Abstract
The formation of polyglutamine-containing aggregates and inclusions are hallmarks of pathogenesis in Huntington's disease that can be recapitulated in model systems. Although the contribution of inclusions to pathogenesis is unclear, cell-based assays can be used to screen for chemical compounds that affect aggregation and may provide therapeutic benefit. We have developed inducible PC12 cell-culture models to screen for loss of visible aggregates. To test the validity of this approach, compounds that inhibit aggregation in the PC12 cell-based screen were tested in a Drosophila model of polyglutamine-repeat disease. The disruption of aggregation in PC12 cells strongly correlates with suppression of neuronal degeneration in Drosophila. Thus, the engineered PC12 cells coupled with the Drosophila model provide a rapid and effective method to screen and validate compounds.
Huntington's disease (HD) is a late-onset neurodegenerative disease characterized by a movement disorder, psychiatric symptoms, and cognitive dysfunction (1). It is one of several neurodegenerative diseases, including the spinocerebellar ataxias and Kennedy's disease, caused by the expansion of a CAG repeat encoding an endogenous polyglutamine (polyQ) tract within the corresponding disease protein (1–5).
Mouse, rat, Caenorhabditis elegans, Drosophila, yeast and numerous cell-culture systems (for review see refs. 1, 3, and 4) have been developed for the polyQ-repeat diseases to model different aspects of disease. A transgenic HD mouse model expressing human huntingtin (Htt) exon 1 protein with ≈150 polyQs demonstrated that truncated mutant Htt was sufficient to cause an HD-like phenotype (6). This mouse model led to the discovery of a major pathogenic feature of the disease, the appearance of neuronal intranuclear inclusions containing mutant Htt (7), which suggests a role for nuclear localization and aggregation in HD.
Amino-terminal fragments of mutant Htt protein were subsequently identified in neuronal intranuclear inclusions in HD brain tissue (8) and in the neuronal processes (neuropil) of both HD patient and transgenic mouse brain tissues (9). They have been found in brain tissue from transgenic mouse models and human patient material for almost every polyQ-repeat disease investigated (1, 10), and cytosolic and nuclear aggregates have been demonstrated in most model systems (3, 11–13). In vitro filter assays have shown that the formation of these aggregates depends on polyQ repeat length and protein concentration (14, 15). The rate and degree of aggregation, a highly specific process in cells (16, 17), is influenced by cellular processes such as aging (18), proteasome function (19–21), and chaperone activity (22–25).
Although the primary role of expanded polyQ repeats in the disease process is undisputed, the contribution of aggregation and visible inclusion formation to neurodegeneration is not clear (5). Abnormal protein folding and/or proteolytic cleavage of the mutant Htt protein are likely to contribute to this process (1, 3, 26). In addition to the polyQ-repeat diseases, the phenomenon of protein aggregation is a common histopathologic hallmark of many other neurodegenerative diseases including Alzheimer's and Parkinson's diseases (25, 27).
To study pathogenic mechanisms we have developed inducible rat phaeochromocytoma (PC12) cell model systems that recapitulate many of the early events in HD progression including inclusion formation and transcriptional dysregulation (3, 28). One set of inducible cell lines produces high numbers of visible aggregates. Because inclusion formation is an easily assayable phenotype for polyQ-repeat disease and represents a major disease hallmark, a screen designed to monitor disruption of inclusion formation in the presence of chemical compounds has been developed for these lines. The disruption of aggregation in this inducible cell-culture model correlates with the in vivo rescue of neuronal degeneration in Drosophila, thus providing an effective approach for high-throughput screening in cells and validation of candidates in vivo.
Materials and Methods
Plasmid Constructs.
Constructs encode either truncated exon 1 (first 17 aa plus polyQ repeat), complete exon 1 (with proline-rich region), or the first five exons (584 aa) of Htt, contain a range of polyQ repeats encoded by alternating CAG/CAA repeats (25–300 Qs), and are fused in frame to the coding sequence for the enhanced GFP (EGFP) tag at the carboxyl terminus (29, 30). An amino-terminal FLAG epitope tag is present in some constructs. Exons 2–5 (bases 581–2,074, GenBank accession no. L12392) were amplified with gene-specific primers and subcloned into plasmids encoding Htt exon 1. All constructs were subcloned into pIND (Invitrogen) and sequence-verified.
Inducible PC12 Cells.
PC12 cells were stably transfected with a hybrid ecdysone receptor (VgRXR, Invitrogen) and a clonal line, PC12ec, that expresses high levels of the receptor, selected. pIND constructs were stably transfected into PC12ec and selected with G418 (Omega Scientific, Tarzana, CA). Pools and clones were flow-sorted (MoFlo cell sorter, Cytomation, Fort Collins, CO) for maximal EGFP expression (upper 2–5%) after a 12-h pulse of induction with 5 μM ponasterone (PA, Invitrogen). Cells were routinely induced with 5 μM PA or muristerone (MA, Invitrogen). Unless noted otherwise, PC12 cells were maintained in complete media with continued selection. Cell lines were differentiated by growing cells on collagen (Becton Dickinson Biosciences)-coated plates in DMEM with 1% horse serum containing 50 ng/ml nerve growth factor (Harlan Bioproducts, Indianapolis) for 48 h or as indicated. Subclonal cell lines were further selected for a high percentage of cells with aggregates after 48 h by consecutive single-colony selections.
