Abstract
The lemurs of Madagascar provide an excellent model for exploring evolutionary diversification. This study investigates genetic divergence among most extant lemur taxa in relation to potential geographical boundaries to gene flow. For this purpose, ≈2,400 bp of mitochondrial DNA (part of the COIII gene; ND3, ND4L, and ND4 genes; and five tRNAs) were sequenced in a total of 131 lemurs from 5 families, 12 genera, 25 species, and 18 subspecies to reconstruct phylogenetic relationships among them. The comprehensive range of taxa makes this a particularly suitable molecular data set to examine lemur evolution. Those data clearly reveal that the Betsiboka River acts as an isolating barrier between populations of lemurs in north-western Madagascar. The Tsiribihina River similarly serves as a barrier to gene flow between northern and southern populations of lemurs in central western Madagascar, whereas the Mahavavy River does not seem to lead to genetic isolation of lemur populations. Several discrepancies among molecular data, current taxonomy, and geographic distribution along the western coast emerged. Examination of geographical distribution of the taxa concerned in comparison with distribution boundaries of other lemur taxa in that region yielded explanations for these inconsistencies. Eulemur fulvus and Eulemur mongoz are the only lemur taxa that also occur outside Madagascar, on the Comoro Islands. Genetic data show no significant differentiation between Malagasy and Comorian populations of these species, supporting the interpretation that both were introduced only recently to the Comoro Islands.
Evolution of the lemurs (infraorder Lemuriformes) is a spectacular example of adaptive radiation among primates, providing an excellent model for studies of evolutionary diversification (1, 2). The island of Madagascar furnished the natural experimental context for this exemplary radiation, the diversity of which equals that of the anthropoid primates from Asia, Africa, or South America. Madagascar is the world's fourth-largest island and has a diverse geology, climate, and vegetation. Its flora and fauna are highly endemic. Madagascar can be divided into eight major zones of species distribution, each with distinctive climatic and vegetational characteristics and/or delimited by physical barriers (1, 2). Those climatic, vegetational, and physical factors are important in understanding the phylogeography of the Malagasy lemurs.
The infraorder Lemuriformes is now commonly allocated to the primate suborder Strepsirrhini, along with the Loriformes (3). With five extant endemic lemuriform families including 14 genera, at least 32 species, and 50 distinct taxa, Madagascar's diversity ranks third highest in the world for primates (4). In addition to the extant lemurs, at least 17 species of recently extinct lemurs have been found on Madagascar (5), documenting a significantly larger lemur fauna just a few hundred years ago. Locomotor adaptations, differences in body size, and feeding adaptations are all highly variable features among lemurs, highlighting the impressive diversity of this group. Apart from the lemurs, Madagascar's mammalian fauna is relatively impoverished (6), and the number of bird species is also low. In contrast to birds and mammals, however, the reptilian and amphibian faunas (with the notable exception of salamanders) are rich compared with those in mainland Africa.
Classification within the Lemuriformes remains highly controversial. Several different taxonomic schemes have been proposed (3, 4, 6–11). At present, a tentative consensus accepts four genera (Eulemur, Hapalemur, Lemur, and Varecia) in the family Lemuridae, which includes at least 10 species. The Cheirogaleidae are currently classified into five genera (Allocebus, Cheirogaleus, Microcebus, Mirza, and Phaner), containing at least 13 species. The family Indridae includes at least seven species in three genera (Avahi, Indri, and Propithecus). The family Daubentoniidae contains only one extant lemur species (Daubentonia madagascariensis). Lepilemur is the only genus in the family Lepilemuridae and is currently divided into a maximum of seven species.
