Abstract
Introduction and expression of multiple transgenes is frequently required for basic and applied studies. However, at present, multigene transformation is very difficult due to technical limitations of existing methods. Here, we describe a vector system for efficient multigene assembly and transformation. The system consists of a transformation-competent artificial chromosome (TAC)-based acceptor vector together with two donor vectors. By exploiting the Cre/loxP recombination system and homing endonucleases, multiple rounds of gene assembly cycling were carried out with alternate use of the donor vectors, and multiple genes were sequentially delivered into the TAC vector. With this system, we created constructs containing as many as 10 foreign DNA fragments. Multiple genes, including six resistant genes stacked in a construct, were transferred into rice genome by Agrobacterium-mediated transformation. This system extends the repertoire of molecular genetic studies and biotechnological endeavors by enabling simultaneous manipulation of multiple genes.
Over the past decade, the rapid progress in biotechnology has made it possible to improve organisms with respect to agronomic/industrial traits through genetic engineering. The majority of experiments reported to date involve the manipulation of single genes. However, many important traits, as well as complex metabolic pathways, depend on interactions among a number of genes, and so genetic engineering must now advance from manipulation of single-gene traits to manipulation of polygenic traits, multiple traits, and multiple gene products. However, genetic transformation with multiple genes is encumbered by technical limitations of the present methods. Therefore, new and effective technologies are urgently needed.
A number of approaches have been used to introduce multiple genes into plant genomes and then to coordinate transgene expression (1). These approaches include sexual crossing between plants carrying separate transgenes (2, 3), sequential retransformation (4), and cotransformation with multiple plasmids (5–8) or with single plasmids on which several transgenes are linked (9–14). Each approach has its specific problems. For example, genetic crossing between transgenic plants and sequential retransformation are very time-consuming and/or require the use of different selectable marker genes whereas the efficiency of cotransformation with multiple plasmids decreases progressively with increasing plasmid number. Cotransformation with separate multiple plasmids is a by-chance event; thus, the inserted copy numbers and the relative arrangement among transgenes cannot be controlled. In addition, multiple plasmids cotransferred by biolistics are often integrated into a few chromosome loci at high copy number, which is not favorable for expression of transgenes (5, 8, 15, 16). Cotransformation with linked transgenes in single vectors is a conventional and reliable approach. However, this approach is technically demanding. The lack of unique restriction cloning sites, loss of direct selection, as well as the relatively low efficiency for ligation of inserts into larger vectors, become the technical hindrances when three or more foreign genes are subcloned into a transformation vector by using existing cloning methods. The binary bacterial artificial chromosome (BIBAC) and transformation-competent artificial chromosome (TAC) vector systems are effective for transfer of large DNA fragments into plant genomes by Agrobacterium-mediated transformation (17–19); however, these vectors are not suitable for combining multiple separate DNA fragments together (20).
In this paper, we present a multigene assembly vector system that enables stacking of numerous genes from different sources into a TAC-based vector for subsequent transfer into plant genomes. By exploiting the Cre/loxP site-specific recombination system (21) and homing endonucleases (22), multiple rounds of gene assembly can be carried out indefinitely through alternate use of two donor vectors to deliver foreign genes sequentially into the TAC vector. We describe the technology and show the delivery of as many as 10 genes and functional DNA fragments into the vector and the transfer of the genes into rice genome by Agrobacterium-mediated transformation.
Materials and Methods
Vector Construction.
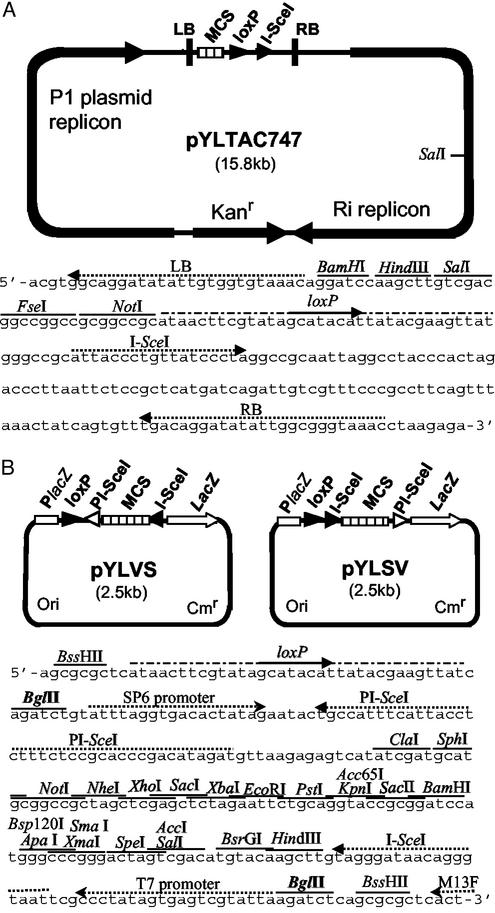
The TAC vector backbone sequence (15,680 bp) was isolated from pYLTAC17 (18) by PCR with two primers: P1, 5′-ACCGGATCCTGTTTACACCACAATATATCCTGCCACGTTAAAGACTTCATGATGTCGGAC-3, which contains a T-DNA (portion of the tumor-inducing plasmid that is transferred to plant cells) left border (LB) sequence; and P2, 5′-AGGTCTAGACCCTTAATTCTCCGCTCATGATCAGATTGTCGT-3′. The sequence was digested with BamHI and XbaI to produce cohesive ends, and ligated with a fragment (5′-gGATCCAAGCTTGTCGACGGCCGGCCGCGGCCGCATAACTTCGTATAGCATACATTATACGAAGTTATGGGCCGCATTACCCTGTTATCCCTAGGCCGCAATTAGGCCTACCCACTAGt-3′), which contains a BamHI end, HindIII/SalI/FseI/NotI sites, a loxP site, an I-SceI site, and an SpeI end (see Fig. 1 for the sites). The resulting plasmid was designated pYLTAC747 and used as an acceptor vector.