Protein Expression.
Cells (1 × 106) were plated in 10-cm plates, grown overnight, and treated with PA for 48 h before harvesting. Cells were lysed in hypotonic buffer containing 20 mM Hepes, pH 7.5, 5 mM NaCl, 10 mM NaF, 2 mM EDTA, 1% Nonidet P-40, 1 mM sodium orthovanadate, and protease inhibitors (1 mM PMSF/10 μg/ml leupeptin/10 μg/ml aprotinin). Western blotting was performed as described (31).
Immunocytochemistry.
Immunocytochemical analyses of cells expressing Htt–EGFP fusion polypeptides has been described (29). Cells were grown on UV-treated cover slips to ≈50% confluency and induced with 0.25–10 μM MA for 12 h to 6 days, fixed in 2% formaldehyde, and incubated for 2 min with 0.1% Triton X-100 in PBS. Fixed cells were 4′,6-diamidino-2-phenylindole-stained, and tubulin immunoreactivity was detected with a mouse monoclonal anti-α-tubulin antibody at 1:500 (Sigma) followed by secondary FluoroLink Cy3 antibody at 1:2,000 (Amersham Pharmacia). Epifluorescent microscopy was performed on a Zeiss Axioplan II equipped with a Quantix charge-coupled device camera (Photometrics, Tucson, AZ) and SPECTRUM imaging software (Scanalytics, Billerica, MA). Sectioning and confocal immunofluorescence analysis of eye discs was performed as described (32).
Quantitation of Aggregates.
Visual counts of aggregates and EGFP-positive cells were performed by using fluorescent microscopy. At least 300 cells were counted from five to six fields in two independent experiments for each data point (×10 magnification). Aggregation is expressed as the percentage of cells with aggregates versus total number of EGFP-positive cells.
Cell-Viability Assays, Estimation of Neurite Outgrowth, and Proteasome Inhibition.
To evaluate cell death, three methods were used: (i) trypan-blue staining, (ii) DNA fragmentation with either a DNA laddering kit (Trevigen, Gaithersburg, MD) or 4′,6-diamidino-2-phenylindole staining of cells, and/or (iii) Live/Dead kit (Molecular Probes). The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide assays [CellTiter 96 nonradioactive cell proliferation assay (Promega)] and neurite-outgrowth experiments were performed on a subset of the cell lines. Cells with at least one visible process equal to or greater than one cell body were counted as positive for neurite formation.
Chemical Compound Screens in PC12 Cells.
Congo red was solubilized in DMSO at 25 mM, cystamine bitartrate in water at 10 mM, and minocycline hydrochloride in 50/50 (vol/vol) DMSO/water at 20 mM. Compounds (Sigma) were added directly to cell-culture media at final concentrations of 1–50 μM, and aggregation was calculated. Chemical compounds were assessed for cellular toxicity by evaluating cell morphology, Htt expression, expression of endogenous proteins (e.g., Hsp70, nucleolin, p53, and α-tubulin), and cell viability.
Chemical Compound Screens in Drosophila.
Flies were mated at 25°C as described (31). Eggs were collected and transferred into vials at 29°C containing standard food supplemented with the compound to be tested, and adults were transferred to vials containing fresh food every 3 days after eclosion. Eyes were monitored 3, 7, and 10 days after eclosion by using the pseudopupil technique (31). The transgenic polyQ line used was P{UAS-Q48myc}42 (31).
To assay the motor behavior of the flies, 10 flies were placed in a vial, and the time required for half of them to cross a line 6 cm from the bottom was determined. This process was repeated five times on each population sample (33).
Results and Discussion
Development of Ecdysone-Inducible PC12 Cells Expressing Htt with Normal and Expanded polyQ Repeats.
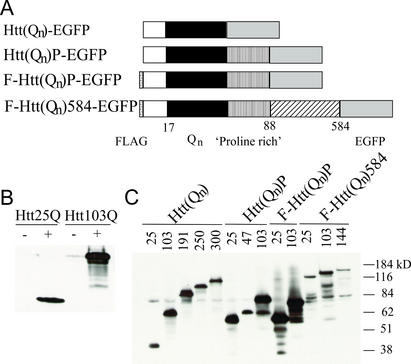
PC12 cells, expressing a hybrid ecdysone receptor, were stably transfected with ecdysone-inducible plasmids encoding epitope-tagged human Htt cDNAs with normal range (25 Qs) and expanded (47–300 Qs) polyQ repeats (Fig. 1A). Basal and induced expression levels of Htt fusion proteins were examined by Western blot analysis (Fig. 1 B and C) and fluorescence microscopy (Fig. 2 A and B). Maximal induction was observed at 5 μM PA after 48 h, with expression detected as early as 12 h. Low basal expression of fusion proteins before the addition of inducer is observed (Fig. 1B, representative Western). Clones and pools of stably transfected cells were selected (Fig. 1C).
Figure 1.
