Abstract
In an effort to identify tumor suppressor gene(s) associated with the frequent loss of heterozygosity observed on chromosome 6q25–q27, we constructed a contig derived from the sequences of bacterial artificial chromosome/P1 bacteriophage artificial chromosome clones defined by the genetic interval D6S1581–D6S1579–D6S305–D6S1599–D6S1008. Sequence analysis of this contig found it to contain eight known genes, including the complete genomic structure of the Parkin gene. Loss of heterozygosity (LOH) analysis of 40 malignant breast and ovarian tumors identified a common minimal region of loss, including the markers D6S305 (50%) and D6S1599 (32%). Both loci exhibited the highest frequencies of LOH in this study and are each located within the Parkin genomic structure. Whereas mutation analysis revealed no missense substitutions, expression of the Parkin gene appeared to be down-regulated or absent in the tumor biopsies and tumor cell lines examined. In addition, the identification of two truncating deletions in 3 of 20 ovarian tumor samples, as well as homozygous deletion of exon 2 in the lung adenocarcinoma cell lines Calu-3 and H-1573, supports the hypothesis that hemizygous or homozygous deletions are responsible for the abnormal expression of Parkin in these samples. These data suggest that the LOH observed at chromosome 6q25–q26 may contribute to the initiation and/or progression of cancer by inactivating or reducing the expression of the Parkin gene. Because Parkin maps to FRA6E, one of the most active common fragile sites in the human genome, it represents another example of a large tumor suppressor gene, like FHIT and WWOX, located at a common fragile site.
Tumorigenesis is the result of a multistep process resulting in genetic alterations that drive the progressive transformation of normal cells into malignant derivatives (1). Tumor suppressor genes (TSGs) are defined as genetic elements whose loss or mutational inactivation allows cells to acquire a neoplastic phenotype (2). Frequent loss of heterozygosity (LOH) within genetically defined chromosomal regions is considered an indication of the presence of a putative TSG (3, 4). Deletion in the long arm of chromosome 6 is associated with several solid tumors, including carcinomas of the ovary (5, 6), breast (7), kidney (8), lung (9), melanoma (10), and hematological cancers such as acute lymphoblastic leukemia (11), Burkitt's lymphoma (12), and non-Hodgkin's B cell lymphoma (13). In addition, microcell-mediated transfer of human chromosome 6 has been shown to suppress tumorigenicity in two breast cancer cell lines (14) and to reduce the tumorigenicity of several melanoma cell lines (15). Similarly, introduction of an intact human chromosome 6 into the breast cancer cell line MCF-7 (14) and into the mouse BK virus-transformed cell line pRPcT1ss1 restores their ability to senesce (16). All of these studies provide support for the existence of a TSG(s) or a senescence-related gene(s) on chromosome 6q.
LOH analysis of the long arm of chromosome 6 identified several regions of loss: 6q21–q23 (17), 6q25.1–q25.2 (18), and 6q25–q27 (6, 19). Moreover, deletions at 6q27 are present in benign ovarian tumors (20), suggesting that alterations in one or more genes mapping to this region represent an early event in ovarian tumorigenesis. In this study we performed LOH analysis to define regions of allelic loss at human chromosome 6q25–q27 in breast and ovarian carcinomas. Physical mapping combined with LOH analysis identified Parkin as a gene that is frequently targeted by hemizygous deletion and inactivation in both malignant tumors and tumor-derived cell lines.
Parkin is the target in individuals affected by autosomal recessive juvenile parkinsonism (AR-JP) (21). Functional analysis has shown Parkin activity to be that of an E3 ubiquitin ligase, the specific involvement of which in protein degradation is under study (22).
Furthermore, we report evidence for homozygous deletions (HDs) at the Parkin genomic locus in two tumor-derived cell lines. These data suggest that the Parkin gene at 6q25–q27 is a strong candidate TSG and may play an important role in the development of mammary and ovarian tumors.
Materials and Methods
Tumor Specimens and Cell Lines.
As part of a Fox Chase Cancer Center Institutional Review Board-approved protocol, tumor samples and matching peripheral blood and/or normal adjacent tissue specimens were obtained from patients undergoing cancer surgery. Tumor and normal tissue specimens were snap-frozen, while blood was separated and DNA was isolated from lymphocytes. A portion of the tissue specimens was assessed for tumor content by histology, and only tissue with >60% tumor cells was used. All cell lines were purchased from the American Type Culture Collection. Cells were maintained in either McCoy's 5A medium containing 10% FBS, or DMEM containing 10% FBS (GIBCO) and each was supplemented with 100 μg/ml gentamicin (BioWhittaker).
LOH Analysis.
Matched normal and tumor genomic DNA pairs were analyzed for LOH by the amplification of dinucleotide or tetranucleotide repeats by using fluorescently end-labeled primers derived from the chromosome 6q25–q27 region. Primer sequences for each highly polymorphic (>60%) microsatellite marker (Table 1) are available at the National Center for Biotechnology Information database (www.ncbi.nlm.nih.gov/). PCRs and fragment analysis were performed as described (25). LOH was defined for those samples that had XLOH values <0.7 or an allelic loss of ≈40%. Allelic loss was scored by two independent observers and confirmed at least twice for each marker.
Table 1.