Figure 1.
Physical maps of the acceptor vector pYLTAC747 and donor vectors pYLVS and pYLSV. (A) Structural features of the pYLTAC747 and sequence between the T-DNA LB and RB. Four unique restriction sites, a loxP site, and a recognition site for I-SceI were placed between LB and RB. The plasmid backbone contains a kanamycin-resistance gene (Kanr, modified NPTI), a P1 plasmid replicon for replication in E. coli, and an Ri replicon for replication in Agrobacterium. (B) Structural features of the donor vectors and a part of sequence of pYLVS showing the key sites. The orientation of the segment between the two BglII sites is reversed in the two constructs. Cmr denotes the chloramphenicol-resistance gene and Ori denotes the ColE1 replicon.
A donor vector pYLSV was constructed by linking three fragments. The first fragment (1,660 bp), encompassing the replication origin and the lacZ marker gene, was obtained from pBluescript SK (Stratagene) by PCR with two primers: P3, (5′-GTTCTCGCGGTATCATTG-3′); and P4, (5′-CCATTCGCCATTCAGGCTG-3′). The chloramphenicol-resistant gene was amplified from pCAMBIA1200 with two primers: P5, (5′-CTTCAATATTACGCAGCA-3′; and P6, (5′-GAGCAATATTGTGCTTAG-3′). After treatments with T4 DNA polymerase and T4 polynucleotide kinase, the two fragments were ligated together to yield an intermediate plasmid. Multiple rounds of oligonucleotide-extension with eight primers produced the third fragment, LSV. LSV contained two BssHII sites, one at each end, a loxP site, an I-SceI site, a multicloning site (MCS) consisting of 22 unique restriction sites, and a PI-SceI site in order. The intermediate plasmid was digested with BssHII to excise the MCS fragment from pBluescript SK, and ligated with BssHII-digested LSV to produce the donor vector pYLSV. Another donor vector, pYLVS, was derived from pYLSV by digestion with BglII and religation to make the BglII-fragment containing the I-SceI/MCS/PI-SceI sites insert back into the vector in opposite orientation.
Subcloning of Target Genes to Donor Vectors.
The target genes were subcloned into the donor vectors by conventional methods. An SphI/SpeI fragment (1.2 kb) of the matrix attachment region (MAR) sequence from pGEM-T-MAR was inserted into pYLVS to create pYLVS-MAR. An XbaI/KpnI fragment (5.2 kb) containing RSs1promoter/GNA/Nos3′ from pRSSGNA1 was inserted into pYLSV to obtain pYLSV-GNA. An HindIII fragment (3.0 kb) of PinII5′/PinII/Nos3′ gene was isolated from pTW-α and subcloned into pYLVS to create pYLVS-PinII. An ApaI/NotI fragment (6.5 kb) containing Act15′/RCH10/Nos3′ and Act15′/RAC22/Nos3′ from pABC2 was inserted into pYLSV to create pYLSV-ABC2. A KpnI fragment (9.6 kb) containing the Xa21 gene from pB822 was ligated into pYLVS to obtain pYLVS-Xa21. A PmlI fragment (1.8 kb) containing P35S/Bar/Nos3′ from pTAC37 was inserted into pYLSV to create pYLSV-Bar. A fragment (3.0 kb) composed of RB/P35S/GUS/Nos3′/LB was obtained from pCAMBIA1305.2 by PCR with two specific primers containing T-DNA right border (RB) and LBs, respectively. This fragment was ligated into pYLSV to produce pYLSV-LB/GUS/RB.
Plasmid Cointegration and Vector-Backbone Removal.
For plasmid cointegration, a mixture of acceptor (≈5.0–10.0 ng) and donor (≈1.0–2.0 ng) plasmids was electroporated together into Escherichia coli strain NS3529 (23) that expresses the Cre recombinase. Colonies were selected on LB-agar plates supplemented with kanamycin (20 mg/liter) and chloramphenicol (15 mg/liter). The resulting plasmid population composed of both cointegrated and independent parental types was isolated, and introduced into E. coli strain DH10B. On the selection plates containing kanamycin (20 mg/liter) and chloramphenicol (15 mg/liter), colonies bearing cointegrates exhibited a more transparent appearance than those bearing separate plasmids, and were selected for further analysis.
