Abstract
The plant mitochondrial genome is retained in a multipartite structure that arises by a process of repeat-mediated homologous recombination. Low-frequency ectopic recombination also occurs, often producing sequence chimeras, aberrant ORFs, and novel subgenomic DNA molecules. This genomic plasticity may distinguish the plant mitochondrion from mammalian and fungal types. In plants, relative copy number of recombination-derived subgenomic DNA molecules within mitochondria is controlled by nuclear genes, and a genomic shifting process can result in their differential copy number suppression to nearly undetectable levels. We have cloned a nuclear gene that regulates mitochondrial substoichiometric shifting in Arabidopsis. The CHM gene was shown to encode a protein related to the MutS protein of Escherichia coli that is involved in mismatch repair and DNA recombination. We postulate that the process of substoichiometric shifting in plants may be a consequence of ectopic recombination suppression or replication stalling at ectopic recombination sites to effect molecule-specific copy number modulation.
Argument for the mitochondrion as a central regulator of cellular functions has become increasingly persuasive in the past several years, as information expands detailing cell metabolic functions (1, 2), programmed cell death (3), and intracellular signaling (4). In higher plants, mitochondrial functions and behavior have clearly been influenced by the unique context of the plant cell. Coevolution of mitochondria and chloroplasts has permitted economy of function via protein dual-targeting (5, 6), genome capacity and coding have been altered (7), and the mitochondrial genomes of plants have acquired structural and maintenance features distinct from their animal counterparts.
The plant mitochondrial genome seems to be organized as a collection of small circular and large circularly permuted linear molecules (8, 9), not unlike what has been postulated for yeast (10, 11). DNA replication may be conducted by a rolling circle mechanism, and experimental difficulties identifying replication origins have led to the suggestion of recombination-mediated replication initiation (12). In fact, a distinct feature of plant mitochondrial genome organization is the prominent role of recombination.
High-frequency inter- and intramolecular recombination is detected within the higher plant mitochondrial genome at large repeated sequences that can be readily identified by physical mapping (13). Their presence in direct orientation permits the subdivision of the genome into a collection of molecules, each containing only a portion of the genetic information. Most intriguing, however, is the common observation in plants of intragenic ectopic recombination events that can occur at sites containing as few as seven nucleotides of homology (14). Ectopic recombination results in expressed gene chimeras that cause cytoplasmic male sterility, plant variegation, and other aberrant phenotypes (15, 16).
A phenomenon rendering the plant mitochondrial genome unusually variable in structure is termed substoichiometric shifting. First reported in maize (17) as the stable presence of subgenomic mitochondrial DNA molecules within the genome at nearly undetectable levels, the process seems to be highly dynamic. Mitochondrial genomic shifting involves rapid and dramatic changes in relative copy number of portions of the mitochondrial genome over the time of one generation (18). These substoichiometric forms have been estimated at levels as low as one copy per every 100–200 cells (19). Generally, the rapid shifting process involves only a single subgenomic DNA molecule, often containing recombination-derived chimeric sequences, and the process is apparently reversible (18, 20). Genomic shifting can alter plant phenotype because the process activates or silences mitochondrial sequences located on the shifted molecule. Observed phenotypic changes have included plant tissue culture properties (20), leaf variegation and distortion (16), and spontaneous reversion to fertility in cytoplasmic male sterile crop plants (18, 21). It has been postulated that substoichiometric shifting may have evolved to permit the species to create and retain mitochondrial genetic variation in a silenced but retrievable form (22).
Mitochondrial substoichiometric shifting has been shown in at least two cases to be under nuclear gene control, involving the Fr gene in Phaseolus vulgaris (23) and the CHM gene in Arabidopsis (24, 25). Mutation of the nuclear CHM gene results in a green-white leaf variegation that, in subsequent generations, displays maternal inheritance (25). The appearance of the variegation phenotype is accompanied by a specific rearrangement (24) that includes amplification of a mitochondrial DNA molecule encoding a chimeric sequence (16). Genetic analysis suggests that the wild-type form of CHM actively suppresses copy number of the subgenomic molecule carrying the chimeric sequence. Loss of proper function of the CHM gene, characterized by two available ethyl methanesulfonate-derived mutant alleles, chm1-1 and chm1-2 (25), and a tissue culture-derived mutant allele, chm1-3 (24), results in rapid and specific copy number amplification of the subgenomic molecule, producing the consequent leaf variegation. It is not clear whether the copy number amplification or suppression of a single subgenomic molecule occurs by differential replication or a recombination mechanism. To investigate further this unusual plant phenomenon, we have cloned the nuclear CHM gene in Arabidopsis that effects mitochondrial substoichiometric shifting. This gene, located on chromosome III, was shown to encode a protein that is targeted to mitochondria and that has high homology to a MutS protein. The identified locus seems to represent a plant counterpart to the yeast mitochondrial MutS homolog (MSH1) and was designated AtMSH1.
