Abstract
Retrotransposons and retroviruses integrate nonrandomly into eukaryotic genomes. For the yeast retrotransposon Ty5, integration preferentially occurs within domains of heterochromatin. Targeting to these locations is determined by interactions between an amino acid sequence motif at the C terminus of Ty5 integrase (IN) called the targeting domain, and the heterochromatin protein Sir4p. Here we show that new Ty5 integration hot spots are created when Sir4p is tethered to ectopic DNA sites. Targeting to sites of tethered Sir4p is abrogated by single amino acid substitutions in either IN or Sir4p that prevent their interaction. Ty5 target specificity can be altered by replacing the IN-targeting domain with other peptide motifs that interact with known protein partners. Integration occurs at high efficiency and in close proximity to DNA sites where the protein partners are tethered. These findings define a mechanism by which retrotransposons shape their host genomes and suggest ways in which retroviral integration can be controlled.
Retroviruses and retrotransposons integrate their cDNA into host genomes using retroelement-encoded integrase (IN). Integration is essential for retroviral proliferation and has significantly shaped eukaryotic genome organization. For example, endogenous retroviruses and retrotransposons constitute over one-half of the genomes of human and some plant species such as maize (1, 2). Retroelement insertions are not randomly distributed within genomes, and they are often enriched in heterochromatin or other gene-poor regions. This distribution may be due, in part, to selection against integration within gene-rich euchromatin. Alternatively, retroelements may actively select targets such as heterochromatin where insertion will not compromise host fitness (3, 4).
Integration site choice is clearly the rule for the well studied Ty elements of Saccharomyces cerevisiae. Ty1 and Ty3 both integrate preferentially upstream of genes transcribed by RNA polymerase III (Pol III; e.g., tRNA genes), which are harmless sites because they are gene-poor and integration does not disrupt Pol III transcription (5, 6). The precision of integration, however, differs for these two yeast retroelements. Ty3 typically integrates within 1–2 bases of Pol III transcription start sites, whereas Ty1 inserts within a 750-bp window upstream of target genes. Pol III complexes are required for both Ty1 and Ty3 integration specificity, and for Ty3, transcription factor (TF)IIIB is the major determinant of target choice (7–10). This suggests that target sites are selected through interactions between Ty1 and Ty3 integration complexes and the Pol III transcription machinery.
In contrast to Ty1 and Ty3 and like many other eukaryotic retrotransposons, Ty5 insertions are found within heterochromatic regions of the yeast genome (11). This distribution is due to target site selection because 95% of de novo Ty5 transposition events occur within heterochromatin found at yeast telomeres and the silent mating loci (HML and HMR) (12, 13). Several lines of evidence suggest that the heterochromatin protein Sir4p is the major determinant of Ty5 integration specificity (14, 15). A 6-aa motif at the C terminus of Ty5 IN (the targeting domain; TD) interacts with Sir4p (15), and mutations in the TD that abrogate Sir4p interactions randomize Ty5 integration patterns (15, 16). Similarly, Ty5 integrates randomly in strains that lack Sir4p (14).
A recent study (17) revealed that HIV integration occurs preferentially at sites of active transcription. Furthermore, HIV IN interacts with Ini1, a homolog of the yeast transcriptional activator Snf5p (18). These observations, coupled with the data for the Ty retrotransposons, suggest a general model wherein interactions between IN and DNA-bound proteins mediate retroelement target choice. By further defining the determinants of Ty5 integration specificity and by engineering Ty5 elements with altered target site preference, we demonstrate that this model describes the mechanism by which Ty5 selects integration sites.
Methods
Plasmid Constructs.
