. pp. 191–218. [Google Scholar]
- 24.Michaloud G, Michaloud-Pelletier S, Wiebes J T, Berg C C. Proc K Ned Akad Wet C. 1986;88:93–119. [Google Scholar]
- 25.Michaloud G, Carriere S, Kobbi M. J Biogeogr. 1996;23:513–520. [Google Scholar]
- 26.Rasplus J-Y. In: The Biodiversity of African Plants. van der Maesen L J G, van der Burgt X M, van Medenbach de Rooy J M, editors. Dordrecht, The Netherlands: Kluwer; 1994. pp. 639–649. [Google Scholar]
- 27.Molbo D, Krieger M J B, Herre E A, Keller L. Mol Ecol Notes. 2002;2:440–442. [Google Scholar]
- 28.Molbo D. Ph.D. thesis. Lausanne, Switzerland: University of Lausanne; 2002. [Google Scholar]
- 29.Wiebes J T. Proc K Ned Akad Wet. 1995;98:167–183. [Google Scholar]
- 30.Swofford D L. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 31.Caccone A, Sbordoni V. Evolution (Lawrence, Kans) 2001;55:122–130. doi: 10.1111/j.0014-3820.2001.tb01278.x. [DOI] [PubMed] [Google Scholar]
- 32.Farrell B D. Mol Phylogenet Evol. 2001;18:467–478. doi: 10.1006/mpev.2000.0888. [DOI] [PubMed] [Google Scholar]
- 33.Brower A V. Proc Natl Acad Sci USA. 1994;91:6491–6495. doi: 10.1073/pnas.91.14.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buckley T R, Simon C, Chambers G K. Evolution (Lawrence, Kans) 2001;55:1395–1407. doi: 10.1111/j.0014-3820.2001.tb00661.x. [DOI] [PubMed] [Google Scholar]
- 35.Nei M, Li W-H. Proc Natl Acad Sci USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kishino H, Hasegawa M. J Mol Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 37.Goldman N, Anderson J P, Rodrigo A G. Syst Biol. 2000;49:652–670. doi: 10.1080/106351500750049752. [DOI] [PubMed] [Google Scholar]
- 38.Lopez-Vaamonde C, Dixon D J, Cook J M, Rasplus J-Y. Zool J Linn Soc. 2002;136:637–683. [Google Scholar]
- 39.Compton S G. S Afr J Sci. 1990;86:39–40. [Google Scholar]
- 40.Weiblen G D. Annu Rev Entomol. 2002;47:299–330. doi: 10.1146/annurev.ento.47.091201.145213. [DOI] [PubMed] [Google Scholar]
- 41.Jousselin E, Rasplus J-Y, Kjellberg F. Evolution. Kans.: Lawrence; 2003. , in press. [DOI] [PubMed] [Google Scholar]
- 42.Bull J J, Rice W R. J Theor Biol. 1991;149:63–74. doi: 10.1016/s0022-5193(05)80072-4. [DOI] [PubMed] [Google Scholar]
- 43.Leigh E G., Jr . Tropical Forest Ecology. New York: Oxford Univ. Press; 1999. pp. 211–226. [Google Scholar]
- 44.Maynard Smith J, Szathmary E. The Major Transitions in Evolution. Oxford: Freeman; 1995. [Google Scholar]
- 45.Yu D W. Biol J Linn Soc Lond. 2001;72:529–546. [Google Scholar]
- 46.Pellmyr O, Leebensmack J, Huth C J. Nature. 1996;380:155–156. doi: 10.1038/380155a0. [DOI] [PubMed] [Google Scholar]
- 47.Herre E A, Knowlton N, Mueller U G, Rehner S A. Trends Ecol Evol. 1999;14:49–53. doi: 10.1016/s0169-5347(98)01529-8. [DOI] [PubMed] [Google Scholar]
- 48.Pellmyr O. Ann M Bot Gard. 2003;90:35–55. [Google Scholar]
- 49.Compton S, Holton K C, Rashbrook V K, van Noort S, Vincent S L, Ware A B. Biotropica. 1991;23:188–194. [Google Scholar]
- 50.Rowan R, Knowlton N, Baker A, Jara J. Nature. 1997;388:265–269. doi: 10.1038/40843. [DOI] [PubMed] [Google Scholar]
- 51.Husband R, Herre E A, Turner S L, Gallery R, Young J P W. Mol Ecol. 2002;11:2669–2678. doi: 10.1046/j.1365-294x.2002.01647.x. [DOI] [PubMed] [Google Scholar]
- 52.Weiblen G D, Bush G L. Mol Ecol. 2002;11:1573–1578. doi: 10.1046/j.1365-294x.2002.01529.x. [DOI] [PubMed] [Google Scholar]
- 53.Axelrod R, Hamilton W D. Science. 1981;211:1390–1396. doi: 10.1126/science.7466396. [DOI] [PubMed] [Google Scholar]
- 54.Kathuria P, Greeff J M, Compton S G, Ganeshaiah K N. Oikos. 1999;87:520–530. [Google Scholar]
- 55.Kinoshita M, Kasuya E, Yahara T. Oikos. 2002;96:31–35. [Google Scholar]
- 56.Hamilton W D. Narrow Roads of Geneland I: Evolution of Social Behaviour. Oxford: Freeman; 1996. [Google Scholar]