Expression of inducible Htt transgenes in PC12 cells. (A) Expression constructs. All constructs contain a carboxyl-terminal EGFP epitope tag, and some constructs contain an amino-terminal FLAG (F) tag as indicated. Htt(Qn)-EGFP, a truncated exon 1 encoding the first 17 aa of Htt with 25, 103, 191, 250, or 300 Qs; Htt(Qn)P-EGFP, the region of Htt encoded by exon 1 with 25, 47, or 103 Qs containing the proline-rich region (P); F-Htt(Qn)P-EGFP, exon 1 protein (25 or 103 Qs) with an amino-terminal FLAG tag; F-Htt(Qn) 584-EGFP, an amino-terminal FLAG-tagged construct encoding 584 aa of Htt [numbering is consistent with GenBank accession no. P42858 with 23 Qs and including consensus caspase cleavage sites (52)], with 25, 103, or 144 Qs. (B and C) Western blots of total cell lysates. Proliferating cells were induced with 5 μM PA for 48 h, and 100 μg of total protein was electrophoresed on 10% SDS gels, blotted, and incubated with CAG53b antibody. The apparent higher level of the Htt103Q polypeptide may be due to the fact that the antibody preferentially recognizes proteins containing longer polyQ stretches. (B) Htt(Qn)-EGFP cells with 25 or 103 Qs plus and minus PA induction. (C) Pools of cells containing the indicated Htt constructs were induced with 5 μM PA for 48 h. Molecular mass markers are shown to the right of the figure.
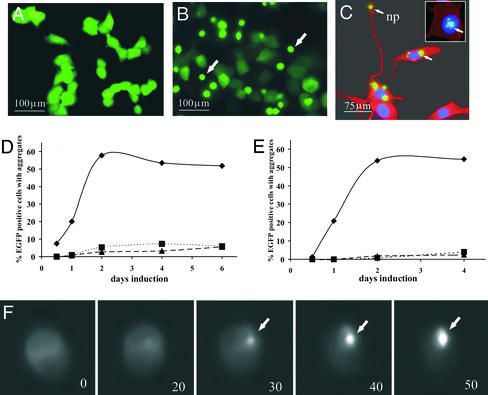
Figure 2.
Characterization of aggregation in inducible PC12 cells expressing Htt transgenes with fluorescent microscopy. (A) Diffuse EGFP fluorescence in pools of proliferating cells expressing normal-length polyQs tagged with EGFP (Htt25Q-EGFP). (B) Aggregates in proliferating cells expressing Htt103Q-EGFP. Cells have dim, diffuse fluorescence of soluble polyQs and bright EGFP fluorescent aggregates. The images shown in A and B were taken of live cells at ×10 magnification. (C) Aggregates in cytoplasm and neuropils and in the nuclei (Inset) of differentiated Htt103Q-EGFP. The images shown in C were taken from fixed cells at high magnification (×40). Aggregation is expressed as the percentage of cells with aggregates versus the total number of EGFP-positive cells. Nuclei were visualized by 4′,6-diamidino-2-phenylindole staining (blue), and cytoplasm (red) was visualized by probing with anti-α-tubulin antibodies as described in Materials and Methods. (D and E) Time course of aggregate appearance in proliferating (D) and differentiated (E) cells. ♦, Htt14A2.5; ▪, F-Htt103QP EGFP; ●, F-Htt103QP EGFP. (F) Kinetics of aggregation in proliferating cells using high-resolution video microscopy. The nucleation site of aggregation became evident after 20 min of filming (frame 20), followed by rapid polymerization, which led to complete aggregation in 30 min (frames 30, 40, and 50). Frames were taken at 10-min intervals. (B, C, and F) Htt14A2.6 clonal line. (D and E) Htt14A2.5 clonal line. The arrows indicate aggregates. np, neuropil.
PC12 cells can be stimulated with growth factors to stop dividing and differentiate into neuronal-like structures. Expression of the Htt exon 1 proteins was compared in proliferating cells and differentiated cells (data not shown). Similar levels of transgene expression were detected by Western blot analysis, a consideration important for the use of differentiated cells in secondary chemical compound screens.
Neurite Outgrowth of PC12 Cells Is Not Altered Significantly by Expression of Expanded Repeat Htt.
To determine whether the expression of the Htt transgene influenced formation of neurites, a subset of cell lines were evaluated for neurite outgrowth. F-Htt25Q-EGFP clones (nos. 17 and 20), F-Htt103QP-EGFP clones (nos. 14 and 19), Htt14A2.5 (described below), and the parental PC12ec line each were induced with PA for 24 h before nerve growth factor treatment. Neurites were quantitated 2 and 6 days after the addition of growth factor (34). No significant difference in neurite formation was detected in the presence of the normal or mutant Htt protein relative to uninduced controls (P > 0.05, unpaired two-tailed Student's t tests). These results are in contrast to studies in other PC12 cell lines that show impairment of neurite outgrowth after expression of mutant Htt proteins constitutively (35) or at 6 days after induction of the mutant protein (34), whereas differentiation appears unaffected in a SCA3 PC12 cell model (22). Because of differences in the origins of the PC12 clonal lines used to make the transgenic lines and differences in levels of mutant Htt expression, a threshold may exist for differentiation effects.