DNA fragments encoding various regions of the Sir4p C terminus (SIR4C) were amplified by the PCR (19) from plasmid pRS316-SIR4 (gift of Jasper Rine, University of California, Berkeley). The amplification products were cloned into the EcoRI–BamHI sites of the LexA-expressing vector pBTM116 (20) or a derivative with a LEU2 marker gene (pYZ275). PCR mutagenesis was used to substitute an alanine at residues 971–976 of Sir4p (19). Additional LexA fusion constructs were generated by PCR-amplifying the FHA1 domain of Rad53p (amino acids 14–154) from yeast genomic DNA and the coding region for Npw38 from a human cDNA clone (ATCC5806979). PCR products were cloned into the EcoRI and BglII sites of pYZ275. The C terminus of Ty5 IN (amino acids 934–1131) was amplified by PCR from pNK254 (21) or a variant with the S1094L-TD mutation (16). The amplification products were cloned into the XmaI–PstI sites of pGAD-C1 (22) to generate fusions with the Gal4p transcriptional activation domain.
The donor plasmids carry either a WT Ty5 element or one with the S1094L mutation. Both Ty5 elements are under transcriptional control of the GAL1–10 promoter and carry a his3AI selectable marker gene (23). The 6-aa-TD of Ty5 (LDSSPP) was replaced with a motif of Rad9p (SLEVTEADATFVQ) (24) to generate Ty5-Rad9p or a motif of NpwBP (PRLLPPFPPPGR) (25) to generate Ty5-NpwBP. These modifications were made by using a two-step PCR replacement method (19). The target plasmid was generated by inserting 3-kb and 4-kb DNA fragments from Arabidopsis into the EcoRI and SacI sites of pRS424 (26), which was modified to carry a Chlr gene. One to four copies of overlapping, double LexA operators (20) were inserted into the BamHI site.
Tethered Integration and Two-Hybrid Assays.
YPH499 or its sir derivatives were transformed with a Ty5 donor plasmid, a target plasmid, and a plasmid expressing a LexA fusion protein. Yeast cells were grown as patches on synthetic complete media lacking tryptophan, leucine, and uracil (SC-T-L-U) and incubated at 30°C for 2 days. The cells were then replica plated onto the same selective media supplemented with 2% galactose and incubated at room temperature for 2 days. Finally, the cells were replica plated onto synthetic complete media that lacked histidine (SC-T-L-U-H) and incubated at 30°C for 3 days. Cells were scraped from the plates and washed twice with water, and total DNA was prepared (19). The DNA was used to transform competent Escherichia coli cells with a hisB mutation (strain eDW335, D. A. Wright and D.F.V., unpublished data). After transformation, the E. coli cells were incubated in rich media at 37°C for 3 h and washed twice with water to remove the residual histidine. One-tenth of the cells were plated onto rich media with 20 μg/μl chloramphenicol and nine-tenths were plated on M9 minimal media lacking histidine and supplemented with 20 μg/μl chloramphenicol. The plates were incubated at 37°C for 1–3 days before counting the colonies.
The two-hybrid assays used strain L40 (27) or its sir derivatives that express LexA-SIR4C and Gal4p transcriptional activation domain–IN C terminus (GAD-INC) fusion proteins. A single colony was inoculated into 2 ml of SC-T-L liquid media and grown at 30°C for 24 h. The yeast cells were spotted (10-fold serial dilutions) onto solid SC-T-L-H media that was supplemented with 1–5 mM 3-amino 1,2,4-triazol (3-AT). As controls, cells were also spotted onto SC-T-L media. Plates were incubated at 30°C for 3–4 days before being imaged to record their growth.