Cointegrative plasmid (≈300 ng) purified with a plasmid purification kit (Qiagen-tip20; Qiagen, Valencia, CA) was digested completely with 5–10 units of I-SceI or PI-SceI (VDE) (New England Biolabs) in 30 μl for 1 h. An aliquot (3 μl, ≈30 ng) of the digested plasmid was directly added to a ligation reaction (10 μl) containing 0.1–0.01 pmol (not more than these amounts) of a phosphorylated linker LS (for I-SceI ends) or LV (for PI-SceI ends) and about 30 units (0.25 Weiss unit) of T4 DNA ligase (Takara Shuzo, Kyoto). The recognition site of I-SceI is 5′-TAGGGATAACAGGGTAAT-3′, and that of PI-SceI is 5′-ATCTATGTCGGGTGCGGAGAAAGAGGTAATGAAATGGCA-3. Thus, the two cutting ends of opposite orientation were linked as follows: I-SceI end/LS(lowercase)/I-SceI end, 5′-TAGGGATAAgcggccgcttatCCCTA-3′, 3′-ATCCCtattcgccggcgAATAGGGAT-5′; PI-SceI end/LV(lowercase)/PI-SceI end, 5′-ATCTATGTCGGGTGCgcggccgcgcacCCGACATAGAT-3′, 3′-TAGATACAGCCcacgcgccggcgCGTGGGCTGTATCTA-5′
The ligated product was dialyzed against 0.3× TE (10 mM Tris/1 mM EDTA, pH 8.0) and electroporated into DH10B. Transformants were selected on LB-agar plates containing kanamycin (25 mg/liter), and then tested for sensitivity to chloramphenicol (20 mg/liter).
Rice Transformation.
The construct pYLTAC747H-GMBXRRPGM was introduced into Agrobacterium tumefaciens strain EHA105 by electroporation. Before plant transformation, the intact of the plasmid in Agrobacterium was confirmed as described (18). The Agrobacterium cells were cocultured with embryogenic calli induced from mature seeds of Japonica rice varieties Zhonghua11 and Ishikari Shiroke as described (24). Regenerating calli were selected in the presence of 50 mg/liter hygromycin and subsequently transferred to rooting medium containing hygromycin. After further culturing for 3 to 4 wk, transformed plants were transferred to soil in a greenhouse.
Southern Blot Analysis.
Genomic DNAs isolated from transgenic plants were digested with HindIII, fractionated on a 0.7% agarose gel, and blotted onto Hybond-N+ nylon membranes (Amersham Pharmacia). Transgene fragments were labeled with [α-32P]dCTP and used as hybridization probes, and hybridization signals were detected and analyzed by using an FX Molecular Imager System (Bio-Rad).
RT-PCR.
Total RNA was prepared from rice leaf tissue by using TRIzol reagent (Invitrogen). Reverse transcription was performed by using SuperScript II reverse transcriptase (Invitrogen) with a polyT primer. Gene-specific primers for RT-PCR were: 5′-CATGGATGTTCACAAGGAAGTT-3′ and 5′-CTCTCCTTCACATACAAACTTG-3′ for PinII; 5′-GCTAAGGCAAGTCTCCTCATTT-3′ and 5′-TCACAAGCTTTATCTTTCCAGC-3′ for GNA; 5′-GCACCATCGTCAACCACTACA-3′ and 5′-GTCCAGCTGCCAGAAACCCT-3′ for Bar; 5′-CTGCCTCTGCTGGAGCCAGT-3′ and 5′-CGCCGTTGATGATGTTGGTGA-3′ for RCH10; 5′-GCGATGTTCGAGTCGATGCTG-3′ and 5′-CTGAGTAAGCTGTATGGGTCCT-3′ for RAC22; and 5′-GTCTTGCCTTGCACTTCTGCACGA-3′ and 5′-ATTGAATAATTCACTGGGTATTGG-3′ for Xa21. The Xa21 primers are specific only for the transgene but not for the endogenous Xa21-homologous sequence of cultivated rice. PCR amplification was performed with 35 cycles of 94°C for 30 s, 52–60°C (according to genes) for 60 s, and 72°C for 90 s.
Results
Vector Design and Construction.
The Cre recombinase catalyzes reversible site-specific recombination reactions between loxP sites; hence, two plasmids each carrying a loxP site can undergo cointegration via the Cre/loxP system. Theoretically, genes carried on separate plasmids can be linked together indefinitely by repeated plasmid cointegration events. However, for this purpose, several technical issues must first be resolved: (i) the backbone sequence of the donor vector, which contains a loxP site, a replication origin, and a bacterial selective marker, must be removed from the cointegrate before each subsequent round of plasmid cointegration; and (ii) if endonucleases are used for this removal, appropriately positioned digestion sites in the vectors must be available that do not occur elsewhere within the backbone of acceptor vector and its growing insert(s).
We designed and constructed a vector system that enables the stacking of multiple genes within a transformation vector by means of gene-assembly cycling. The vector system consists of an acceptor vector, pYLTAC747, and two donor vectors, pYLVS and pYLSV (Fig. 1). The acceptor vector was derived from the binary TAC vector (18, 19) and contained a loxP site and a digestion site for the homing endonuclease I-SceI between the RBs and LBs. The vector is able to accept a large number of genes by Cre-mediated recombination as described below, stably maintain that extensive gene tract in both E. coli and A. tumefaciens, and transfer the genes into plant genomes via Agrobacterium-mediated transformation. The ColE1-based donor vectors pYLVS and pYLSV contain a well-designed synthetic fragment encompassing a loxP site, an MCS, and two recognition sites for the homing endonucleases (meganucleases) I-SceI and PI-SceI (VDE). The arrangement of the sites differs critically between the two donor vectors: loxP/PI-SceI/MCS/I-SceI for pYLVS, and loxP/I-SceI/MCS/PI-SceI for pYLSV. Homing endonucleases are very-rare-cutting enzymes occurring in nature as inteins or encoded by introns (22). For example, the two enzymes I-SceI and PI-SceI used in this study recognize asymmetric sites of 18 bp and 39 bp, respectively. Therefore, neither the acceptor vector nor inserted genes are digested when these enzymes are used to remove the backbone sequence of either donor vector. Although six different homing endonucleases are commercially available, one of the key points of the system is that the gene-assembly cycling is carried out indefinitely through alternating use of just two of the enzymes. In addition, a large number of 22 unique restriction enzyme sites was incorporated into the MCS of the donor vectors for subcloning of target genes. We chose chloramphenicol-resistance (Cmr) for our donor vectors to facilitate subcloning from more widely used vectors that generally use ampicillin resistance for selection.