Expanded Repeat Htt Expression Does Not Reduce Viability Significantly.
Several HD cell culture models show reduced viability after expression of mutant Htt proteins (20, 34–36), whereas others show no toxicity after expression of mutant Htt independent of additional cellular insults (21). To test polyQ-mediated toxicity, proliferating or differentiated Htt-expressing lines were evaluated for altered viability after induction with PA. No obvious reduction in viability of these cells was detected in any line after induction of the transgene for 2 or 6 days [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide; P > 0.05, unpaired two-tailed Student's t tests]. Based on gene-expression profiling of the Htt14A2.5 line (described below), classes of genes up-regulated after induction of the transgene include those encoding proteins involved in cellular stress responses (unpublished results). Therefore, it is possible that these PC12 cells can mount a stress response that may aid in protecting them from subsequent cell death. Alternatively, these cells are representative of neurons in early stages of differentiation that can take decades to progress to cell death in HD patients. Mechanisms for differences in susceptible and nonsusceptible lines remain to be elucidated.
Aggregation in Proliferating and Differentiated Cells.
A hallmark feature of polyQ-repeat diseases is the presence of inclusions in the cytosol and nuclei of neurons. Therefore, the cellular distribution of the Htt fusion proteins containing normal-range or expanded glutamine repeats was characterized by fluorescent microscopy in proliferating and differentiated cells. In proliferating cells expressing Htt proteins with normal-range repeats, diffuse EGFP fluorescence was observed throughout the cell (Fig. 2A). Lines expressing 103 Qs contained both diffuse EGFP fluorescence and brightly fluorescent aggregates (high aggregate line, Htt14A2.5, is shown in Fig. 2B). However, in general the percentage of cells containing aggregates was low (1–10%, data not shown). Very few aggregates were observed in lines expressing Htt103Q584-EGFP (<1%), presumably because of the lower expression level and the longer Htt protein sequence. Two subclones of an Htt103Q-EGFP line, Htt14A2 (Htt14A2.5 and Htt14A2.6), were isolated by further selection for a high percentage of cells containing visible aggregates after induction (≈50–80%). Inclusions were detected in both the cytoplasm and nuclei (Fig. 2B). Inclusion formation was analyzed over time in the Htt14A2.5 line and F-Htt103QP-EGFP (clones 14 and 19) lines; small aggregates could be detected 12 h after induction with 5 μM PA and reached a maximum at 36–48 h over a 6-day time course (Fig. 2D).
Similar results were obtained in differentiated cells over a 4-day time course. For each line, nuclear and cytosolic aggregates were observed as well as aggregates in neuritic processes (Fig. 2C shows representation of each type). Aggregates appeared more slowly in differentiated cells than in dividing cells (Fig. 2E) possibly because of a significant increase in the cytoplasmic volume of differentiating cells and consequently a relative decrease in polyQ concentration.
A small fraction of the total aggregates were nuclear in the Htt14A2.5 and Htt14A2.6 clonal lines after a 48-h induction (≈6% of cells with aggregates). As shown in other systems (1), nuclear aggregates appear more slowly than cytoplasmic aggregates. For example, when induction of proliferating Htt14A2.6 cells with 10 μM MA was extended to 72 h, the percentage of nuclear aggregates increased to ≈15%, whereas the percentage of cytoplasmic aggregates did not change significantly. Typically, expanded polyQ aggregates in the cytoplasm formed a single or a few large, bright inclusions (see Fig. 2C in differentiated cells) with little soluble protein remaining, whereas nuclear aggregates frequently appear as round, multiple bodies, often with soluble EGFP fluorescence still visible in the cytoplasm.
By using high-resolution video microscopy, the process of aggregation in proliferating PC12 cells was captured (Fig. 2F). Aggregation of expanded polyQ protein was complete within 30–40 min of initial nucleation. As in transient transfection assays (16, 32, 37), a nucleation step followed by rapid polymerization was observed. Most of the soluble cytoplasmic polyQs aggregated in one inclusion, suggesting either active cellular transport to the aggregation site or, more likely, transient protein interaction of soluble polyQ in the cell. Once nucleation occurred, the kinetics of aggregation appeared to be similar in proliferating and differentiated cells. These results are consistent with studies suggesting that nucleation is the rate-limiting step followed by rapid growth of the aggregate (14, 16, 37).
PC12 Cell-Based Screen for Chemical Inhibitors of Aggregation.
A primary goal in establishing inducible PC12 cell lines was to develop a cell-based assay to screen chemical compounds for their ability to inhibit expanded polyQ protein interactions and/or inclusion formation, particularly in the absence of obvious cellular toxicity. For these screens, a visual aggregation assay was developed. To establish assay conditions and characterize the potential efficacy of a PC12 cell-based system, we tested the effect of three previously characterized chemical inhibitors of polyQ-mediated toxicity upon aggregation: Congo red, cystamine, and minocycline. These chemical inhibitors each target different cellular processes. Congo red, a histological dye commonly used to detect amyloid deposits, inhibits HD aggregate formation in vitro and in cell culture (15) and in HD transgenic hippocampal slice cultures (38). Cystamine, a putative transglutaminase inhibitor, promotes extended survival and improved motor function in HD transgenic mouse models (39, 40). Transglutaminases catalyze a calcium-dependent covalent linkage of glutamine residues to lysine residues. When cystamine treatment is initiated before inclusions are present, it markedly prevents their formation (40). Treatment after inclusions are present does not seem to affect them significantly (39). Minocycline, an inhibitor of caspase 1 and caspase 3 activity and of inducible nitric-oxide synthetase production, delays mortality in a transgenic HD mouse model (41), whereas aggregate formation appears unaffected.