Results and Discussion
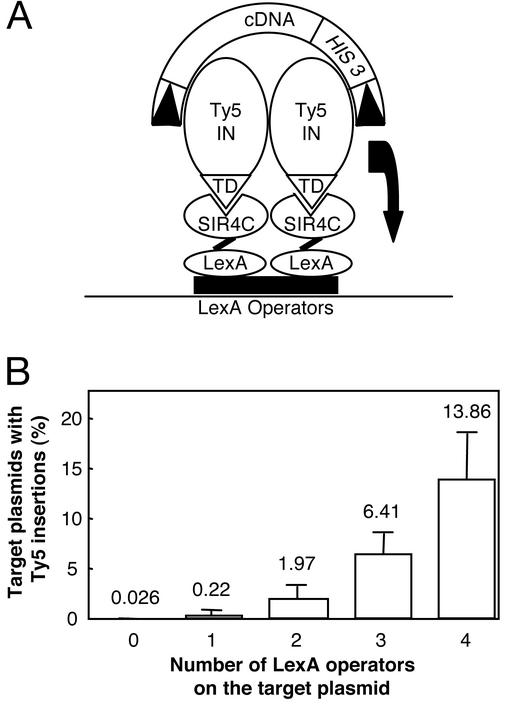
To test whether the interaction between Ty5 IN and Sir4p is the primary determinant of Ty5 target site choice, integration was measured at DNA sites to which Sir4p is tethered (Fig. 1A). The Sir4p C terminus (SIR4C) (amino acids 951–1358), which interacts with Ty5 IN in two-hybrid and in vitro-binding assays (15), was expressed as a fusion protein with the LexA DNA-binding domain (LexA-SIR4C). LexA-SIR4C was tethered to a target plasmid through LexA operators, which, in turn, were flanked by 3–4 kb of Arabidopsis DNA that serve as a landing site for Ty5 and prevent insertions from compromising plasmid function. To measure targeted integration, the plasmid was introduced into a yeast strain with a galactose-inducible Ty5 element (12). After growth on galactose, transposition events were selected by plating cells onto media lacking histidine. Ty5 carries a his3AI marker gene, and splicing of the Ty5 mRNA removes an inactivating intron, thereby reconstituting a functional HIS3 gene upon reverse transcription and cDNA integration (28). Total DNA was prepared from His+ yeast cells and used to transform a hisB E. coli strain. The HIS3 gene within Ty5 complements the E. coli hisB mutation (29). Because the target plasmid also carries a chloramphenicol resistance gene, plasmids with Ty5 insertions confer a ChlrHis+ phenotype to E. coli. The ratio of ChlrHis+ colonies (target plasmids with Ty5) to Chlr colonies (target plasmids) measures the efficiency of integration to the target plasmid.
Figure 1.
Ty5 integrates at sites of tethered Sir4p. (A) A tethered integration assay. The target plasmid carries LexA operators that bind LexA fusion proteins. LexA-SIR4C is shown interacting with the targeting domain (TD) of Ty5 IN that is complexed to its cDNA. Arabidopsis DNA flanks the operators to create a Ty5 landing pad. The target plasmid also has selectable markers and replication origins that function in yeast (TRP1, 2μ) and E. coli (chloramphenicol resistance gene, ColE1). Details of the tethered integration assay are described in the text. In brief, after induction of Ty5 transposition, target plasmids are recovered in E. coli by selecting either for chloramphenicol resistance (Chlr) or for chloramphenicol resistance and histidine prototrophy (ChlrHis+). The latter selection determines the number of target plasmids that carry Ty5 with its HIS3 marker. The ratio of ChlrHis+ to Chlr indicates the efficiency with which Ty5 integrates into the plasmid. (B) Monitoring integration to sites of tethered Sir4p. The efficiency of Ty5 integration to the target plasmid correlates with the number of LexA operators.
LexA-SIR4C created a strong Ty5 integration hot spot when tethered to the target plasmid. With four copies of the LexA operator, ≈14% of the recovered target plasmids carried Ty5 insertions (Fig. 1B). Targeting displayed a strict dependence on the number of LexA operators, suggesting that targeting efficiency is determined by the amount of SIR4C tethered to the plasmid. Ty5 insertions into the target plasmid were true integration events, as evidenced by target site duplications flanking several characterized insertions (data not shown).