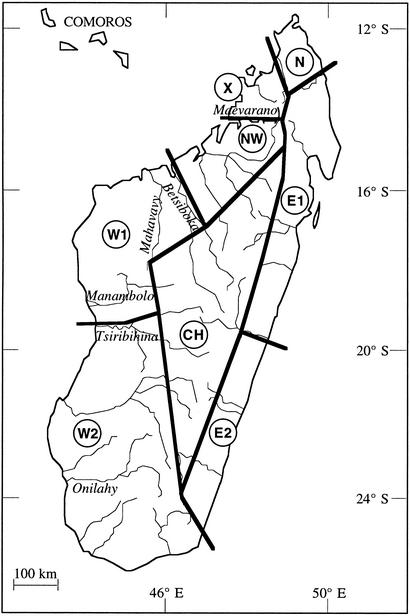
Madagascar is divided into two major ecological zones, a relatively humid eastern region and a dry western region. Each zone provides a wide range of habitats. Analyzing distribution patterns of all species and subspecies for which sufficient data were then available, Martin (1) divided Madagascar into seven biogeographical zones (Fig. 1). Along the western coast, three zones (NW, W1, and W2) were defined, all bounded by the highlands to the east and the Mozambique Channel to the west. NW covers the western coast north of the Betsiboka River, W1 includes the area between the Betsiboka and Tsiribihina rivers, and W2 covers the western coast south of the Tsiribihina. The zone in the north (N) and the two eastern zones (E1 and E2) are also separated from each other by large rivers. In the southeast, the Anosy hill chain possibly isolates zone W2 from zone E2. The central highland area (CH), with its high altitude and relatively low temperatures in the winter, separates the east from the west (1). An additional small zone (X) in the northwest was suggested subsequently because of biological and climatic affinities with zone E1 (2).
Figure 1.
Map of Madagascar showing the eight main areas of lemur distribution according to Martin (1, 2). E1 and E2, east coast zones; W1, W2, and NW, west coast zones; X, zone in the northwest; N, north coast zone; CH, central highland zone.
In this study, a mtDNA sequence data set including as many taxa (genera, species, and subspecies) as possible was generated to reconstruct phylogenetic relationships among lemurs. For several taxa, individuals from two or all three western coast zones were included, yielding information on genetic differentiation among lemur taxa across zoogeographic zones of the western coast. The data set thus allows examination of effects exerted by rivers on gene flow in various lemur taxa.
Methods
This study includes 131 lemurs representing 12 genera, 25 species, and 18 subspecies of all five lemur families. The origins of most samples have been reported (12–17). Two bush babies (Otolemur crassicaudatus and Galago senegalensis) were sequenced to provide an outgroup.
The 2,387-bp segment of mtDNA investigated in this study includes part of the COIII gene, complete sequences for the NADH-dehydrogenase subunits 3, 4L, and 4 (ND3, ND4L, and ND4), as well as the tRNAGly, tRNAArg, tRNAHis, tRNASer, and partial tRNALeu genes. DNA extraction, amplification, and direct sequencing of the PCR product were performed as reported (12). The new mtDNA sequences generated for taxa examined have been deposited in the GenBank database (AF224512–AF224644).
Sequences were aligned by eye and analyzed with PAUP* 4.0b2 (18) by using maximum parsimony and neighbor-joining methods. Kimura two-parameter distances (19) were used for neighbor-joining analyses, and heuristic searches by random addition (1,000 replicates) were used in parsimony analyses. Gaps were considered as a fifth character state in parsimony analyses. Bootstrap analyses (20) of 1,000 replicates were performed to assess relative support of each relationship in the resulting topologies.
Results and Discussion
Phylogeny of the Malagasy Lemurs.
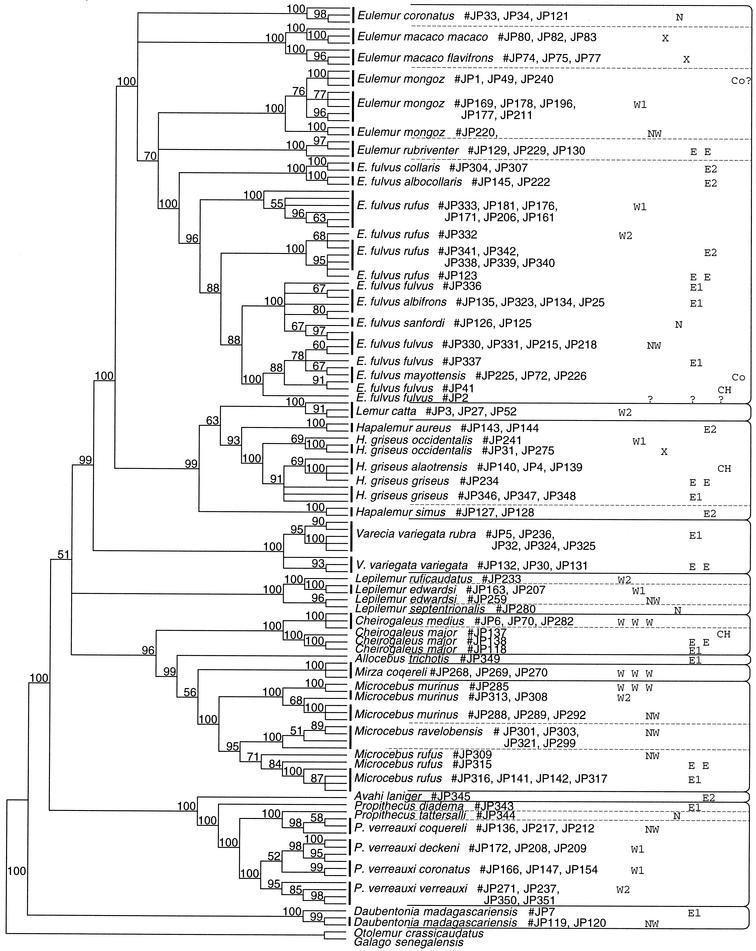