LOH frequency at the 6q25–q27 locus in breast and ovarian tumors
| Marker, percent informative cases | LOH, % (cases showing LOH/informative cases)
|
||
|---|---|---|---|
| Ovarian tumors | Breast tumors | Total | |
| D6S1581 (93) | 33 (6/18) | 21 (4/19) | 27 (10/37) |
| D6S1579 (68) | 43 (6/14) | 8 (1/13) | 26 (7/27) |
| D6S305 (95) | 45 (9/20) | 56 (10/18) | 50 (19/38) |
| D6S1599 (70) | 53 (8/15) | 8 (1/13) | 32 (9/28) |
| D6S1008 (65) | 8 (1/12) | 0 (0/14) | 3 (1/26) |
Northern Blotting.
Multiple-tissue Northern blots and normal tissue poly(A)+ RNA were purchased from CLONTECH. Poly(A)+ RNA from cultured cell lines was extracted by using the MACS mRNA isolation kit (Miltenyi Biotec, Auburn, CA) according to the manufacturer's instructions. Four micrograms of each poly(A)+ RNA was electrophoretically resolved on denaturing 0.8% agarose gels and transferred to a nylon membrane in 20× SSC. Blots were hybridized with a 1.5-kb cDNA probe labeled with [α-32P]dCTP by random priming (Stratagene). The cDNA probe was synthesized by RT-PCR using forward primer ParkFw (5′-CCAGTGACCATGATAGTGTT-3′) and reverse primer ParkORFRw (5′-TGAAGGTAGACACTGGGTAT-3′). Prehybridization and hybridization were carried out as described (25). All Northern blots were stripped in 0.5% SDS at 100°C and reprobed with a β-actin control probe (CLONTECH).
Western Blotting.
Cell proteins were extracted with a RIPA (150 mM NaCl/1% Triton X-100/1% sodium deoxycholate/0.1% SDS/50 mM Tris · HCl, pH 7.5/2 mM EDTA) lysis buffer and were quantified by using the BCA kit (Pierce). Normal human tissue lysates were purchased (DNA Technologies, Gaithersburg, MD). Fifty micrograms of cellular proteins was size fractionated on Tris-glycine SDS/12%/PAGE gels and electrotransferred onto nitrocellulose membranes (Bio-Rad). The membranes were blocked overnight in 5% nonfat dried milk in TBST (50 mM Tris · HCl/150 mM NaCl/0.1% Tween 20, pH 7.5) and incubated with a polyclonal Parkin antibody (Cell Signaling Technology, Beverly, MA) for 1–3 h in blocking solution. Expression was detected by using ECL Western blotting detection reagents (Amersham Pharmacia) according to the manufacturer's protocol. Subsequently, membranes were stripped and incubated with the MAB-374 antibody for human GAPDH (Chemicon) to verify equal protein loading.
Mutation Analysis.
Total RNA was isolated from 5 × 106 cells or homogenized tumor material by using the TRI Reagent (Molecular Research Center, Cincinnati) and was reverse transcribed with the SuperScript First-Strand Synthesis System (Invitrogen), each according to the manufacturer's instructions. The PCRs were performed as described (24). PCR products were resolved on TBE/2% agarose and gel purified by using the Qiagen gel extraction kit (Qiagen, Valencia, CA) for direct sequencing. Primers designed to amplify three overlapping fragments of the Parkin coding sequence were used for amplification and sequencing. Primer pairs are as follows: ParkFw/Park659Rw (5′-AACATCATCCCAGCAAGATG-3′); Park508Fw (5′-GTCCAGCAGGTAGATCAATC-3′)/Park1046Rw (5′-GTACCGGTTGTACTGCTCTT-3′); and Park929Fw (5′-GTTTGTTCACGACCCTCAAC-3′)/ParkORFRw. Primers sequences specific for each Parkin exon are available on request. DNA sequencing was carried out as described (25) and all sequence analysis and alignments were performed using the sequencer program (Gene Codes, Ann Arbor, MI).
Semiquantitative RT-PCR.
Total RNA obtained from frozen tumor biopsies was reverse transcribed as described above. Five microliters of cDNA was amplified in a 25-μl reaction mixture containing 0.04 unit of Taq DNA polymerase (Roche Applied Science), 0.4 mM dNTPs, 1× buffer containing 1.5 mM MgCl2 (Roche Applied Science), 0.18 μM each primer specific for Parkin (Park Fw/Park659Rw) and 0.03 μM primers specific for β-actin (Applied Biosystems).
PCR amplifications were performed for 25 cycles (94°C for 30 sec, 58°C for 30 sec, and 72°C for 30 sec) followed by an extension of 7 min at 72°C. Primers (ParkFw/Park659Rw) were designed to specifically amplify transcripts of the human Parkin gene. In addition, a set of primers specific for the β-actin gene (Applied Biosystems) was included in each reaction as an internal control. PCR products were separated on 2.0% agarose gels and transferred by blotting to nylon membranes under standard conditions (22). Membranes were hybridized with a fragment corresponding to the full-length Parkin cDNA labeled with [α-32P]dCTP by random priming, using the Prime It II labeling kit (Stratagene). In addition, blots were rehybridized with a β-actin-specific probe. Parkin and β-actin RT-PCR products were quantified with the Personal Densitometer SI (Molecular Dynamics) and imagequant software, Version 5 (Molecular Dynamics). The relative values of Parkin expression were determined by calculating the ratio of normalized expression levels with that of the corresponding normal tissue. The criteria for overexpression or reduced expression was a ratio of >1.5 or <0.5, respectively.