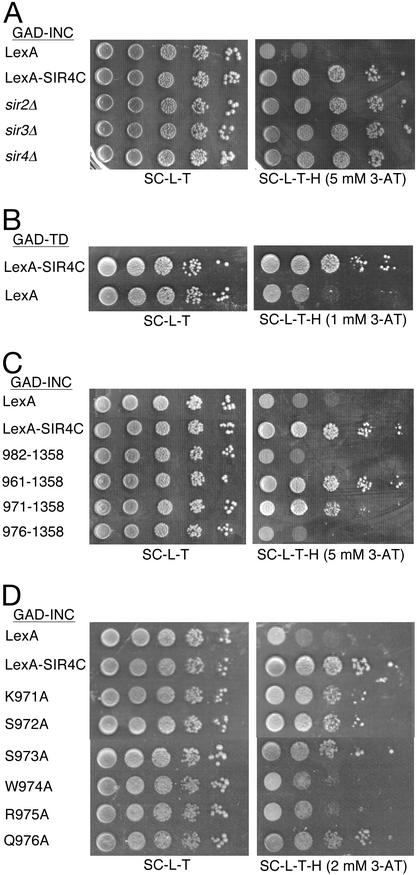
Sir4p interacts with a number of proteins, including Sir2p and Sir3p, and loss of Sir proteins significantly decreases Ty5 target specificity (14). To test whether SIR4C requires other components of yeast heterochromatin for its interaction with IN, two-hybrid interactions were measured between IN and SIR4C in the absence of Sir proteins. These assays used LexA-SIR4C and a fusion protein generated between the Ty5 INC and GAD (15). The 6-aa-TD is located within INC and corresponds to positions 1092–1097 in the Ty5 polyprotein. The strength of the INC–SIR4C two-hybrid interaction was determined by expression of a HIS3 reporter with upstream LexA operators (27). HIS3 expression confers growth on media without histidine and with the inhibitor 3-amino 1,2,4-triazol (3-AT). The INC–SIR4C two-hybrid interaction was not significantly affected by loss of Sir2p, Sir3p, or Sir4p (Fig. 2A). In addition, SIR4C interacts with fusion proteins generated between GAD and nine aa of Ty5 IN that encompass the 6-aa-TD (GAD-TD, Fig. 2B). These results, coupled with previous in vitro-binding studies (15), support the conclusion that INC and SIR4C interact directly through the 6-aa-TD.
Figure 2.
Defining the determinants that mediate IN–Sir4p interactions. Two-hybrid assays measure the ability of LexA-SIR4C fusion proteins to interact with a fusion protein generated between GAD and Ty5 IN. The GAD-INC construct has 258 aa from the C terminus of Ty5 IN. The GAD-TD construct has 9 aa from IN that encompass the 6-aa-TD. The strength of interaction is assayed by the expression of a HIS3 reporter gene under the control of upstream LexA operators. Tenfold serial dilutions of cells were spotted onto media that selects either for the plasmids expressing the fusion proteins (SC-L-T) or for both the plasmids and HIS3 expression [SC-T-L-H plus the inhibitor 3-amino 1,2,4-triazol (3-AT)]. (A) Sir-independence of the IN–Sir4p interaction. Each of the sir strains expresses LexA-SIR4C. (B) Sufficiency of the TD for Sir4p interactions. (C) The IN-interacting domain of Sir4p. As indicated on the left side of C, various N-terminal truncations of SIR4C were tested for their ability to interact with GAD-INC. Residues 971–975 of Sir4p are required for the INC–SIR4C interaction. (D) Amino acid residues of SIR4C required for the IN–Sir4p interaction. Substitutions with alanine at tryptophan 974 (W974A) and arginine 975 (R975A) significantly disrupted the INC–SIR4C interaction.