Multigene Assembly Cycling.
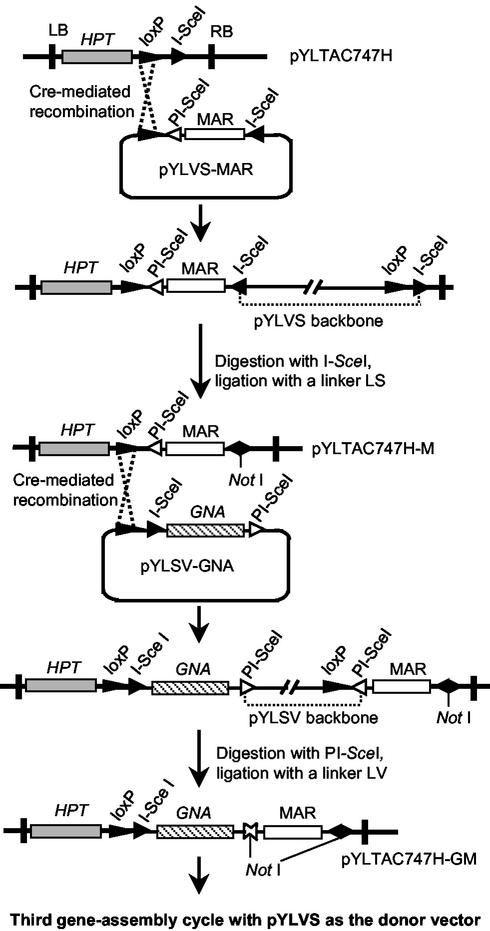
Our strategy for multigene assembly was to alternate use of the two donor vectors to stack genes of interest into the acceptor vector by multiple rounds of site-specific recombination. We first inserted a plant-selectable hygromycin-resistant marker gene (P35S/HPT/Nos3) by conventional ligation into a NotI site in the acceptor vector pYLTAC747 to yield plasmid pYLTAC747H. This plasmid was then used as the starting acceptor vector for multiple rounds of gene-assembly. Fig. 2 shows a schematic diagram for the first and second cycles of this assembly process.
Figure 2.
Schematic diagram of the multigene assembly process. Only two assembly cycles are shown: the first to deliver the MAR and the second to add the GNA gene into the acceptor vector pYLTAC747H. The linkers LS and LV were prepared with phosphorylated oligonucleotides 5′-gcggccgcttat-3′ and 5′-gcggccgcgcac-3′, respectively, whose self-pairing cores form a NotI site with compatible ends extending outward for their intended ligation partners.
A MAR sequence (25) was subcloned into the first donor vector pYLVS to yield pYLVS-MAR. Plasmid integration was carried out in vivo by cotransfer of pYLVS-MAR and pYLTAC747H into the E. coli host NS3529 (23) that expresses Cre recombinase. Because the recombination reaction is reversible, three types of plasmids, i.e., the cointegrate along with the parents, were present in the host cells. The plasmids were isolated and retransferred into DH10B, a host strain lacking the Cre gene. Transformants bearing the cointegrate were obtained by double-selection with kanamycin and chloramphenicol, and plasmid was purified. To remove the pYLVS backbone, the plasmid was subjected to digestion with I-SceI. Cotransformation of DH10B with both the donor plasmid and cointegrate was observed in some cases. However, this cotransformation impaired neither the homing endonuclease digestion nor the subsequent ligation. Because the two asymmetric I-SceI sites on the cointegrative plasmid are arranged in reverse orientation, the protruding 3′ ends are not complementary to each other (see Materials and Methods), and plasmid circularization requires the aid of a compatible double-stranded linker LS. The resulting joining site on the plasmid is no longer recognized by the enzyme during subsequent gene-assembly cycles. Even though plasmid circularization required the linker, the ligation efficiency was high and comparable to that of self-ligating of single fragments with compatible ends. The ligated product was transferred again into DH10B, and the desired constructs were obtained by screening clones that were kanamycin-resistant and chloramphenicol-sensitive. The homing endonuclease site I-SceI present in the starting vector pYLTAC747H was replaced by PI-SceI in the new construct pYLTAC747H-M (Fig. 2), adapting it for use with the second donor vector pYLSV in the following cycle of gene assembly.