Generally, there is good resolution among genera, species, and subspecies across all taxa examined by maximum parsimony (Fig. 2) or neighbor joining (data not shown). Additionally, there are very high bootstrap supports with respect to branching order of genera, species, or subspecies. Monophyly is well supported for all five lemur families in all analyses. Daubentonia is unambiguously the first genus to diverge among lemurs. However, the mtDNA sequence data failed to yield clear resolution of phylogenetic relationships among the four remaining families: Cheirogaleidae, Indridae, Lemuridae, and Lepilemuridae. These results agree with those from a previous study that used mtDNA sequences (21). Phylogenetic relationships among subspecies, species, or genera within the families Lemuridae, Cheirogaleidae, and Indridae have been discussed in detail elsewhere (12–14, 16, 17). The overview of mtDNA data for all lemur families presented here allows investigation of potential geographic barriers to gene flow for a comprehensive range of lemur taxa.
Figure 2.
Strict consensus tree from maximum parsimony heuristic search with all characters weighted equally. Bootstrap values were obtained in 101 replicates. To the right of each taxon the geographical zone is indicated (see Fig. 1 for abbreviation descriptions; Co, Comoro Islands). Continuous horizontal lines separate genera, and dashed lines separate species.
Evolutionary Rate of Lemurs.
In interpreting the molecular results, we directly compare genetic distances between taxa. To ensure that all lemurs evolve at a similar rate, hence justifying such comparisons, a relative-rate test was conducted (22). We used Eulemur mongoz (Lemuridae), Microcebus murinus (Cheirogaleidae), Propithecus verreauxi coquereli (Indridae), and Lepilemur edwardsi (Lepilemuridae) as representatives for each family, and Daubentonia (Daubentoniidae) was always used as the outgroup. In most cases, the members of the two families tested proved to evolve at a similar rate (P > 0.05). The molecular evolutionary-clock hypothesis was rejected only twice (P < 0.05 for Propithecus–Microcebus and Propithecus–Lepilemur). The genus Propithecus evidently evolved at a slower rate than other lemurs. However, even if we were to increase the genetic distances between Propithecus populations by 30% to compensate for the slower evolutionary rate, all conclusions would remain the same.
Betsiboka River.
The Betsiboka River is considered to be a major physical barrier separating the two geographic zones NW and W1 (Fig. 1). Current taxonomy and distribution indicate that the Betsiboka River forms the boundary between the subspecies Eulemur fulvus fulvus and Eulemur fulvus rufus and between Propithecus verreauxi coronatus and P. v. coquereli (Table 1). However, there has been no previous indication that the Betsiboka has served as a boundary for populations of E. mongoz, Hapalemur griseus, L. edwardsi, M. murinus, or Cheirogaleus medius.
Table 1.
Comparison between the current consensus taxonomy and the taxonomy as indicated by mtDNA sequence data of lemur taxa on opposite sides of three rivers
| River | Current taxonomy | Relative genetic distances, % | mtDNA taxonomy |
|---|---|---|---|
| Betsiboka (NW and W1) | E. mongoz | 0.63–0.72 | 1 species |
| E. f. fulvus–E. f. rufus | 2.33–2.41 | 2 subspecies | |
| P. v. coquereli–P. v. coronatus | 5.25–5.50 | 2 species | |
| L. edwardsi | 11.06–11.23 | 2 species | |
| Mahavavy (in W1) | E. mongoz | 0.34–0.42 | 1 species |
| E. f. rufus | 0.21–0.42 | 1 subspecies | |
| P. v. coronatus–P. v. deckeni | 0.25–0.85 | 1 subspecies | |
| Tsiribihina (W1 and W2) | E. f. rufus | 2.28–2.54 | 2 subspecies |
| P. v. deckeni–P. v. verreauxi | 0.85–1.02 | 1 subspecies | |
| Betsiboka and Mahavavy* | H. g. occidentalis | 1.99–2.03 | 2 subspecies |
| Mahavavy and Tsiribihina* | L. edwardsi–L. ruficaudatus | 6.02–6.19 | 2 species |
| All three rivers* | M. murinus | 2.54–2.62 | 2 subspecies |
Because of incomplete taxon sampling, the genetic divergence spans more than one river.