To further map the region of SIR4C that interacts with INC, a series of SIR4C truncations were fused to the LexA DNA-binding domain (Fig. 2C and data not shown). One SIR4C truncation (amino acids 982–1358) lost its ability to interact with INC (Fig. 2C). This construct differs by only 30 amino acids from SIR4C constructs used in the previous experiments (amino acids 951–1358). A series of additional constructs were made with SIR4C N-termini corresponding to positions 961, 971, and 976. Of these, only the construct beginning at residue 976 failed to interact with INC, indicating that the region spanning residues 971–975 is critical for the INC–SIR4C interaction. To pinpoint essential residues, alanine was substituted at each of these amino acid positions. Only the W974A and R975A substitutions disrupted the two-hybrid interaction (Fig. 2D). In no case was failure of the two-hybrid interaction due to differences in expression of the various LexA-SIR4C constructs; all were expressed at comparable levels as measured by immunoblotting experiments performed with a LexA-specific Ab (data not shown).
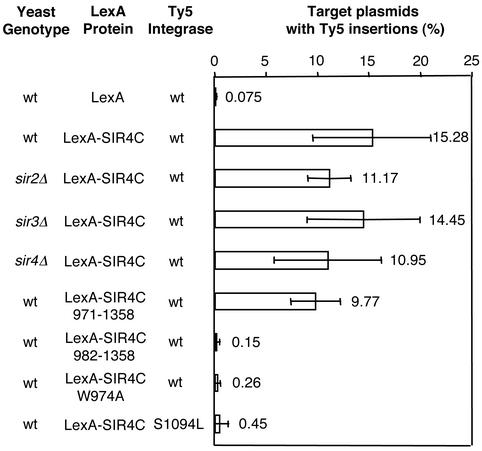
We tested whether the requirements for the INC–SIR4C two-hybrid interaction correlated with requirements for targeted integration. This was invariably the case. Loss of Sir proteins affected neither two-hybrid interactions nor targeting efficiency of Ty5 to sites of tethered SIR4C (Fig. 3). When the various SIR4C derivatives were tested, only those fusion proteins that supported two-hybrid interactions created integration hot spots. For example, the construct beginning at residue 971 of Sir4p created an integration hot spot comparable to SIR4C, whereas no significant targeting was observed with the construct beginning at residue 982. Furthermore, the W974A sir4 mutation, which abrogated two-hybrid interactions, also failed to target integration. In a complementary experiment, the tethered integration assay was performed with a Ty5 element with a mutation in its TD (S1094L). This mutation randomizes genomic integration patterns and disrupts interactions with SIR4C in both two-hybrid and in vitro-binding assays (15, 16). The Ty5 S1094L mutation also prevented integration to sites of tethered SIR4C (Fig. 3).
Figure 3.
Tethered integration requires interaction between IN and Sir4p. The tethered integration assay was performed with LexA-SIR4C in the absence of Sir2p, Sir3p, and Sir4p. As with the two-hybrid assays, no effect on tethered integration was observed because of the absence of Sir proteins. Deletions or mutations in SIR4C that abrogate two-hybrid interactions (i.e., LexA-SIR4C 982–1,358, W974A) prevent tethered integration. Similarly, mutations in the Ty5 TD (S1094L) that randomize chromosomal integration patterns (15, 16) do not support integration to sites of tethered SIR4C.
Having defined the targeting determinants of IN and its interacting partner, Sir4p, we next asked whether we could engineer Ty5 elements with altered target specificity. To accomplish this, we replaced the 6-aa Ty5 TD with peptide motifs with known protein ligands. In one construct (Ty5-Rad9p), the TD was swapped with a 13-aa motif from Rad9p, which interacts with the two forkhead-associated domains of Rad53p (FHA1 and FHA2) (30, 31). In a second construct (Ty5-NpwBP), the TD was replaced with a 12-aa, proline-rich motif from the human nuclear protein NpwBP (25). This motif interacts with the WW domain of a second nuclear protein, Npw38. The modified INs were initially tested in two-hybrid assays with their respective protein partners, and interactions were comparable to the INC–SIR4C two-hybrid interaction (data not shown).