Southern Blotting.
Genomic DNA was extracted according to standard procedures (25) and normal human genomic DNA was purchased from Roche. Seven micrograms of each genomic DNA was digested overnight with restriction endonucleases EcoRI or BglII (Roche Applied Science) and was electrophoresed on a 0.7% agarose gel in 1× TBE. Gels were transferred onto Hybond N+ membranes (Amersham Pharmacia) and were UV crosslinked and hybridized with either a 1.5-kb Parkin cDNA probe or a Parkin exon 2-specific cDNA probe. The exon 2-specific probe was generated by PCR of normal genomic DNA by using the primers Ex2Fw (5′-ATGTTGCTATCACCATTTAAGGG-3′) and Ex2Rw (5′-AGATTGGCAGCGCAGGCGGCATG-3′). PCR fragments were gel purified and labeled with [α-32P]dCTP as described above. Membranes were prehybridized and hybridized in the PerfectHyb-Plus solution (Sigma), washed, and exposed overnight to X-Omat autoradiographic film (Kodak) at −80°C.
Results
Mapping of Chromosome 6q25–q27 Region.
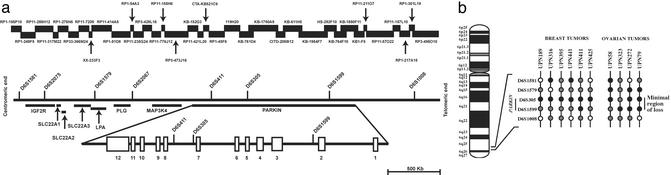
Sequences from bacterial artificial chromosome/P1 bacteriophage artificial chromosome clones mapping to chromosome 6q25–q27 were obtained by searching the National Center for Biotechnology Information database (www.ncbi.nlm.nih.gov), allowing us to assemble a 3.5-Mb sequence contig encompassing a genomic interval defined by eight microsatellite markers (Fig. 1a and Table 1). No gaps are present within the contig and the localization of each microsatellite marker was verified by sequence alignment. blast (www.ncbi.nlm.nih.gov/blast) analysis of the assembled contig identified the position of both known and predicted genes relative to the location of each microsatellite marker. To date, the following eight genes have been mapped to this chromosomal region: mannose-6-phosphate/insulin-like grow factor 2 receptor (M6P/IGF2R) (26), apolipoprotein-A (LPA) (27), plasminogen (PLG) (28), mitogen-activated protein kinase 4 (MAP3K4) (29), Parkin (30), and solute carrier 22 members 1, 2, and 3 (SLC22A1, SLC22A2, and SLC22A3, respectively) (31). Interestingly, the Parkin locus spans a large genomic region of ≈1.5 Mb and consists of 12 exons and large introns with an average size of 125 kb (32). Markers D6S411 and D6S305 localize within Parkin intron 7, whereas marker D61599 localizes within Parkin intron 2 (Fig. 1a).
Figure 1.
Physical map of the D6S1581–D6S1008 genomic interval. (a) All microsatellite markers and bacterial artificial chromosome/P1 bacteriophage artificial chromosome clone sequences were obtained from the National Center for Biotechnology Information database. The positions and alignment of known genes identified in this region and the markers used for LOH analysis are indicated. The Parkin genomic structure according to ref. 32 is also shown (Lower). (b) Summary of allelic loss in six breast and four ovarian malignant tumors with partial deletions at 6q25–q27. Each vertical line represents a single case and black, gray, and white circles represent LOH, retention of heterozygosity, and noninformative results, respectively. The shared minimal region of loss among informative cases is indicated on the right side between markers D6S1599 and D6S305.
LOH Analysis and Identification of a Common Minimal Region of Loss at 6q25–q27.
Five polymorphic microsatellite markers used to anchor our sequence contig of the 6q25–q27 region (Fig. 1a) were used to test for LOH in 20 breast and 20 ovarian normal/tumor DNA pairs. Overall, 11 of 20 (55%) breast and 11 of 20 (55%) ovarian samples showed LOH in at least one locus in the region examined (Fig. 1b and Table 1). The number of markers at which a single tumor displayed LOH ranged from one to four, whereas none of the tumors demonstrated LOH at all loci. The percentage of LOH across each of the five markers ranged from 0% (D6S1008) to 56% (D6S305) in breast tumors, 8% (D6S1008) to 53% (D6S1599) in ovarian tumors, and 3% (D6S1008) to 50% (D6S305) in the combined analysis. Tumors showing LOH at one or more loci, but retaining heterozygosity at flanking markers, were used to define a shared minimal region of loss between markers D6S305 and D6S1599 (Fig. 1b). Partial loss on the chromosome 6q25–q27 region was detected in six breast and four ovarian tumors with LOH in at least one locus in the D6S305–D6S1599 interval. Ovarian tumors unique patient number (UPN) 79 and UPN 272 defined the centromeric and telomeric boundaries of the deleted region, respectively, and case UPN 323 allowed us to identify an even smaller common region of LOH around marker D6S1599. Results from the LOH analysis of the breast tumors showed a commonly deleted region between markers D6S305 and D6S1599 defined by tumors UPN 425 (centromeric end) and UPN 411 (telomeric end), and this region of loss was more precisely defined by sample UPN 395, which demonstrated LOH at marker D6S305 while retaining heterozygosity at the D6S1579 and D6S1599 loci.