When absolute genetic distances between populations of various lemur taxa west (Anjamena, zone W1) and east (Ampijoroa, zone NW) of the Betsiboka River are examined, very different levels of genetic divergence can be observed (Table 1).
E. mongoz individuals at Ampijoroa differ by only 0.63–0.72% from their relatives at Anjamena. This finding matches the current taxonomy, which does not distinguish subspecies.
Pairwise distances between E. f. fulvus and E. f. rufus are 2.33–2.41%. This degree of genetic divergence lies in the range for comparisons among other lemur subspecies and thus confirms the current taxonomy.
Genetic distances between P. v. coronatus and P. v. coquereli reach 5.25–5.50%, which exceeds the level of genetic differentiation otherwise seen among lemur subspecies. The molecular data hence suggest that the Betsiboka River serves as a barrier to gene flow and that speciation between those two populations may already be complete.
The greatest genetic distances (11.06–11.23%) across the Betsiboka River are found among L. edwardsi individuals. DNA sequence data clearly support the inference that the Betsiboka River serves as boundary between two different Lepilemur species. This is an unexpected result, because current taxonomy does not distinguish those Lepilemur populations even at the subspecific level.
Classification, notably nomenclature and taxonomy, of Lepilemur has been the subject of much controversy and discussion (2, 10, 23). The three L. edwardsi individuals sequenced for this study consistently fail to resolve into one monophyletic clade. The subclade containing one L. edwardsi individual (JP259, from Ampijoroa, zone NW) and Lepilemur septentrionalis has strong bootstrap support (96–99%), as does the subclade containing the other two L. edwardsi individuals (JP163 and JP207 from Anjamena, zone W1) and Lepilemur ruficaudatus (100%). Branch lengths in the neighbor-joining phylogram indicate that both clades and the two taxa within each clade are all deeply separated. The high degree of divergence among the L. edwardsi individuals clearly lies in the range for comparisons among other species of Lepilemur (6.02–11.65%) and among species of Eulemur, Hapalemur, Microcebus, or Propithecus.
Mahavavy River.
The Mahavavy River was not found to serve as a barrier between major zones (Fig. 1), consistent with the general view that this river is not an isolating boundary between different taxa in current taxonomy. The only current taxonomic exception is provided by the subspecies P. v. coronatus and Propithecus verreauxi deckeni (Table 1).
The data set presented here includes individuals from Anjamena (east of the Mahavavy River) and Anadabomandry (west of the Mahavavy) for E. mongoz, E. f. rufus, and P. verreauxi. Within each taxon, genetic distances between populations of those two localities never exceed 0.85% (Table 1). The molecular data set thus confirms the current taxonomy in failing to distinguish between different subspecies of E. mongoz or E. fulvus west and east of the Mahavavy. Likewise, the mtDNA sequence data, in contrast to external pelage coloration (24), do not support subspecific differentiation among populations of P. verreauxi east and west of the Mahavavy River. P. v. deckeni and P. v. coronatus even fail to form two monophyletic clades (Fig. 2). Therefore, the Mahavavy River indeed represents no major physical barrier for lemurs, as predicted in Martin's model (1).
Tsiribihina River.
The Tsiribihina River is thought to isolate zone W1 from W2 (Fig. 1). According to current taxonomy, this river separates the subspecies P. v. verreauxi from P. v. deckeni or P. v. coronatus (25). By contrast, populations of E. fulvus, M. murinus, or C. medius north and south of the Tsiribihina have not generally been considered to be taxonomically distinct units. The persistent assumption that the Tsiribihina River separates L. edwardsi and L. ruficaudatus was not confirmed by studies of chromosomes, allozymes, and random amplified polymorphic DNAs (26, 27).
P. v. deckeni individuals from Anadabomandry (far north of the Tsiribihina River) differ by only 0.85–1.02% from P. v. verreauxi populations south of the Tsiribihina (Table 1). Molecular data do not support a subspecific distinction between those populations and thus indicate that the Tsiribihina River does not currently serve as an efficient genetic barrier for Propithecus.