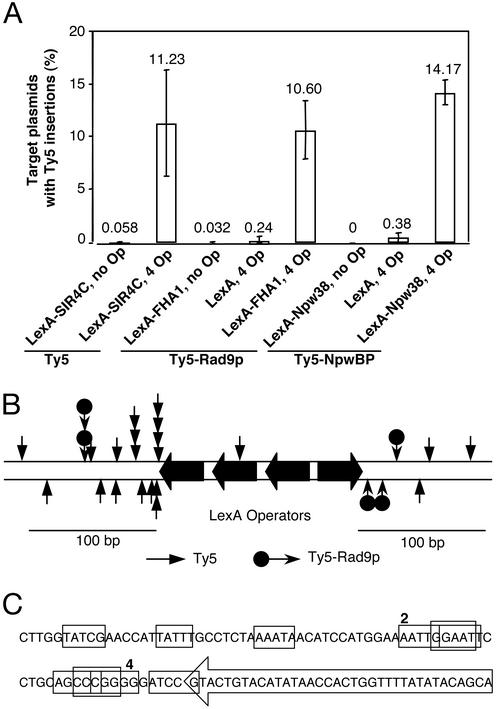
Neither of the TD modifications compromised IN function, as transposition frequencies of Ty5-Rad9p and Ty5-NpwBP were comparable to those of a WT element (data not shown). Remarkably, the efficiencies with which modified elements targeted to sites of tethered LexA-FHA1 and LexA-Npw38 were comparable to the efficiency with which the WT element targeted to sites of tethered SIR4C (Fig. 4A). Targeting required both the LexA operators and either the tethered Npw38 or the FHA1 domain. These results indicate that Ty5 target specificity can be altered and suggest that Ty5 variants with a range of integration specificity can be generated by substituting the TD with peptide aptamers that recognize different chromosomal proteins.
Figure 4.
Altering specificity and precision of Ty5 integration. (A) Ty5-Rad9p and Ty5-NpwBP integrate at sites of tethered FHA1 and Npw38, respectively. For Ty5-Rad9p, the TD of Ty5 was replaced with a peptide motif of Rad9p that interacts with the FHA1 domain of Rad53p (24). Similarly for Ty5-NpwBP, the TD was replaced with a proline-rich motif of NpwBP that interacts with Npw38 (25). Targeting was measured to sites of tethered LexA-FHA1 and LexA-Npw38 and requires both the LexA operators and LexA fusion proteins. (B) Precision of integration to sites of tethered proteins. Ty5 insertions (26) generated by both WT and modified Ty5 elements were characterized by DNA sequencing. The insertions occurred within 120 bases of the LexA operators in the target plasmid. Ty5 insertions on the top strand are oriented 5′–3′; insertions in the bottom strand are in the opposite orientation. (C) DNA sequence of 14 insertion sites adjacent to the leftmost LexA operator depicted in B. The sequence of the LexA operator is enclosed by an arrowhead. Target sites are boxed, and numbers adjacent to a given box denote the number of independent insertions that occurred at that site.
To characterize where Ty5 integrates on the target plasmid, 26 insertions generated by both WT Ty5 and Ty5-Rad9p were analyzed by DNA sequencing (Fig. 4 B and C). All 26 insertions occurred within 120 bases of the nearest LexA operator. No orientation preference was observed and no obvious sequence consensus defined the insertion sites. The insertions that clustered near the leftmost LexA operator (as depicted in Fig. 4B) displayed a regular periodicity of ≈10–12 bp, suggesting that they occurred on the same face of the DNA helix. For 18 of the 26 insertions, DNA sequences were obtained from both ends of the element, and all 18 were flanked by five-base target site duplications.