Parkin Expression Analysis.
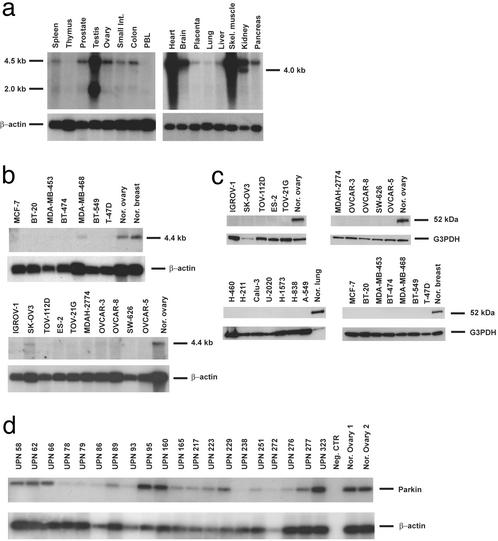
Expression levels of Parkin mRNA were analyzed in 24 tumor-derived cell lines and in a panel of normal tissues (Fig. 2 a and b). A major transcript of 4.5 kb was detected in all human tissues examined, with the exceptions of thymus and peripheral blood lymphocytes. In addition, two smaller transcripts of 2.0 and 4.0 kb were identified in testis and kidney, respectively. mRNA from tumor-derived cell lines exhibited various levels of Parkin expression from absent, 18 of 24 (75%) cases, to almost normal levels (MDA-MB-468, SK-OV-3, and H-211), relative to each normal tissue. One exception was the lung adenocarcinoma cell line, H-460, where an elevated amount of the Parkin transcript was observed. Because of the recent characterization of the Parkin promoter (32) and the abnormal epigenetic regulation of gene expression observed in cancer (33), we examined the methylation status of the Parkin gene. We found, however, no correlation with methylation of the Parkin promoter and gene expression (data not shown).
Figure 2.
Northern and Western analysis of Parkin expression in normal human tissues and tumor-derived cell lines. (a and b) Multiple normal tissue blots (CLONTECH; a) and poly(A)+ RNA (b) from several tumor-derived cell lines were hybridized with a 1.5-kb cDNA probe containing the entire Parkin ORF. (c) Western blot analysis of Parkin protein levels in tumor-derived cell lines. (d) Parkin expression analysis in malignant ovarian and breast tumors by semiquantitative RT-PCR. Representative results are shown for each.
To investigate whether the expression of Parkin mRNA levels correlates with the amount of cellular protein, we analyzed lysates extracted from tumor-derived cell lines and normal tissues by Western blotting (Fig. 2c). All normal tissues exhibited a 52-kDa band corresponding to the predicted Parkin protein. None of the breast, ovarian, and lung cell lines analyzed was found to contain this isoform as shown in Fig. 2c. In addition, we analyzed Parkin expression in a series of ovarian and breast tumors by semiquantitative RT-PCR (Fig. 2d). Twenty-seven of 38 (71%) tumor tissues showed decreased or no expression of Parkin transcript relative to normal ovary or breast tissue. Nine of 38 samples (24%) showed nearly identical levels of expression, whereas 2 of 38 (5%) cases demonstrated an increased level of expression (Table 2).
Table 2.
Semiquantitative RT-PCR analysis of Parkin mRNA expression in breast and ovarian cancer
| Case no. (breast) | TB/NB | Case no. (ovary) | TOV/NOV |
|---|---|---|---|
| UPN 316 | 0.1 | UPN 58 | 0.6 |
| UPN 411 | ND | UPN 62 | 1.3 |
| UPN 405 | 0.6 | UPN 66 | 0.3 |
| UPN 73 | ND | UPN 78 | ND |
| UPN 307 | ND | UPN 79 | ND |
| UPN 304 | 0.1 | UPN 86 | ND |
| UPN 413 | ND | UPN 89 | 0.6 |
| UPN 409 | 0.5 | UPN 93 | ND |
| UPN 407 | 0.2 | UPN 95 | 4.0 |
| UPN 425 | ND | UPN 160 | 3.0 |
| UPN 410 | ND | UPN 165 | 0.8 |
| UPN 404 | 0.1 | UPN 217 | 0.7 |
| UPN 395 | 0.2 | UPN 223 | 0.9 |
| UPN 392 | 0.1 | UPN 229 | 1.0 |
| UPN 441 | ND | UPN 238 | ND |
| UPN 330 | 0.1 | UPN 251 | 0.2 |
| UPN 196 | ND | UPN 272 | ND |
| UPN 189 | 0.3 | UPN 276 | 0.1 |
| UPN 277 | 0.5 | ||
| UPN 323 | 1.0 |
Expression levels of Parkin in tumor cDNAs were calculated as described in Materials and Methods. ND, not detected; TB/NB, mRNA expression in breast tumors related to normal breast tissue. TOV/NOV, mRNA expression in ovarian tumors related to normal ovarian tissue.
Mutation Analysis.