The range of pairwise distances between E. f. rufus individuals from Anadabomandry or Maintirano (north of the Tsiribihina, zone W1) and E. f. rufus individuals from Morondava (south of the Tsiribihina, zone W2) is 2.28–2.54%. In contrast to current taxonomy, genetic data thus clearly support differentiation at the subspecific level between E. fulvus populations south and north of the Tsiribihina. This is supported by pelage coloration; female E. f. rufus from zone W1 have a consistently different appearance from individuals from zone W2 (28).
Further Genetic Differentiation Along the Western Coast.
For some taxa, samples could not be obtained from sites across all three western coast zones. However, even if more than one river lies between two populations investigated, genetic divergences can provide valuable information on gene flow along the western coast.
For Lepilemur, no sample could be obtained from the area between the Mahavavy and the Tsiribihina rivers. However, the data set includes one L. ruficaudatus sample. Although the exact locality of this sample is unknown, the distribution area of L. ruficaudatus certainly includes a part of zone W2 south of the Tsiribihina River, probably to the Onilahy River, where it is replaced by Lepilemur leucopus. Pairwise distances between this L. ruficaudatus and L. edwardsi from Anjamena (zone W1) are 6.02–6.19% (Table 1). Genetic divergence thus supports differentiation at the species level in current taxonomy. However, taxon sampling in this study is inadequate to test whether the Tsiribihina, Manambolo, or Mahavavy rivers act as the genetic barrier. Because the Mahavavy River has failed to impede gene flow among all other lemur populations investigated in this study, it seems more likely that the Tsiribihina River would form the isolating barrier between L. edwardsi and L. ruficaudatus. However, previous studies with karyotypes (26) or allozyme and random amplified polymorphic DNA markers (27) have shown clearly that L. ruficaudatus occurs north and south of the Tsiribihina River. Because of incomplete sampling in all studies to date, no definite conclusion can be drawn regarding taxonomic boundaries for Lepilemur.
One sample (JP241) of Hapalemur griseus occidentalis sequenced in this study was collected from the Tsiombikibo forest (west of the Mahavavy River, zone W1), and the other two samples (JP31 and JP275) originate from Ambato (north of the Betsiboka and Maevarano rivers, zone X). The range of absolute pairwise distances between those two localities is 1.99–2.03% (Table 1). Genetic divergence thus clearly lies in the range for other lemur subspecies. This finding is inconsistent with current taxonomy, which recognizes only one subspecies of H. griseus along the western coast. Because the Mahavavy River does not act as an efficient barrier to gene flow in other lemur taxa, it is more likely that those two H. griseus subspecies are isolated by the Betsiboka River. Thus far, no Hapalemur has been reported between the Betsiboka and Maevarano rivers (zone NW), which indicates that in this case the combination of both rivers may separate the two populations.
M. murinus purportedly is distributed along the whole western coast (9, 29). The data set presented here includes samples from Ampijoroa (east of the Betsiboka River, zone NW) and Kirindy (south of the Tsiribihina River, zone W2). Pairwise comparisons among individuals of those two populations yield distance values of 2.54–2.62%. Such marked genetic divergence indicates, in addition to the already recognized second full species Microcebus myoxinus in W1 (29, 30), the existence of two different M. murinus subspecies along the western coast that are not reflected in current taxonomy. Because three large rivers lie between the two sample localities concerned, the actual genetic barrier between those two subspecies cannot be identified.
C. medius also occurs along the western coast (zones NW, W1, and W2). The samples investigated in this study show considerable genetic variation (2.79–3.17%), which suggests that three different subspecies might exist. However, because nothing is known regarding the origin of the captive animals used in this study, it is not possible either to allocate individuals to taxa newly proposed or resurrected by Groves (31) or to examine any putative isolating boundaries. Nevertheless, in light of information on other lemur taxa along the western coast, it seems likely that either the Betsiboka or Tsiribihina River might serve as genetic barriers in C. medius.
Evolution of Lemurs Along the Western Coast.