The second cycle of gene assembly was carried out to deliver a snow drop lectin gene (GNA) (26) to pYLTAC747H-M. The procedures of this cycle were the same as those described above, except that the homing endonuclease PI-SceI was used for digestion of the cointegrative plasmid, and a new linker LV compatible to the ends of the PI-SceI-digested sequence was used for the circularization ligation. On completion of this cycle, the resulting construct pYLTAC747H-GM carried the GNA gene along with an I-SceI site adjacent to the loxP site, just as in the starting acceptor vector.
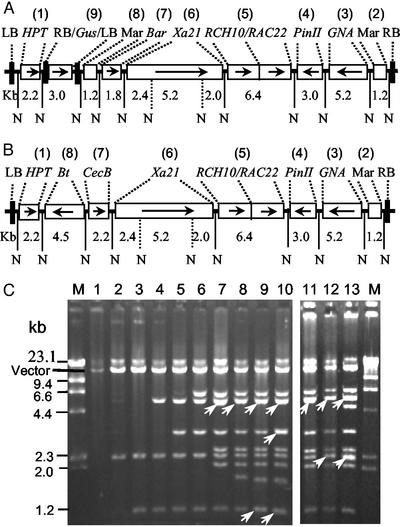
By alternating repetition of the process described above with additional target genes, we obtained a number of constructs carrying different numbers of foreign genes and functional DNA sequences (Fig. 3). These genes and DNA fragments included MAR, GNA, potato proteinase inhibitor II gene (PinII) (27), rice acidic chitinase-22, and rice basic chitinase-10 genes (RAC22-RCH10) (28, 29), rice bacterial blight resistant gene (Xa21) (30), phosphinotricine acetyltransferase gene (Bar), β-glucuronidase gene (GUS), cecropin B gene (CecB) (31), and δ−endotoxin gene of Bacillus thuringiensis (Bt) (32). One construct, pYLTAC747H-GMBXRRPGM, was obtained by eight cycles of the gene assembly, and carried 10 foreign genes or functional sequences with a total of 33.6 kb. In this construct, two T-DNA regions were generated by insertion of the LBs and RBs together with the GUS gene (Fig. 3A). Another construct pYLTAC747H-BCXRRPGM contained nine genes or functional sequences.
Figure 3.
Structure and restriction analysis of recombinant constructs. (A) Structure of a construct pYLTAC747H-GMBXRRPGM containing two LB- and RB-sandwiched T-DNA regions. The hygromycin-resistant gene (P35S/HPT/Nos3′) occupied its own T-DNA region, whereas the second T-DNA region carried the genes/functional sequences MAR, GNA (RSs15′/GNA/Nos3′), PinII (PinII5′/PinII/PinII3′), RAC22 (Act5′/RAC22/Nos3′), RCH10 (Act5′/RCH10/Nos3′), Xa21(rice genomic sequence), and Bar (P35S/Bar/Nos3′). The GUS gene (P35S/GUS/Nos3′) flanked by LB and RB in pYLSV-LB/GUS/RB was inserted between the second MAR and HPT gene to create two T-DNA regions, and so the GUS gene was located between the two T-DNA regions. Arrows denote the transcription directions of the genes. The numbers in parentheses indicate the order of the sequences inserting into the vector. N denotes NotI sites. (B) Structure of pYLTAC747H-BCXRRPGM, another construct that contained two additional pathogen-resistant genes, i.e., the cecropin B gene (Ubi5′/CecB/Nos3′) and δ-endotoxin gene of B. thuringiensis (Act5′/Bt/Nos3′), in addition to the five resistant genes shown in A. (C) Restriction analysis of a series of TAC constructs containing different numbers of genes. The pYLTAC747 plasmid (lane 1) and the derived constructs were digested with NotI and separated on 0.7% agarose gels. Lanes 2–9 and lanes 11 and 12 are intermediate products created during the construction of pYLTAC747H-GMBXRRPGM (lane 10) as depicted in A and pYLTAC747H-BCXRRPGM (lane 13) as depicted in B, respectively. Lane 11 is the same digest as shown in lane 7. Arrows indicate bands with two comigrating fragments.
Multigene Transformation of Rice.
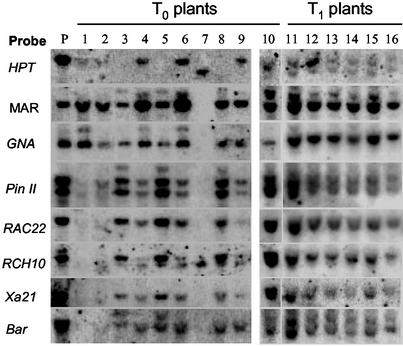
The multigene construct pYLTAC747H-GMBXRRPGM contained two T-DNA regions, one carrying the HPT marker gene and the other, multiple resistant transgenes including the Bar gene conferring resistance to the herbicide Basta. This arrangement was designed to obtain HPT-free transformants by cotransformation of two T-DNAs generated from a binary vector followed by locus segregation (33). The GUS gene was positioned between the two T-DNA regions and could be used as a probe or reporter for analysis of T-DNA insertion patterns among transformants. This construct was introduced into Agrobacterium strain EHA105 by electroporation, and used to test multigene transformation of rice. Embryogenic calli from two Japonica rice varieties, Zhonghua11 and Ishikari Shiroke, were cocultivated with Agrobacterium cells containing the TAC construct according to established transformation procedures (24). Hygromycin was used for the selection of transformed calli. More than 50 T0 transformants were obtained, and ≈50% were resistant to both hygromycin and Basta, indicating that both T-DNAs had integrated into the genome. The remaining transformants were resistant only to hygromycin. Southern blot analysis of the T0 plants and their T1 progenies was performed by using the transgenes as probes. The results indicate that, in most transgenic plants, all transgenes present in the same T-DNA region were transferred together into the rice genome, and were inherited stably in the progeny (Fig. 4). However, it was observed in two transformants that only part of the T-DNA was integrated into the rice genome, and four genes (RAC22, RCH10, Xa21, and Bar) adjacent to the LB side were missed during the T-DNA transfer (Fig. 4, lanes 1 and 2). The sizes of the hybridized bands of the transgenes were identical with that of the plasmid (lane P), demonstrating the intact of the transgenes in the rice genome.