The narrow integration window next to the LexA operators contrasts with the integration pattern observed for chromosomal Ty5 insertions, which typically occur within a 3 kb window flanking the HM silencers or the subtelomeric X repeats (12, 13). The tethered Ty5 integration patterns more closely resemble those of Ty3, which occur within 1–2 bases upstream of genes transcribed by RNA Pol III (5). Ty3 integration specificity TFIIIB (9), and like LexA-SIR4C, TFIIIB occupies a well-defined chromosomal site. Collectively, these targeting patterns suggest that the precision of integration is determined primarily by the physical location of the protein or protein complex recognized by the integration machinery.
Retroviral vectors are widely used for gene delivery in gene therapy, in part because viral integration generates stable, defined, chromosomal insertions (32). The randomness of retroviral integration, however, is potentially hazardous and could have deleterious genetic effects, e.g., by creating loss-of-function mutations or by activating oncogenes. A previous approach to control retroviral integration has been to fuse sequence-specific DNA-binding domains to retroviral INs (33–36). This approach has proven effective in in vitro integration assays, but because the IN modifications often compromise viral replication, this approach has not been successfully used in vivo. The findings described here suggest an alternative approach for controlling retroviral integration, wherein retroviral INs are modified to carry small peptide aptamers that recognize proteins bound to chromosomal target sites. In addition, the results have relevance for understanding eukaryotic genome organization. The successful proliferation of retrotransposons is thought to be due to their ability to identify safe havens in the genome where integration is not harmful to their hosts (3, 4). The widespread association of eukaryotic retrotransposons with heterochromatin suggests that these gene-poor domains are one such safe-haven (3). If the strategy of Ty5 for selecting integration sites is used by other retrotransposons, then targeted integration may have significantly shaped eukaryotic genome organization.
Acknowledgments
This study is dedicated to the memory of Francis J. Voytas (father of D.F.V.). The work was supported by American Cancer Society Grant RPG9510106MBC and National Institutes of Health Grant GM61657.
Abbreviations
- IN
integrase
- INC
IN C terminus
- TD
targeting domain
- SIR4C
Sir4p C terminus
- GAD
Gal4p transcriptional activation domain
- Pol III
RNA polymerase III
- SC-T-L-U
synthetic complete media lacking, tryptophan, leucine, and uracil
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 5586.
References
- 1.Medstrand P, Van De Lagemaat L N, Mager D L. Genome Res. 2002;12:1483–1495. doi: 10.1101/gr.388902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.SanMiguel P, Tikhonov A, Jin Y K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer P S, Edwards K J, Lee M, Avramova Z, et al. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 3.Boeke J D, Devine S E. Cell. 1998;93:1087–1089. doi: 10.1016/s0092-8674(00)81450-6. [DOI] [PubMed] [Google Scholar]
- 4.Craig N L. Annu Rev Biochem. 1997;66:437–474. doi: 10.1146/annurev.biochem.66.1.437. [DOI] [PubMed] [Google Scholar]
- 5.Chalker D L, Sandmeyer S B. Genes Dev. 1992;6:117–128. doi: 10.1101/gad.6.1.117. [DOI] [PubMed] [Google Scholar]
- 6.Devine S E, Boeke J D. Genes Dev. 1996;10:620–633. doi: 10.1101/gad.10.5.620. [DOI] [PubMed] [Google Scholar]
- 7.Kirchner J, Connolly C M, Sandmeyer S B. Science. 1995;267:1488–1491. doi: 10.1126/science.7878467. [DOI] [PubMed] [Google Scholar]