Along the western coast, three major distribution zones (NW, W1, and W2) were defined on the assumption that they are separated by the Betsiboka (NW–W1) and Tsiribihina (W1–W2) rivers (1, 2). The present molecular data set clearly supports the subdivision of western Madagascar into these three major zones. Genetic divergence among taxa along the western coast shows that both the Betsiboka and Tsiribihina rivers act as efficient barriers to gene flow. This finding confirms that those two rivers serve as effective major physical barriers between the western distribution zones
Depending on the taxa considered, the level of genetic divergence between populations separated by the Betsiboka or Tsiribihina River varies markedly. The results indicate that the Tsiribihina does not act as a major barrier to gene flow for P. verreauxi. In E. fulvus, however, the Tsiribihina separates the distribution areas of two subspecies. The Betsiboka separates populations at all taxonomic levels. Genetic divergence is highest among Lepilemur populations on both sides of the river, but Propithecus populations also show genetic distances at the species level. E. fulvus is separated at the subspecific level by the Betsiboka, whereas in E. mongoz no genetic differentiation exists. Thus far, there is no evidence for genetic separation between the lemur taxa examined through an isolating effect of the Mahavavy River. The Mahavavy originates in the Bongolava Massif, which may serve as the contact zone between different subregions within the W1 region (25).
The complexity of lemur evolution is evident from the pattern of genetic divergences along the western coast. If the great genetic divergences observed among Lepilemur species are any indication, these taxa have been restricted to the three different zones in the west for some considerable time, possibly from an age when a different biogeographical pattern prevailed. Such an interpretation is suggested by occurrence of two different Lepilemur species within region W2.
The large degree of genetic differentiation of Lepilemur compared with other lemur taxa on the western coast indicates that Lepilemur radiated first over the three zones and then was followed by the other taxa. The low genetic divergence among E. mongoz populations west and east of the Betsiboka River suggests that either E. mongoz has only recently extended its distribution area by crossing the Betsiboka or that E. mongoz is the only taxon for which this river does not act as an efficient barrier. Such different levels of separation were in fact predicted by Martin (1), who noted that, because of the sequential dynamics of speciation among Malagasy lemurs, the situation is necessarily extremely complex.
Central Highlands.
The central highland region is supposed to act as a physical barrier between lemurs from eastern and western Madagascar. Examination of the distribution of all lemur taxa accepted by current taxonomy reveals that only two E. fulvus subspecies occur on both sides of the central highland.
E. f. fulvus individuals from both northwestern and eastern Madagascar were sequenced. Pairwise distances between populations of the western and eastern coast do not exceed 0.25%. This small degree of genetic divergence supports the inference that both populations represent the same subspecies.
The genetic analyses indicate that E. f. rufus represents two subspecies. One occurs in the northwest (W1), and the other occurs on the eastern coast (E1) and south of the Tsiribihina River on the western coast (W2). Genetic distances between the western and eastern populations (0.04–0.21%) clearly lie in the range of differentiation within a subspecies.
The data set presented here confirms that the central highland region generally acts as an efficient barrier to gene flow between lemur taxa. However, two E. fulvus subspecies show no significant differentiation between western and eastern populations, agreeing with current taxonomy.
Lemurs on the Comoro Islands.
Most lemurs are endemic to the island of Madagascar. The only two exceptions are E. fulvus and E. mongoz, which are also found on the Comoro Islands. E. fulvus occurs on Mayotte and E. mongoz on Anjouan and Mohéli. Both species are generally thought to have been introduced to the Comoros from Madagascar relatively recently by humans. The E. fulvus population on the Comoros is recognized as a subspecies endemic to Mayotte (Eulemur fulvus mayottensis), but it was hitherto left open whether Malagasy and Comorian populations of E. mongoz are distinct lineages.
The E. f. fulvus and E. f. mayottensis individuals analyzed in this study do not form distinct monophyletic lineages (Fig. 2). Furthermore, genetic distances between Malagasy E. f. fulvus and Comorian E. f. mayottensis populations are not equivalent to those otherwise found between subspecies.
The current study included samples of 10 mongoose lemurs from four populations that can be distinguished with mtDNA sequence data (Fig. 2). The three Malagasy populations, Anadabomandry (JP177 and JP211), Anjamena (JP169, JP178, and JP196), and Ampijoroa (JP220 and JP221), are separated by large rivers (Mahavavy and Betsiboka). Three samples were obtained from unrelated captive animals. Although only one was unquestionably descended from a Comorian founder, it is assumed that the captive population is derived mainly from animals of Comorian origin. The three captive animals (JP1, JP49, and JP240) share a very similar mtDNA sequence (0–0.04%), suggesting provenance from the same population and thus supporting all three having Comorian or closely related ancestors. Pairwise distances between all four investigated mongoose lemur populations are approximately the same (0.34–0.80%). There is no increase in genetic distance between the Malagasy and captive populations (0.38–0.80%), and distances are smaller than the range observed at subspecific level in other Eulemur taxa.