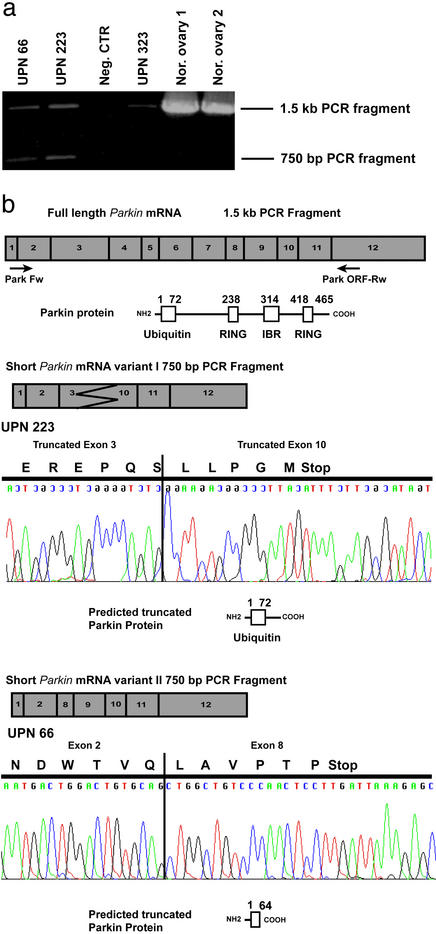
RT-PCR followed by direct sequencing of the full-length Parkin cDNA was performed in all of the tumor-derived cell lines and in each of the invasive breast tumors and ovarian adenocarcinomas available for analysis. Aberrant RT-PCR products were identified in 3 of 20 (15%) ovarian adenocarcinomas (Fig. 3a). In each of these cases, the aberrant transcript was accompanied by the wild-type RT-PCR product, which may be derived from normal cells present in the tumor samples. Sequence analysis of the aberrant transcripts revealed the absence of sequences between exons 2 and 10, whereas the expected products did not show any abnormalities. Although the size of the aberrant bands was apparently similar in all of the cases analyzed, two different types of altered transcripts were observed. The more common of the two is a combination of a partial deletion of exon 3, a deletion of exons 4–9, and a partial deletion in the 5′ of exon 10. The second aberrant transcript contained a deletion of exons 2–7. Both alterations result in the disruption of the Parkin ORF, causing premature termination of the predicted protein sequence (Fig. 3b).
Figure 3.
Identification of tumor-specific Parkin transcripts. (a) PCR amplification of tumor and normal ovarian cDNA. Samples UPN 66 and UPN 223 possess both wild-type and aberrant transcripts, whereas UPN 323 and normal ovaries 1 and 2 possess only wild-type Parkin. (b) Sequence analysis of aberrant Parkin transcripts. A schematic representation of each variant is shown, along with the resulting truncated protein isoform. Also indicated is the location of primer pairs used for both analyses.
Identification of Hemizygous and Homozygous Deletions in the Parkin Gene Locus.
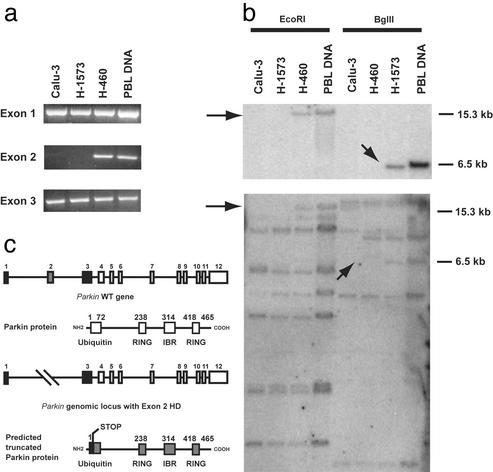
Our initial attempts to amplify the full-length Parkin transcript in the lung adenocarcinoma cell lines Calu-3 and H-1573 were unsuccessful. Subsequently, RT-PCR using three overlapping primer sets specific for the 5′ and 3′ regions of the Parkin coding sequence resulted in the failure to amplify nucleotides 93–659 (data not shown).
To further analyze what appeared to be a deletion at the 5′ end of the Parkin genomic structure, primer pairs specific for each Parkin exon were used for PCR. Products for each exon were detected, with the exception being exon 2 in both the Calu-3 and H-1573 genomic DNA (Fig. 4a). The deletion of exon 2 was confirmed by Southern analysis (Fig. 4b). The absence of the exon 2-specific EcoRI and BglII fragments indicates that it is homozygously deleted in these cell lines, thereby resulting in a frameshift mutation causing the predicted premature termination of the parkin-encoding sequence (Fig. 4c).
Figure 4.
Analysis and mapping of HDs in the Parkin locus. (a) PCR analysis of exons 1–3 in Calu-3, H-1573, and H-460 cell lines and in normal human genomic DNA. (b) Southern analysis of Parkin gene in DNA digested with EcoRI or BglII. Blots were hybridized with either a Parkin exon 2-specific probe (Upper) or the full-length Parkin cDNA (Lower). Arrows indicate the absence of the expected 15.3- and 6.5-kb EcoRI and BglII fragments, respectively. PBL, human peripheral blood leukocyte. (c) A schematic representation of exon 2 deletions observed in the lung adenocarcinoma cell lines, Calu-3 and H-1573.