Figure 4.
Southern blot analysis of the T0 and T1 rice plants obtained by transformation with the construct pYLTAC747H-GMBXRRPGM. HindIII-digested genomic DNAs and the construct (lane P) were separated on 0.7% agarose gels, blotted onto nylon membranes, and hybridized with the transgenes as probes. Lanes 1–9 are T0 plants of variety Zhonghua11; lanes 10–16 represent a T0 plant (lane 10) of variety Ishikari Shiroke together with six plants of its T1 progeny (lanes 11–16).
Expression of the Transgenes.
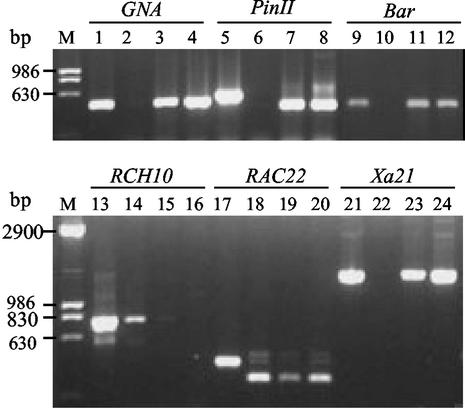
Preliminary study of the expression of the transgenes in two T1 transgenic plants was carried out by RT-PCR. The predicted cDNA fragments of all transgenes except the RCH10 gene, that were linked in the same T-DNA region, could be amplified (Fig. 5). Cosuppression of the transgenic and endogenous copies of the RCH10 gene was observed (Fig. 5, lanes 15 and 16). The function of the transferred Bar gene was also investigated by herbicide-resistant test. The T0 and T1 transformants hybridized positively by the Bar gene probe were all Basta-resistant. These results indicate that most of the linked transgenes were coexpressed. Detailed analyses will be necessary to investigate the T-DNA insertion patterns and the resistance of the transgenic plants to pathogens and insects.
Figure 5.
Analysis of the expression of the transgenes by RT-PCR. Lanes 1, 5, 9, 13, 17, and 21 are PCR products of the genes amplified from a plasmid pYLTAC747H-MBXRRPGM with gene-specific primers; lanes 2, 6, 10, 14, 18, and 22 are controls with untransformed plants; the other lanes are cDNA fragments amplified from two T1 plants. Lanes 14 and 18 show the expression of the endogenous RCH10 and RAC22 genes, respectively, and lanes 15 and 16 show the cosuppression of the transgenic and endogenous copies of the RCH10 gene. The fragments of the PinII (lane 5) and RAC22 (lane 17) amplified from the plasmid contain an intron, respectively, and show sizes larger than those of the cDNA fragments.
Discussion
Although site-specific recombination systems are generally used for gene subcloning, existing methods can perform only one or two rounds of recombination (34). Here, we described an efficient multigene assembly vector system that overcomes the drawbacks of existing cloning methods and permits the performing of many rounds of gene recombination. In this study, we stacked as many as 10 foreign DNA fragments into the vector. But this demonstration does not exploit the system to its limits, because the cloning capacity of the TAC-based vector can be larger than 100 kb, whereas the number of gene-assembly cycling is not a limiting factor within a reasonable range. The vector system is flexible and versatile, and the placement and orientation of transgenes in the vector can be freely designed and readily achieved in a reliable step-by-step process. Therefore, the system provides a powerful tool for manipulating multiple genes for either academic or applied purposes.
DNA engineering based on homologous recombination systems (recombinogenic engineering) also can be used to manipulate DNA constructs, and recent advances have generated much excitement (35). However, for the purpose of stacking multiple genes from different sources into a vector, the method we describe here using a site-specific recombination system should be more robust, especially for manipulation of constructs that carry sequence repeats, whether by design or happenstance in genome-derived sequences. In designing a multigene construct, one might wish to reuse the same elements such as promoters, enhancers, or polyA addition signal sequences. For example, the battery of resistant genes carried by pYLTAC747H-GMBXRRPGM was flanked with directed repeats of the MAR sequence. The entire gene battery would have been prone to deletion if a homologous recombination system had been used for making the construct.
In this study, we carried out the Cre-mediated recombination in vivo to produce the plasmid cointegrate. Alternatively, this step can be carried out in vitro by using purified Cre recombinase. Recently, there has been developed a Cre-mediated irreversible recombination system (Y.-G.L., unpublished data). Further modification of the vector system should be conducted with this irreversible recombination system. With this modification, the steps of plasmid cointegration and removal of donor vector backbone in each gene-assembly cycle can be completed in a single recombination reaction. The donor vectors also can be modified as BAC-based vectors by replacing the ColE1 Ori with replicon of the miniF factor, so that larger DNA fragments can be cloned and delivered into the acceptor vector.