The level of genetic divergence indicates that E. fulvus and E. mongoz populations on the Comoros are recently derived from their relatives in northwest Madagascar. Genetic data thus support the interpretation that both species were introduced to the Comoros by humans at some time within the past several hundred years. There is no support for recognition of distinct subspecies of either species on the Comoro Islands.
Outlook
In this study, genetic divergence of several lemur taxa across potential geographic boundaries to gene flow was investigated. Discrepancies between the molecular data and current taxonomy have been discussed in light of the geographical distribution of a variety of lemur taxa occurring in the same region. Comparison of the molecular phylogeny, taxonomy, and geographical distribution of several lemur taxa in the same area allowed an initial insight into the complexity of lemur evolution.
Despite the heavy investment required for such investigations, the evolution of lemurs on Madagascar provides an ideal model for studying evolutionary biology. For future work, samples from all lemur taxa present in each geographically isolated subregion should be collected and analyzed. Although thorough sampling would primarily serve to complete an overview of the distribution of lemurs remaining on Madagascar, it would also allow further examination of their genetic diversity and differentiation. DNA sequence data would allow determination of genetic divergences among populations from each subregion and thus permit additional inferences about the efficacy of geographical barriers (e.g., rivers or mountains) as isolating mechanisms, notably in the hitherto little-studied eastern region of Madagascar. Evolutionary patterns from different taxa could be compared both within and across regions, which would yield critical insights into the various evolutionary patterns exhibited by the different taxa and possibly into earlier biogeographical patterns. By using a combination of nuclear and mtDNA markers, a reliable time frame for the radiation of lemurs could be estimated.
Ultimately, the unique primate fauna of Madagascar can yield insights not only into the evolutionary history of lemurs but also into the mechanisms and outcomes of evolutionary processes in primates in general. In the short term, such information is critically needed for conservation efforts. In the long term, it would serve to increase our understanding of the pattern and process of evolution in this unique endemic group. The biogeographical zones recognized for lemurs and further confirmed in this study may well apply to other groups of organisms in Madagascar. Furthermore, a dynamic process of successive waves of speciation across semipermeable barriers separating zones with distinctive climatic features may explain the evolution of many endemic organisms in Madagascar.
Acknowledgments
We thank M. Forstner for valuable comments throughout this study and A. Yoder for critical comments on the manuscript. Thanks also go to D. Curtis, P. Ehresmann, T. Geissmann, H. Hirai, P. Kappeler, Y. Kawamoto, A. Kitchener, P. Moisson, C. Nievergelt, C. Rabarivola, G. Rakotoarisoa, M. Rasmussen, C. Reber, C. Roos, U. Ruempler, Y. Rumpler, W. Scheffrahn, N. Vasey, B. Wimmer, and E. Zimmermann for providing samples for this study. We also thank G. Anzenberger, D. Armitage, T. Geissmann, M. Lavaux, C. Rabarivola, U. Radespiel, W. Scheffrahn, and Y. Wyner for help in collecting samples. Acquisition of samples in Madagascar took place under an “Accord de Coopération” between the Anthropological Institute (University of Zürich, Zürich) and the Départment des Sciences de la Terre (University of Mahajanga, Mahajanga, Madagascar). We thank the governmental institutions of Madagascar (Commission Tripartite) for research permission. We are most grateful for support during field work from M. Clark, D. Curtis, J. Gerson, A. Müller, C. Rabarivola, O. Raheliarisoa, W. Scheffrahn, M. Waters, and A. Zaramody. Financial support from the Julius Klaus and A. H. Schultz Foundations is gratefully acknowledged.