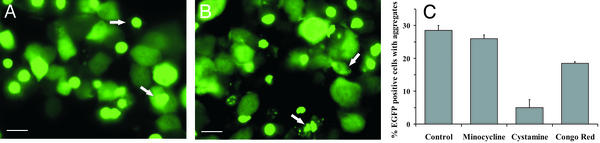
Htt14A2.5 and Htt14A2.6 clonal lines, specifically selected for high levels of aggregation, were used to develop an assay system that would detect both weak and strong effects of compounds after inclusion formation. We reasoned that poor inhibitors of aggregation might fail to significantly block the process under conditions that strongly favor aggregation such as high polyQ-expression levels and/or a high percentage of cells with visible aggregates. Aggregation in these PC12 cell lines depends on inducer concentration: e.g., visible aggregates form in 50–80% of the cells induced for 48 h with a high concentration of MA (5 μM) but at lower concentrations ranges from 0.5 to 1.0 μM, inclusions were detected in only 20% and 40% of the Htt14A2.5 and Htt14A2.6 EGFP-positive cells, respectively. Because the process of aggregation is also time-dependent, aggregate formation was assessed in the presence of compounds at 12-h intervals after induction. Congo red, minocycline, or cystamine bitartrate were added to the media at a concentration range of 1–50 μM together with 1 or 5 μM MA, and the percentage of EGFP-positive cells containing visible aggregates was determined (Fig. 3 A and B). Under conditions of low polyQ expression, Congo-red treatment resulted in an ≈33% inhibition, and cystamine treatment resulted in an ≈67% inhibition of aggregation (Fig. 3C). In contrast, minocycline only slightly inhibited aggregation (Fig. 3C). All three compounds were toxic at higher concentrations (≈100 μM), and therefore effects on aggregation at these concentrations were not determined. At high inducer concentrations, the suppression of aggregation observed with low inducer concentration (e.g., 1 μM MA), is reduced significantly for cystamine, and is no longer observed with minocycline or Congo red (data not shown).
Figure 3.
Chemical compound screen in inducible PC12 cells expressing Htt103Q-EGFP. Aggregation in PC12 cells expressing Htt103Q-EGFP (Htt14A2.5) in the presence of DMSO (A) or treated with 10 μM Congo red in DMSO (B) is shown. The altered morphology of aggregates in the presence of Congo red is indicated with arrows. (Scale bar, 50 μm.) (C) Quantification of aggregation in the Htt14A2.5 cell line with Congo red (10 μM), minocycline hydrochloride (50 μM), and cystamine bitartrate (50 μM). Expression of truncated exon 1 was induced for 48 h with 1 μM MA, and compounds were added to cells simultaneously with an inducer.
For use as a secondary screen, differentiated inducible PC12 cells were tested for visible aggregate production in the presence and absence of cystamine. Inhibition of aggregation proceeded in a concentration-dependent manner with maximal inhibition and minimal toxicity at 50 μM cystamine. These results suggest that the PC12 cell lines represent a sensitive screening tool for compounds that alter the process of aggregation.
Chemical Inhibitors of Aggregation in Cell Culture Can Suppress Pathogenic polyQ-Mediated Phenotypes in Drosophila.
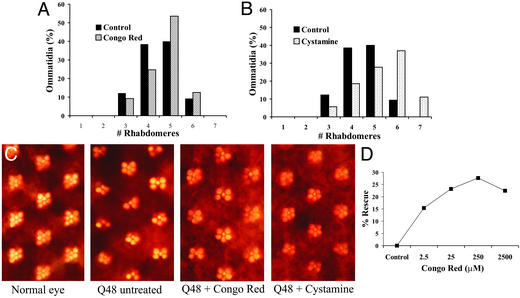
To determine whether the reduction of aggregation observed in cell culture in the presence of chemical inhibitors can translate into a reduction of polyQ pathogenesis in vivo, we fed Congo red or cystamine to Drosophila expressing an expanded polyQ-repeat polypeptide independent of disease-gene context. These peptides are intrinsically cytotoxic and cause neuronal degeneration and reduced viability when expressed in Drosophila neurons (42, 43), and chemical compounds can be fed directly to these flies (31). The neurodegeneration is observed most easily in the fly compound eye, in which photoreceptor cells produce a repeating trapezoidal arrangement of seven visible rhabdomeres (subcellular light-gathering structures) (42–46). As a primary assay, the effect of these compounds on the integrity of photoreceptor neurons in the compound eye was evaluated. The expression of a polypeptide with 48 glutamines (Q48) leads to a progressive loss of rhabdomeres. Congo red and cystamine were tested at concentrations of 2.5–2,500 μM and 1–500 μM, respectively, based on initial toxicity tests in wild-type flies. Both compounds suppressed degeneration of photoreceptor neurons, increasing the average number of visible rhabdomeres of 10-day-old flies from 4.4 to 4.7 for 250 μM Congo red and to 5.3 for 100 μM cystamine (Fig. 4 A–C). At high doses >250 μM Congo red, toxicity is evident, and suppression of pathogenesis is reduced (Fig. 4D). In hippocampal slice cultures from HD transgenic mice (38), Congo red produces a more complex dose-response curve, but again lower concentrations proved more effective than higher ones. Aggregation in eye discs was also evaluated. Minimal effects on inclusion formation in vivo were observed after feeding either Congo red or cystamine, presumably because of the low visible inclusion formation in eye discs of the Q48 flies during larval development and consequent lower sensitivity of the assay. The effect of minocycline was also tested in the Q48 polyQ model; however, minimal improvement was observed at the concentrations tested.