Discussion
Chromosomal region 6q25–q27 has frequently undergone deletions in a wide spectrum of human neoplasms, such as melanoma, ovarian cancer, breast cancer, non-Hodgkin's B cell lymphoma, and several others (9, 10, 13, 17, 19). Because of the recent availability of the human genome draft sequence, we were able to construct a 3.5-Mb bacterial artificial chromosome/P1 bacteriophage artificial chromosome-derived contig encompassing the D6S1581 and D6S1008 interval at human chromosome 6q25–q27 (Fig. 1a). Sequence analysis led to the identification of eight genes aligning to the consensus sequence of this contig, including the previously identified E3 ubiquitin ligase gene, Parkin. Parkin was of particular interest because of its large 1.5-Mb genomic structure covering ≈43% of this chromosomal segment and has been shown to be the target of several characterized familial deletions (21).
In our search for a putative TSG at chromosome 6q25–q27, we genotyped DNA from 20 malignant breast tumors, 20 ovarian tumors, and corresponding nontumor tissues by using five polymorphic microsatellite markers used to anchor our sequence-based contig of this region. Our analysis revealed deletions at the 6q25–q27 locus in 55% of cases analyzed, with percentages of LOH ranging from 0% (D6S1008) to 56% (D6S305) and from 8% (D6S1008) to 53% (D6S1599) in breast and ovarian samples, respectively. Lower frequencies were observed in the loci D6S1579 (26%) and D6S1008 (3%), whereas 10 informative cases exhibiting partial deletions across the D6S1579–D6S1599 interval allowed us to identify a common minimal region of loss between markers D6S305 (50%) and D6S1599 (32%). Because both of these markers are localized within introns 2 and 7, respectively, of the Parkin locus, it prompted us to consider this gene as a likely candidate TSG.
Subsequently, analysis of Parkin gene expression in a variety of human cancers, including several tumor-derived cell lines and malignant ovarian and breast tumors, found transcript levels to be reduced or absent in >70% of the samples examined. Mutation analysis of the tumor-derived cell lines identified only a few previously reported polymorphisms (data not shown). However, aberrant transcripts were found in 3 of 20 (15%) ovarian tumor cDNAs. Sequence analysis of these tumor-specific transcripts revealed two different types of alterations with partial or complete loss of exons 2–10 (Fig. 3b). Interestingly, the common region of loss in our LOH analysis defined by the marker D6S305 and D6S1599 involves Parkin exons 2–10, suggesting that these tumor-specific transcripts are the result of genomic deletions. Translation of each of these altered transcripts, if it occurs, will result in a prematurely terminated parkin protein that does not contain the RING and in-between rings functional domains (34, 35; Fig. 3c).
Biallelic inactivation of a TSG can occur by several mechanisms, including chromosomal deletion of both alleles. Although uncommon, HD is occasionally observed in primary tumors and cell lines from a variety of malignancies. HDs usually span relatively short genomic regions and have been instrumental in the identification of several TSGs, such as FHIT, RB1, and WT1 (36–38). As our LOH studies suggest, HDs may play a role in the inactivation of Parkin gene expression as well as the cause of the several aberrant transcripts observed in the malignant ovarian and breast tumors analyzed. In addition, we have identified HD of Parkin exon 2 in the Calu-3 and H-1573 lung adenocarcinoma cell lines (Fig. 4 a and b). As expected, Parkin expression was absent in these tumor-derived cell lines. In those samples without the above-mentioned alterations, loss of expression may depend either on unidentified alterations of the genomic structure or on alternative epigenetic mechanisms (33, 39). Although the Parkin gene promoter has been described (32), hypermethylation does not seem to account for the silencing of this gene (data not shown). The Parkin gene was initially identified in four Japanese families affected by autosomal juvenile recessive parkinsonism (AR-JP) (30). In AR-JP-affected individuals, Parkin is inactivated by point mutation or more frequently, by exon deletions or amplification (21). Our data suggest that in cancer, Parkin has undergone intragenic deletions, which may contribute to tumor initiation and development.
Parkin has been classified as a RING finger E3 ligase that ubiquitinates itself (35) and is involved in the degradation of the synaptic vesicle-associated protein CDCrel-1 (22) and of the membrane protein Pael-R (40). More recently, a physical and functional interaction between Parkin and CHIP, which contains a multiubiquitin chain-assembling enzyme E4-like activity, has been reported (41). Moreover other E3 RING ligases, such as MdM2 (42) and Parc (43), which has strong structural similarity to parkin, have been implicated in the regulation of p53 function by controlling p53 nuclear export and ubiquitination (42, 44) and subcellular localization (43), respectively. Therefore, to elucidate the role of Parkin in tumorigenesis, it will be necessary to identify substrates of Parkin E3 ubiquitinating activity and their potential relationship to apoptosis and/or cellular proliferation.
Interestingly, chromosomal region 6q25–q27 contains one of the most active chromosomal fragile sites of the human genome, FRA6E (45). Active fragile sites such as FRA3B and FRA16D have been shown to colocalize with the TSGs FHIT and WWOX, respectively (36, 46). In addition, the FHIT and WWOX genetic loci each cover a large genomic region; FHIT is ≈1.8 Mb (47, 48), whereas WWOX is >1.0 Mb (23). Thus, it seems plausible that many common fragile sites may harbor large TSGs involved in a large number of human malignancies.
Our findings suggest that Parkin is a strong candidate TSG located at human chromosome 6q25–q27 and that its reduced expression and inactivation by hemizygous or homozygous deletion may play an important role in ovarian and breast carcinogenesis and other human tumors.