One of our goals is to create transgenic plants that both are resistant to multiple pathogens, insects, and herbicides and are antibiotic marker free. In this study, six resistant genes were linked to a T-DNA region independent of a second T-DNA region carrying the marker gene HPT. Hygromycin-resistant transformants are likely to carry the desired resistant genes as well, but at one or more separate loci that will segregate independently in their progeny. MAR sequences were set on both sides of these resistant genes to improve transgene expression (36). The TAC vectors have been established as an effective means for transfer of large DNA fragments into plants such as Arabidopsis thaliana and rice (18, 19, 37, 38). Therefore, similar performance for large DNA transfer can be expected for the TAC-based acceptor vector developed in this study. Indeed, our results showed that multiple transgenes contained within the same T-DNA region of the TAC vector were all integrated into the rice genome, except that in a few transformants deletion of some transgenes was observed. Our preliminary results show that all but one of the multiple transgenes were expressed as assayed by RT-PCR and by herbicide-resistant test of the Bar gene. In addition, two constructs with multiple genes including the Bar gene were also used to transform Arabidopsis, and a number of Basta-resistant transformants were obtained (unpublished data), showing the usefulness of the system in this important model plant.
Both basic and applied biology now require more complex manipulations for larger or more intricate DNA constructs. The utility of the technology described here is not restricted to plant applications. Using this technology, any appropriate DNA components can be readily assembled in various vectors modified according to this study for different purposes, such as transfer of multiple genes of interest to yeast, insect, or mammalian cells. For example, the functions and interactions of multiple genes related to complex metabolic pathways can be studied by transformation with related genes in various combinations, or by RNA interference through transfer of multiple transcription units that would undergo intracellular processing to short interfering RNAs for targeting of various combinations of endogenous genes. Furthermore, studies on mammalian and plant artificial chromosomes require effective and reliable methods to manipulate such large and complex constructs; our technology including its improved versions should lend itself to these needs.
Acknowledgments
We thank R. Wu, Y. Fan, Y. Xu, G. L. Wang, and K. Tang for providing resistant genes, and R. F. Whittier and H.-M. Lam for critical reading of the manuscript. This research was supported by grants from the Ministry of Science and Technology of China (G1999011603, 2002AA212171, J99-B-012), the Natural Science Foundation of Guangdong Province (B201, 20003023), and the Education Department of Guangdong Province.
Abbreviations
- TAC
transformation-competent artificial chromosome
- LB
left border
- RB
right border
- T-DNA
portion of the tumor-inducing plasmid that is transferred to plant cells
- MCS
multicloning site
- MAR
matrix attachment region
References
- 1.Halpin C, Barakate A, Askari B M, Abbott J C, Ryan M D. Plant Mol Biol. 2001;47:295–310. [PubMed] [Google Scholar]
- 2.Ma J K, Hiatt A, Hein M, Vine N D, Wang F, Stabila P, van Dolleweerd C, Mostov K, Lehner T. Science. 1995;268:716–719. doi: 10.1126/science.7732380. [DOI] [PubMed] [Google Scholar]
- 3.Bizily S P, Rugh C L, Meagher R B. Nat Biotechnol. 2000;18:213–217. doi: 10.1038/72678. [DOI] [PubMed] [Google Scholar]
- 4.Lapierre C, Pollet B, Petit-Conil M, Toval G, Romero J, Pilate G, Leple J C, Boerjan W, Ferret V V, De Nadai V, Jouanin L. Plant Physiol. 1999;119:153–163. doi: 10.1104/pp.119.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen L, Marmey P, Taylor N J, Brizard J P, Espinoza C, D'Cruz P, Huet H, Zhang S, Kochko A, Beachy R N, Fauquet C M. Nat Biotechnol. 1998;16:1060–1064. doi: 10.1038/3511. [DOI] [PubMed] [Google Scholar]
- 6.Xu X P, Hu M, Wei J W, Chen J T, Li B J. Hereditas (Beijing) 1998;20,Suppl.:12–14. [Google Scholar]
- 7.Ye X, Al-Babili S, Klti A, Zhang J, Lucca P, Beyer P, Potrykus I. Science. 2000;287:303–305. doi: 10.1126/science.287.5451.303. [DOI] [PubMed] [Google Scholar]