Figure 4.
Chemical compound screen in Q48-expressing Drosophila. (A and B) Quantitation of the number of rhabdomeres per ommatidium in the presence of 100 μM cystamine (A) or 250 μM Congo red (B) in Q48-expressing flies. (C) Photographs of ommatidia from flies with no transgene expression (normal eye) or Q48 expression alone and from flies fed 250 μM Congo red or 100 μM cystamine. (D) Dose response for Congo red. Flies were fed 2.5–2,500 mM Congo red. Percent rescue was calculated as (percent surviving − percent surviving on solvent alone)/(1 − percent surviving on solvent alone).
A second phenotype associated with expanded polyQ expression is a decrease in motor function. This can be assayed by using an “escape” or climbing assay as described (33). Flies with polyQ-induced neuropathology (Q48) needed an average of 26 sec to climb a defined distance up a vial. No rescue of the motor defect in experiments with cystamine was observed, illuminating the complexity of neuronal dysfunction and phenotypic read-out and the importance of performing multiple tests of phenotypic suppression. However, similar flies fed 250 μM Congo red required only 11 sec to climb the same distance (data not shown), reflecting significant rescue of motor function.
Summary
Many laboratories have developed assays and initiated screens for drugs that prevent aggregation of expanded polyQs (47). In vitro assays include those based on filter retention of SDS-insoluble aggregates (14), incorporation of labeled polyQ from solution into preformed polyQ aggregates immobilized on a solid phase (e.g., growth of aggregates) (48, 49), and screens for compounds that selectively recognize structural features of expanded polyQ protein using fluorescence resonance energy-transfer (FRET) readouts (50). Hippocampal slice cultures have been developed to monitor aggregate formation (38) as well as in vivo C. elegans screens (51). Different classes of aggregation inhibitors may result from these screens depending on whether they inhibit initial protein–protein interactions, nucleation of aggregation, or growth of inclusions, and these differences may translate into separable in vivo effects. The disruption of aggregation in the inducible PC12 cells strongly correlates with suppression of neuronal degeneration in vivo, thus providing a rapid and effective method to screen and validate compounds. The monitoring of visible aggregate formation in proliferating cells is well suited for high-throughput screens of chemical compounds, whereas differentiated cells may represent a valid approach to lower-throughput screens and tests of toxicity of chemical compounds in neuron-like cells. Finally, compounds that show promise can be validated further in Drosophila models of polyQ-repeat pathogenesis before testing in mammalian models.
Acknowledgments
We thank E. Wanker for Htt antibody and the Iowa Hybridoma Bank for anti-elav antibody. This work was supported by a Hereditary Disease Foundation Cure HD Initiative grant (to L.M.T. and J.L.M.), a Hereditary Disease Foundation Lieberman Award (to L.M.T.), the Coalition for the Cure of the Huntington's Disease Society of America (to L.M.T. and S.H.), a Hereditary Disease Foundation postdoctoral fellowship (to N.S.), a Human Frontiers Science Program grant (to L.M.T.), a Sierra Foundation grant (to D.E.H.), and National Institutes of Health Grants HD36081 and HD36049 (to J.L.M.), PO1HL66105 (to D.E.H.), and NS35255 and AT00613 (to S.H.). This work was made possible in part through access to Optical Biology Shared Resource of the Cancer Center Support Grant CA-62203 at the University of California (Irvine) and the National Drosophila Stock Center (Bloomington, IN).
Abbreviations
- HD
Huntington's disease
- polyQ
polyglutamine
- Htt
huntingtin
- EGFP
enhanced GFP
- PA
ponasterone
- MA
muristerone
Note Added in Proof.
Recently, new research has demonstrated that Congo red suppresses pathogenic phenotypes in HD transgenic mice (53).