Footnotes
References
- 1.Martin R D. Philos Trans R Soc London B. 1972;264:295–352. doi: 10.1098/rstb.1972.0013. [DOI] [PubMed] [Google Scholar]
- 2.Martin R D. In: Creatures of the Dark: The Nocturnal Prosimians. Alterman L, Doyle G A, Izard M K, editors. New York: Plenum; 1995. pp. 535–563. [Google Scholar]
- 3.Groves C P. Primate Taxonomy. Washington, DC: Smithsonian Institution Press; 2001. [Google Scholar]
- 4.Mittermeier R A, Tattersall I, Konstant W R, Meyers D M, Mast R B. Lemurs of Madagascar. Washington, DC: Conservation International; 1994. [Google Scholar]
- 5.Godfrey L R, Jungers W L. In: The Primate Fossil Record. Hartwig W C, editor. Cambridge, U.K.: Cambridge Univ. Press; 2002. pp. 97–121. [Google Scholar]
- 6.Harcourt C, Thornback J. Lemurs of Madagascar and the Comoros: The IUCN Red Data Book. Gland, Switzerland: IUCN-The World Conservation Union; 1990. [Google Scholar]
- 7.Hill W C O. Primates: Comparative Anatomy and Taxonomy: Strepsirhini. I. Edinburgh: Edinburgh Univ. Press; 1953. [Google Scholar]
- 8.Petter J-J, Albignac R, Rumpler Y. Mammifères Lémuriens (Primates Prosimiens) Paris: Office de la Recherche Scientifique et Technique Outre-Mer and Centre National de la Recherche Scientifique; 1977. [Google Scholar]
- 9.Tattersall I. The Primates of Madagascar. New York: Columbia Univ. Press; 1982. [Google Scholar]
- 10.Jenkins P D. Catalogue of Primates in the British Museum (Natural History) London: British Museum; 1987. , Part IV. [Google Scholar]
- 11.Groves C P. A Theory of Human and Primate Evolution. Oxford: Oxford Univ. Press; 1989. [Google Scholar]
- 12.Pastorini J, Forstner M R J, Martin R D. Mol Phylogenet Evol. 2000;16:418–429. doi: 10.1006/mpev.2000.0796. [DOI] [PubMed] [Google Scholar]
- 13.Pastorini J, Forstner M R J, Martin R D. Am J Primatol. 2001;53:1–17. doi: 10.1002/1098-2345(200101)53:1<1::AID-AJP1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 14.Pastorini J, Martin R D, Ehresmann P, Zimmermann E, Forstner M R J. Mol Phylogenet Evol. 2001;19:45–56. doi: 10.1006/mpev.2000.0904. [DOI] [PubMed] [Google Scholar]
- 15.Zaramody A, Pastorini J. Lemur News. 2001;6:28–31. [Google Scholar]
- 16.Pastorini J, Forstner M R J, Martin R D. J Hum Evol. 2002;43:463–478. [PubMed] [Google Scholar]
- 17.Pastorini J, Forstner M R J, Martin R D. Evol Anthropol. 2002;11, Suppl. 1:150–154. [Google Scholar]
- 18.Swofford D L. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer; 1999. , Version 4. [Google Scholar]
- 19.Kimura M. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 20.Felsenstein J. Evolution (Lawrence, Kans) 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 21.Yoder A D, Cartmill M, Ruvolo M, Smith K, Vilgalys R. Proc Natl Acad Sci USA. 1996;93:5122–5126. doi: 10.1073/pnas.93.10.5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tajima F. Genetics. 1993;135:599–607. doi: 10.1093/genetics/135.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thalmann U, Ganzhorn J U. In: The Natural History of Madagascar. Goodman S M, Benstead J, editors. Chicago: Univ. of Chicago Press; 2003. pp. 1336–1340. [Google Scholar]
- 24.Thalmann U, Kümmerli R, Zaramody A. Lemur News. 2002;7:11–16. [Google Scholar]
- 25.Thalmann U, Rakotoarison N. Folia Primatol. 1994;63:156–161. doi: 10.1159/000156811. [DOI] [PubMed] [Google Scholar]
- 26.Rumpler Y, Albignac R. J Hum Evol. 1978;7:191–196. [Google Scholar]
- 27.Bachmann L, Rumpler Y, Ganzhorn J U, Tomiuk J. Int J Primatol. 2000;21:853–864. [Google Scholar]
- 28.Hawkins A F A, Durbin J C, Reid D B. Folia Primatol. 1998;69:337–345. doi: 10.1159/000021649. [DOI] [PubMed] [Google Scholar]
- 29.Rasoloarison R M, Goodman S M, Ganzhorn J U. Int J Primatol. 2000;21:963–1019. [Google Scholar]
- 30.Yoder A D, Rasoloarison R M, Goodman S M, Irwin J A, Atsalis S, Ravosa M J, Ganzhorn J U. Proc Natl Acad Sci USA. 2000;97:11325–11330. doi: 10.1073/pnas.200121897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Groves C P. Int J Primatol. 2000;21:943–962. [Google Scholar]