Acknowledgments
We thank Jean Letofsky for expert technical assistance. This work was supported by National Cancer Institute Grants to A.K.G. and C.M.C.
Abbreviations
- TSG
tumor suppressor gene
- LOH
loss of heterozygosity
- HD
homozygous deletion
- UPN
unique patient number
References
- 1.Hanahan D, Weinberg R A. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Hinds P W, Weinberg R A. Curr Opin Genet Dev. 1994;4:135–141. doi: 10.1016/0959-437x(94)90102-3. [DOI] [PubMed] [Google Scholar]
- 3.Lin J C, Scherer S W, Tougas L, Traverso G, Tsui L C, Andrulis I L, Jothy S, Park M. Oncogene. 1996;13:2001–2008. [PubMed] [Google Scholar]
- 4.Knudson A G. Proc Natl Acad Sci USA. 1993;90:10914–10921. doi: 10.1073/pnas.90.23.10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saito S, Sirahama S, Matsushima M, Suzuki M, Sagae S, Kudo R, Saito J, Noda K, Nakamura Y. Cancer Res. 1996;56:5586–5589. [PubMed] [Google Scholar]
- 6.Tibiletti M G, Bernasconi B, Furlan D, Riva C, Trubia M, Buraggi G, Franchi M, Bolis P, Mariani A, Frigerio L, et al. Cancer Res. 1996;56:4493–4498. [PubMed] [Google Scholar]
- 7.Orphanos V, McGown G, Hey Y, Boyle J M, Santibanez-Koref M. Br J Cancer. 1995;71:290–293. doi: 10.1038/bjc.1995.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morita R, Saito S, Ishikawa J, Ogawa O, Yoshida O, Yamakawa K, Nakamura Y. Cancer Res. 1991;51:5817–5820. [PubMed] [Google Scholar]
- 9.Kong F M, Anscher M S, Washington M K, Killian J K, Jirtle R L. Oncogene. 2000;19:1572–1578. doi: 10.1038/sj.onc.1203437. [DOI] [PubMed] [Google Scholar]
- 10.Millikin D, Meese E, Vogelstein B, Witkowski C, Trent J. Cancer Res. 1991;51:5449–5453. [PubMed] [Google Scholar]
- 11.Hayashi Y, Raimondi S C, Look A T, Behm F G, Kitchingman G R, Pui C H, Rivera G K, Williams D L. Blood. 1990;76:1626–1630. [PubMed] [Google Scholar]
- 12.Parsa N Z, Gaidano G, Mukherjee A B, Hauptschein R S, Lenoir G, Dalla-Favera R, Chaganti R S. Genes Chromosomes Cancer. 1994;9:13–18. doi: 10.1002/gcc.2870090104. [DOI] [PubMed] [Google Scholar]
- 13.Gaidano G, Hauptschein R S, Parsa N Z, Offit K, Rao P H, Lenoir G, Knowles D M, Chaganti R S, Dalla-Favera R. Blood. 1992;80:1781–1787. [PubMed] [Google Scholar]
- 14.Negrini M, Sabbioni S, Possati L, Rattan S, Corallini A, Barbanti-Brodano G, Croce C M. Cancer Res. 1994;54:1331–1336. [PubMed] [Google Scholar]
- 15.Trent J M, Stanbridge E J, McBride H L, Meese E U, Casey G, Araujo D E, Witkowski C M, Nagle R B. Science. 1990;247:568–571. doi: 10.1126/science.2300817. [DOI] [PubMed] [Google Scholar]
- 16.Gualandi F, Morelli C, Pavan J V, Rimessi P, Sensi A, Bonfatti A, Gruppioni R, Possati L, Stanbridge E J, Barbanti-Brodano G. Genes Chromosomes Cancer. 1994;10:77–84. doi: 10.1002/gcc.2870100202. [DOI] [PubMed] [Google Scholar]
- 17.Sheng Z M, Marchetti A, Buttitta F, Champeme M H, Campani D, Bistocchi M, Lidereau R, Callahan R. Br J Cancer. 1996;73:144–147. doi: 10.1038/bjc.1996.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colitti C V, Rodabaugh K J, Welch W R, Berkowitz R S, Mok S C. Oncogene. 1998;16:555–559. doi: 10.1038/sj.onc.1201523. [DOI] [PubMed] [Google Scholar]
- 19.Cooke I E, Shelling A N, Le Meuth V G, Charnock M L, Ganesan T S. Genes Chromosomes Cancer. 1996;15:223–233. doi: 10.1002/(SICI)1098-2264(199604)15:4<223::AID-GCC4>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 20.Tibiletti M G, Trubia M, Ponti E, Sessa L, Acquati F, Furlan D, Bernasconi B, Fichera M, Mihalich A, Ziegler A, et al. Oncogene. 1998;16:1639–1642. doi: 10.1038/sj.onc.1201654. [DOI] [PubMed] [Google Scholar]
- 21.Mizuno Y, Hattori N, Mori H, Suzuki T, Tanaka K. Curr Opin Neurol. 2001;14:477–482. doi: 10.1097/00019052-200108000-00008. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Gao J, Chung K K, Huang H, Dawson V L, Dawson T M. Proc Natl Acad Sci USA. 2000;97:13354–13359. doi: 10.1073/pnas.240347797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bednarek A K, Laflin K J, Daniel R L, Liao Q, Hawkins K A, Aldaz C M. Cancer Res. 2000;60:2140–2145. [PubMed] [Google Scholar]