References
- 1.Bates G, Harper P, Jones L. Huntington's Disease. Oxford: Oxford Univ. Press; 2002. [Google Scholar]
- 2.The Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 3.Ross C A. Neuron. 2002;35:819–822. doi: 10.1016/s0896-6273(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 4.Rubinsztein D C. Trends Genet. 2002;18:202–209. doi: 10.1016/s0168-9525(01)02625-7. [DOI] [PubMed] [Google Scholar]
- 5.Zoghbi H Y, Orr H T. Annu Rev Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 6.Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies S W, Bates G P. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 7.Davies S, Turmaine M, Cozens B, DiFiglia M, Sharp A, Ross C, Scherzinger E, Wanker E, Mangiarini L, Bates G P. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 8.DiFiglia M, Sapp E, Chase K O, Davies S W, Bates G P, Vonsattel J P, Aronin N. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 9.Li H, Li S, Cheng A, Mangiarini L, Bates G, Li X. Hum Mol Genet. 1999;7:1227–1236. doi: 10.1093/hmg/8.7.1227. [DOI] [PubMed] [Google Scholar]
- 10.Bates G P, Mangiarini L, Davies S W. Brain Pathol. 1998;8:699–714. doi: 10.1111/j.1750-3639.1998.tb00196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paulson H L. Am J Hum Genet. 1999;64:339–345. doi: 10.1086/302269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor J P, Hardy J, Fischbeck K H. Science. 2002;296:1991–1995. doi: 10.1126/science.1067122. [DOI] [PubMed] [Google Scholar]
- 13.Wanker E. Biol Chem. 2000;381:937–942. doi: 10.1515/BC.2000.114. [DOI] [PubMed] [Google Scholar]
- 14.Scherzinger E, Sittler A, Schweiger K, Heiser V, Lurz R, Hasenbank R, Bates G, Lehrach H, Wanker E E. Proc Natl Acad Sci USA. 1999;96:4604–4609. doi: 10.1073/pnas.96.8.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heiser V, Scherzinger E, Boeddrich A, Nordhoff E, Lurz R, Schugardt N, Lehrach H, Wanker E E. Proc Natl Acad Sci USA. 2000;97:6739–6744. doi: 10.1073/pnas.110138997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajan R, Illing M, Bence N, Kopito R. Proc Natl Acad Sci USA. 2001;98:13060–13065. doi: 10.1073/pnas.181479798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peters P J, Ning K, Palacios F, Boshans R L, Kazantsev A, Thompson L M, Woodman B, Bates G P, D'Souza-Schorey C. Nat Cell Biol. 2002;4:240–245. doi: 10.1038/ncb761. [DOI] [PubMed] [Google Scholar]
- 18.Morley J, Brignull H, Weyers J, Morimoto R. Proc Natl Acad Sci USA. 2002;99:10417–10422. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bence N F, Sampat R M, Kopito R R. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 20.Chai Y, Koppenhafer S, Shoesmith S, Perez M, Paulson H. Hum Mol Genet. 1999;4:673–682. doi: 10.1093/hmg/8.4.673. [DOI] [PubMed] [Google Scholar]
- 21.Ding Q, Lewis J J, Strum K M, Dimayuga E, Bruce-Keller A J, Dunn J C, Keller J N. J Biol Chem. 2002;277:13935–13942. doi: 10.1074/jbc.M107706200. [DOI] [PubMed] [Google Scholar]
- 22.Chai Y, Koppenhafer S L, Bonini N M, Paulson H L. J Neurosci. 1999;19:10338–10347. doi: 10.1523/JNEUROSCI.19-23-10338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warrick J M, Chan H Y, Gray-Board G L, Chai Y, Paulson H L, Bonini N M. Nat Genet. 1999;23:425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- 24.Satyal S H, Schmidt E, Kitagawa K, Sondheimer N, Lindquist S, Kramer J M, Morimoto R I. Proc Natl Acad Sci USA. 2000;97:5750–5755. doi: 10.1073/pnas.100107297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muchowski P J. Neuron. 2002;35:9–12. doi: 10.1016/s0896-6273(02)00761-4. [DOI] [PubMed] [Google Scholar]
- 26.Lunkes A, Lindenberg K, Ben-Haiem L, Weber C, Devys D, Landwehrmeyer G, Mandel J, Trottier Y. Mol Cell. 2002;2:259–269. doi: 10.1016/s1097-2765(02)00602-0. [DOI] [PubMed] [Google Scholar]
- 27.Sherman M Y, Goldberg A L. Neuron. 2001;29:15–32. doi: 10.1016/s0896-6273(01)00177-5. [DOI] [PubMed] [Google Scholar]
- 28.Cha J. Trends Neurosci. 2000;9:387–392. doi: 10.1016/s0166-2236(00)01609-x. [DOI] [PubMed] [Google Scholar]