- 8.Aye M, Dildine S L, Claypool J A, Jourdain S, Sandmeyer S B. Mol Cell Biol. 2001;21:7839–7851. doi: 10.1128/MCB.21.22.7839-7851.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yieh L, Kassavetis G, Geiduschek E P, Sandmeyer S B. J Biol Chem. 2000;275:29800–29807. doi: 10.1074/jbc.M003149200. [DOI] [PubMed] [Google Scholar]
- 10.Yieh L, Hatzis H, Kassavetis G, Sandmeyer S B. J Biol Chem. 2002;277:25920–25928. doi: 10.1074/jbc.M202729200. [DOI] [PubMed] [Google Scholar]
- 11.Zou S, Wright D A, Voytas D F. Proc Natl Acad Sci USA. 1995;92:920–924. doi: 10.1073/pnas.92.3.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou S, Ke N, Kim J M, Voytas D F. Genes Dev. 1996;10:634–645. doi: 10.1101/gad.10.5.634. [DOI] [PubMed] [Google Scholar]
- 13.Zou S, Kim J M, Voytas D F. Nucleic Acids Res. 1996;24:4825–4831. doi: 10.1093/nar/24.23.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Y, Zou S, Wright D, Voytas D. Genes Dev. 1999;13:2738–2749. doi: 10.1101/gad.13.20.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie W, Gai X, Zhu Y, Zappulla D C, Sternglanz R, Voytas D F. Mol Cell Biol. 2001;21:6606–6614. doi: 10.1128/MCB.21.19.6606-6614.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gai X, Voytas D F. Mol Cell. 1998;1:1051–1055. doi: 10.1016/s1097-2765(00)80105-7. [DOI] [PubMed] [Google Scholar]
- 17.Schroder A, Shinn P, Chen H, Berry C, Ecker J, Bushman F. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 18.Kalpana G V, Marmon S, Wang W, Crabtree G R, Goff S P. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 19.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Greene & Wiley; 1987. [Google Scholar]
- 20.Estojak J, Brent R, Golemis E A. Mol Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ke N, Voytas D F. Genetics. 1997;147:545–556. doi: 10.1093/genetics/147.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.James P, Halladay J, Craig E A. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao X, Rowley D J, Gai X, Voytas D F. J Virol. 2002;76:3240–3247. doi: 10.1128/JVI.76.7.3240-3247.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao H, Yuan C, Su M I, Yongkiettrakul S, Qin D, Li H, Byeon I J, Pei D, Tsai M D. J Mol Biol. 2000;304:941–951. doi: 10.1006/jmbi.2000.4291. [DOI] [PubMed] [Google Scholar]
- 25.Komuro A, Saeki M, Kato S. J Biol Chem. 1999;274:36513–36519. doi: 10.1074/jbc.274.51.36513. [DOI] [PubMed] [Google Scholar]
- 26.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 27.Hollenberg S M, Sternglanz R, Cheng P F, Weintraub H. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Curcio M J, Garfinkel D J. Proc Natl Acad Sci USA. 1991;88:936–940. doi: 10.1073/pnas.88.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Struhl K, Davis R W. Proc Natl Acad Sci USA. 1977;74:5255–5259. doi: 10.1073/pnas.74.12.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Z, Hsiao J, Fay D S, Stern D F. Science. 1998;281:272–274. doi: 10.1126/science.281.5374.272. [DOI] [PubMed] [Google Scholar]
- 31.Durocher D, Henckel J, Fersht A R, Jackson S P. Mol Cell. 1999;4:387–394. doi: 10.1016/s1097-2765(00)80340-8. [DOI] [PubMed] [Google Scholar]
- 32.Verma I M, Somia N. Nature. 1997;389:239–242. doi: 10.1038/38410. [DOI] [PubMed] [Google Scholar]
- 33.Goulaouic H, Chow S A. J Virol. 1996;70:37–46. doi: 10.1128/jvi.70.1.37-46.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katz R A, Merkel G, Skalka A M. Virology. 1996;217:178–190. doi: 10.1006/viro.1996.0105. [DOI] [PubMed] [Google Scholar]
- 35.Bushman F D. Proc Natl Acad Sci USA. 1994;91:9233–9237. doi: 10.1073/pnas.91.20.9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bushman F D, Miller M D. J Virol. 1997;71:458–464. doi: 10.1128/jvi.71.1.458-464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]