- 24.Moretti T, Koons B, Budowle B. BioTechniques. 1998;25:716–722. [PubMed] [Google Scholar]
- 25.Sambrook J, Russell D W. Molecular Cloning: A Laboratory Manual. 3rd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 2001. [Google Scholar]
- 26.De Souza A T, Hankins G R, Washington M K, Fine R L, Orton T C, Jirtle R L. Oncogene. 1995;10:1725–1729. [PubMed] [Google Scholar]
- 27.McLean J W, Tomlinson J E, Kuang W J, Eaton D L, Chen E Y, Fless G M, Scanu A M, Lawn R M. Nature. 1987;330:132–137. doi: 10.1038/330132a0. [DOI] [PubMed] [Google Scholar]
- 28.Forsgren M, Raden B, Israelsson M, Larsson K, Heden L O. FEBS Lett. 1987;213:254–260. doi: 10.1016/0014-5793(87)81501-6. [DOI] [PubMed] [Google Scholar]
- 29.Mita H, Tsutsui J, Takekawa M, Witten E A, Saito H. Mol Cell Biol. 2002;22:4544–4555. doi: 10.1128/MCB.22.13.4544-4555.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 31.Koehler M R, Wissinger B, Gorboulev V, Koepsell H, Schmid M. Cytogenet Cell Genet. 1997;79:198–200. doi: 10.1159/000134720. [DOI] [PubMed] [Google Scholar]
- 32.Asakawa S, Tsunematsu K, Takayanagi A, Sasaki T, Shimizu A, Shintani A, Kawasaki K, Mungall A J, Beck S, Minoshima S, Shimizu N. Biochem Biophys Res Commun. 2001;286:863–868. doi: 10.1006/bbrc.2001.5490. [DOI] [PubMed] [Google Scholar]
- 33.Jones P A, Laird P W. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 34.Shimura H, Hattori N, Kubo S, Mizuno Y, Asakawa S, Minoshima S, Shimizu N, Iwai K, Chiba T, Tanaka K, Suzuki T. Nat Genet. 2000;25:302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 35.Imai Y, Soda M, Takahashi R. J Biol Chem. 2000;275:35661–35664. doi: 10.1074/jbc.C000447200. [DOI] [PubMed] [Google Scholar]
- 36.Ohta M, Inoue H, Cotticelli M G, Kastury K, Baffa R, Palazzo J, Siprashvili Z, Mori M, McCue P, Druck T, et al. Cell. 1996;84:587–597. doi: 10.1016/s0092-8674(00)81034-x. [DOI] [PubMed] [Google Scholar]
- 37.Benedict W F, Murphree A L, Banerjee A, Spina C A, Sparkes M C, Sparkes R S. Science. 1983;219:973–975. doi: 10.1126/science.6336308. [DOI] [PubMed] [Google Scholar]
- 38.Gessler M, Poustka A, Cavenee W, Neve R L, Orkin S H, Bruns G A. Nature. 1990;343:774–778. doi: 10.1038/343774a0. [DOI] [PubMed] [Google Scholar]
- 39.Baylin S B, Herman J G. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 40.Imai Y, Soda M, Inoue H, Hattori N, Mizuno Y, Takahashi R. Cell. 2001;105:891–902. doi: 10.1016/s0092-8674(01)00407-x. [DOI] [PubMed] [Google Scholar]
- 41.Imai Y, Soda M, Hatakeyama S, Akagi T, Hashikawa T, Nakayama K I, Takahashi R. Mol Cell. 2002;10:55–67. doi: 10.1016/s1097-2765(02)00583-x. [DOI] [PubMed] [Google Scholar]
- 42.Geyer R K, Yu Z K, Maki C G. Nat Cell Biol. 2000;2:569–573. doi: 10.1038/35023507. [DOI] [PubMed] [Google Scholar]
- 43.Nikolaev A Y, Li M, Puskas N, Qin J, Gu W. Cell. 2003;112:29–40. doi: 10.1016/s0092-8674(02)01255-2. [DOI] [PubMed] [Google Scholar]
- 44.Boyd S D, Tsai K Y, Jacks T. Nat Cell Biol. 2000;2:563–568. doi: 10.1038/35023500. [DOI] [PubMed] [Google Scholar]
- 45.Smith D I, Huang H, Wang L. Int J Oncol. 1998;12:187–196. [PubMed] [Google Scholar]
- 46.Paige A J, Taylor K J, Taylor C, Hillier S G, Farrington S, Scott D, Porteous D J, Smyth J F, Gabra H, Watson J E. Proc Natl Acad Sci USA. 2001;98:11417–11422. doi: 10.1073/pnas.191175898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inoue H, Ishii H, Alder H, Snyder E, Druck T, Huebner K, Croce C M. Proc Natl Acad Sci USA. 1997;94:14584–14589. doi: 10.1073/pnas.94.26.14584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mimori K, Druck T, Inoue H, Alder H, Berk L, Mori M, Huebner K, Croce C M. Proc Natl Acad Sci USA. 1999;96:7456–7461. doi: 10.1073/pnas.96.13.7456. [DOI] [PMC free article] [PubMed] [Google Scholar]