- 29.Kazantsev A, Preisinger E, Dranovsky A, Goldgaber D, Housman D. Proc Natl Acad Sci USA. 1999;96:11404–11409. doi: 10.1073/pnas.96.20.11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steffan J S, Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu Y Z, Gohler H, Wanker E E, Bates G P, Housman D E, Thompson L M. Proc Natl Acad Sci USA. 2000;97:6763–6768. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steffan J S, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol B L, Kazantsev A, Schmidt E, Zhu Y Z, Greenwald M, et al. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 32.Kazantsev A, Walker H A, Slepko N, Bear J E, Preisinger E, Steffan J S, Zhu Y Z, Gertler F B, Housman D E, Marsh J L, Thompson L M. Nat Genet. 2002;30:367–376. doi: 10.1038/ng864. [DOI] [PubMed] [Google Scholar]
- 33.Ganetzky B, Flanagan J R. Exp Gerontol. 1978;13:189–196. doi: 10.1016/0531-5565(78)90012-8. [DOI] [PubMed] [Google Scholar]
- 34.Wyttenbach A, Swartz J, Kitam H, Thykjaer T, Carmichael J, Bradley J, Brown R, Maxwell M, Schapira A, Orntoft T F, et al. Hum Mol Genet. 2001;10:1829–1845. doi: 10.1093/hmg/10.17.1829. [DOI] [PubMed] [Google Scholar]
- 35.Li S-H, Cheng A L, Li H, Li X-J. J Neurosci. 1999;19:5159–5172. doi: 10.1523/JNEUROSCI.19-13-05159.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lunkes A, Mandel J L. Hum Mol Genet. 1998;7:1355–1361. doi: 10.1093/hmg/7.9.1355. [DOI] [PubMed] [Google Scholar]
- 37.Chen S, Ferrone F, Wetzel R. Proc Natl Acad Sci USA. 2002;99:11884–11889. doi: 10.1073/pnas.182276099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith D L, Portier R, Woodman B, Hockley E, Mahal A, Klunk W E, Li X-J, Wanker E E, Murray K D, Bates G P. Neurobiol Dis. 2001;8:1017–1026. doi: 10.1006/nbdi.2001.0438. [DOI] [PubMed] [Google Scholar]
- 39.Karpuj M V, Becher M W, Springer J E, Chabas D, Youseff S, Pedotti R, Mitchell D, Steinman L. Nat Med. 2002;8:143–149. doi: 10.1038/nm0202-143. [DOI] [PubMed] [Google Scholar]
- 40.Dedeoglu A, Kubilus J K, Jeitner T M, Matson S A, Bogdanov M, Kowall N W, Matson W R, Cooper A J, Ratan R R, Beal M F, et al. J Neurosci. 2002;22:8942–8950. doi: 10.1523/JNEUROSCI.22-20-08942.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen M, Ona V, Li M, Ferrante R, Fink K, Zhu S, Bian J, Guo L, Farrell L, Hersch S, et al. Nat Med. 2000;7:797–801. doi: 10.1038/77528. [DOI] [PubMed] [Google Scholar]
- 42.Marsh J L, Walker H, Theisen H, Zhu Y-Z, Fielder T, Purcell J, Thompson L M. Hum Mol Genet. 2000;9:13–25. doi: 10.1093/hmg/9.1.13. [DOI] [PubMed] [Google Scholar]
- 43.Kazemi-Esfarjani P, Benzer S. Science. 2000;287:1837–1840. doi: 10.1126/science.287.5459.1837. [DOI] [PubMed] [Google Scholar]
- 44.Fernandez-Funez P, Nino-Rosales M L, de Gouyon B, She W C, Luchak J M, Martinez P, Turiegano E, Benito J, Capovilla M, et al. Nature. 2000;408:101–106. doi: 10.1038/35040584. [DOI] [PubMed] [Google Scholar]
- 45.Jackson G R, Salecker I, Dong X, Yao X, Arnheim N, Faber P W, MacDonald M E, Zipursky S L. Neuron. 1998;21:633–642. doi: 10.1016/s0896-6273(00)80573-5. [DOI] [PubMed] [Google Scholar]
- 46.Warrick J M, Paulson H L, Gray-Board G L, Bui Q T, Fischbeck K H, Pittman R N, Bonini N M. Cell. 1998;93:939–949. doi: 10.1016/s0092-8674(00)81200-3. [DOI] [PubMed] [Google Scholar]
- 47.Heemskerk J, Tobin A J, Bain L J. Trends Neurosci. 2002;25:494–496. doi: 10.1016/s0166-2236(02)02236-1. [DOI] [PubMed] [Google Scholar]
- 48.Berthelier V, Hamilton J B, Chen S, Wetzel R. Anal Biochem. 2001;295:227–236. doi: 10.1006/abio.2001.5217. [DOI] [PubMed] [Google Scholar]
- 49.Heiser V, Engemann S, Brocker W, Dunkel I, Boeddrich A, Waelter S, Nordhoff E, Lurz R, Schugardt N, Rautenberg S, et al. Proc Natl Acad Sci USA. 2002;99, Suppl. 4:16400–16406. doi: 10.1073/pnas.182426599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hughes R E, Olson J M. Nat Med. 2001;7:419–423. doi: 10.1038/86486. [DOI] [PubMed] [Google Scholar]
- 51.Parker J, Connolly J, Wellington C, Hayden M, Dausset J, Neri C. Proc Natl Acad Sci USA. 2001;98:13318–13323. doi: 10.1073/pnas.231476398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wellington C L, Ellerby L M, Gutekunst C A, Rogers D, Warby S, Graham R K, Loubser O, van Raamsdonk J, Singaraja R, Yang Y Z, et al. J Neurosci. 2002;22:7862–7872. doi: 10.1523/JNEUROSCI.22-18-07862.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanchez I, Mahlke C, Yuan J. Nature. 2003;421:373–379. doi: 10.1038/nature01301. [DOI] [PubMed] [Google Scholar]