- 8.Hadi M Z, McMullen M D, Finer J J. Plant Cell Rep. 1996;15:500–505. doi: 10.1007/BF00232982. [DOI] [PubMed] [Google Scholar]
- 9.Goderis I J, De Bolle M F, Francois I E, Wouters P F, Broekaert W F, Cammue B P. Plant Mol Biol. 2002;50:17–27. doi: 10.1023/a:1016052416053. [DOI] [PubMed] [Google Scholar]
- 10.Dasgupta S, Collins G B, Hunt A G. Plant J. 1998;16:107–116. doi: 10.1046/j.1365-313x.1998.00255.x. [DOI] [PubMed] [Google Scholar]
- 11.Halpin C, Cooke S E, Barakate A, El Amrani A, Ryan M D. Plant J. 1999;17:453–459. doi: 10.1046/j.1365-313x.1999.00394.x. [DOI] [PubMed] [Google Scholar]
- 12.Urwin P, Yi L, Martin H, Atkinsom H, Gilmartin P M. Plant J. 2000;24:583–589. doi: 10.1046/j.1365-313x.2000.00904.x. [DOI] [PubMed] [Google Scholar]
- 13.Decosa B, Moar W, Lee S B, Miller M, Daniell H. Nat Biotechnol. 2001;19:71–74. doi: 10.1038/83559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Gray G, Rajasekaran K, Smith F, Sanford J, Daniell H. Plant Physiol. 2001;127:852–862. [PMC free article] [PubMed] [Google Scholar]
- 15.Gelvin S B. Nat Biotechnol. 1998;16:1009–1010. doi: 10.1038/3452. [DOI] [PubMed] [Google Scholar]
- 16.Maqbool S B, Christou P. Mol Breed. 1999;5:471–480. [Google Scholar]
- 17.Hamilton C M, Frary A, Lewis G, Tanksley S D. Proc Natl Acad Sci USA. 1996;93:9975–9979. doi: 10.1073/pnas.93.18.9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y G, Shirano Y, Fukaki H, Yanai Y, Tasaka M, Tabata S, Shibata D. Proc Natl Acad Sci USA. 1999;96:6535–6540. doi: 10.1073/pnas.96.11.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y G, Liu H, Chen L, Qiu W, Zhang Q, Wu H, Yang C, Su J, Wang Z, Tian D, Mei M. Gene. 2002;282:247–255. doi: 10.1016/s0378-1119(01)00849-6. [DOI] [PubMed] [Google Scholar]
- 20.Shibata D, Liu Y G. Trends Plant Sci. 2000;5:354–357. doi: 10.1016/s1360-1385(00)01689-7. [DOI] [PubMed] [Google Scholar]
- 21.Sternberg N, Hamilton D. J Mol Biol. 1981;150:467–486. doi: 10.1016/0022-2836(81)90375-2. [DOI] [PubMed] [Google Scholar]
- 22.Belfort M, Roberts R J. Nucleic Acids Res. 1997;25:3379–3388. doi: 10.1093/nar/25.17.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sauer B, Henderson N. Gene. 1998;70:331–341. doi: 10.1016/0378-1119(88)90205-3. [DOI] [PubMed] [Google Scholar]
- 24.Hiei Y, Ohta S, Komari T, Kumashiro T. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- 25.Allen G C, Hall G, Jr, Michalowski S, Mowman W, Spiker S, Weissinger A K, Thompson W F. Plant Cell. 1996;8:899–913. doi: 10.1105/tpc.8.5.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rao K V, Rathore D S, Hodges T K, Fu X, Stoger E, Sudhakar D, Williams S, Christou P, Bharathki M, Bown D P, et al. Plant J. 1998;15:469–477. doi: 10.1046/j.1365-313x.1998.00226.x. [DOI] [PubMed] [Google Scholar]
- 27.Duan X, Li X, Xue Q, Abo-el-Saad M, Xu D, Wu R. Nat Biotechnol. 1996;14:494–498. doi: 10.1038/nbt0496-494. [DOI] [PubMed] [Google Scholar]
- 28.Feng D R, Xu X P, Wei J W, Li B J, Yang Q Y, Zhu X Y. Acta Botanica Sinica. 1999;41:1187–1191. [Google Scholar]
- 29.Zhu Q, Lamb C J. Mol Gen Genet. 1991;226:289–296. doi: 10.1007/BF00273615. [DOI] [PubMed] [Google Scholar]
- 30.Song W Y, Wang G L, Chen L L, Kim H S, Pi L Y, Holsten T, Gardner J, Wang B, Zhai W X, Zhu L H, et al. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- 31. Moore, A. J., Beazley, W. D., Bibby, M. C. & Devine, D. A. (1996) 37, 1077–1089. [DOI] [PubMed]
- 32.Wei J W, Xu X P, Chen J T, Zhang L Y, Fan Y L, Li B J. Chin J Biotechnol. 2000;16:603–608. [PubMed] [Google Scholar]
- 33.Komari T, Hiei Y, Saito Y, Murai N, Kumashiro T. Plant J. 1996;10:165–174. doi: 10.1046/j.1365-313x.1996.10010165.x. [DOI] [PubMed] [Google Scholar]
- 34.McCormac A C, Elliott M C, Chen D F. Mol Gen Genet. 1999;261:226–235. doi: 10.1007/s004380050961. [DOI] [PubMed] [Google Scholar]
- 35.Muyrers J P P, Zhang Y, Stewart A F. Trends Biochem Sci. 2001;26:325–331. doi: 10.1016/s0968-0004(00)01757-6. [DOI] [PubMed] [Google Scholar]
- 36.Spiker S, Thompson W F. Plant Physiol. 1996;110:15–21. doi: 10.1104/pp.110.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sawa S, Watanabe K, Goto K, Liu Y G, Shibata D, Kanaya E, Morita E H, Okada K. Genes Dev. 1999;13:1079–1088. doi: 10.1101/gad.13.9.1079. , 2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sato S, Kato T, Kakegawa K, Ishii T, Liu Y G, Awano T, Takabe K, Nishiyama Y, Kuga S, Sato S, et al. Plant Cell Physiol. 2001;42:251–263. doi: 10.1093/pcp/pce045. [DOI] [PubMed] [Google Scholar]