Materials and Methods
Gene Mapping, Cloning, and Sequence Analysis.
A map-based cloning strategy for the isolation of the CHM locus involved the design of PCR-based codominant markers, using the Cereon Arabidopsis Polymorphism Collection (ref. 26; www.arabidopsis.org/cereon/), to distinguish between the Col-0 and Landsberg erecta ecotypes used in the F2 mapping populations. The markers were designed in a 5-Mb region of chromosome III based on information from the classical mapping experiments of CHM (24, 25). The primer sequences for markers are available on request. The F2 mapping population was derived in our laboratory from a cross between the chm1-1 mutant line and Landsberg erecta ecotype (pollen donor). A segregating subpopulation of 172 variegated plants was analyzed. Genomic DNA purification was conducted according to Li and Chory (27). DNA gel blot analysis was conducted by using the protocol of Sambrook et al. (28).
DNA sequencing of the candidate locus in chm1-1, chm1-2, and chm1-3 mutants (24) was conducted in a Beckman Coulter CEQ2000XL eight-capillary DNA sequencer. Two independent PCR samples for each mutant were sequenced. The 5′ RACE analysis was done with the GeneRacer kit (Invitrogen). Mutants chm1-1 and chm1-2 were obtained from the Arabidopsis Biological Resource Center, and mutant chm1-3 was graciously provided to us by C. Somerville (Stanford University, Stanford, CA).
The two sequence-indexed T-DNA [portion of the Ti (tumor-inducing) plasmid that is transferred to plant cells] insertion mutants were identified on The Salk Institute Genomic Analysis Laboratory (SIGnAL) web site at http://signal.salk.edu, and seed for the mutants was obtained from the Arabidopsis Biological Resource Center. The T-DNA insertion positions were confirmed by DNA sequencing of the insertion junctions (data not shown).
Plant Transformation and Biolistic Delivery.
The amino acid sequence of AtMSH1 was analyzed with mitoprot (http://mips.gsf.de/cgi-bin/proj/medgen/mitofilter; ref. 29), and the first 213 nucleotides of the gene were PCR-amplified with the primers MSHtranspFor 5′-GGCCATGGTGTGAATTGCATAGTCGTCG-3′ and MSHtranspRev 5′-GGCCATGGAAA CATCACTTGACGTCTTC-3′. PCR products were ligated to the pGEM-T Easy Vector System (Promega) and digested with NcoI to release the insert. Insert fragments were ligated to the pCAMBIA 1302 vector at the NcoI site that resides at the start of gfp. This vector utilizes the cauliflower mosaic virus 35S promoter. Bombardment experiments used 4-week-old leaves of Arabidopsis (Col-0) with tungsten particles and the Biolistic PDS-1000/He system (Bio-Rad). Particles were bombarded into Arabidopsis leaves by using 900-psi (1 psi = 6.9 kPa) rupture discs under a vacuum of 26 inHg (1 inHg = 3.4 kPa). After the bombardment, Arabidopsis leaves were allowed to recover for 18–22 h on Murashige and Skoog media plates at 22°C in 16 h of daylight. Localization of GFP expression was conducted by confocal laser scanning microscopy with Bio-Rad 1024 MRC-ES by using 488-nm excitation and two-channel measurement of emission, 522 (green/GFP) and 680 (red/chlorophyll) nm.
Results
Positional Cloning of the CHM Locus.
High-resolution mapping of the CHM locus on Arabidopsis chromosome III delimited the gene to an 80-kb interval as shown in Fig. 1. Sequence analysis of the interval revealed a gene candidate with similarity in sequence features to the MutS gene of Escherichia coli (Fig. 2). MutS is a component of the E. coli mismatch repair and DNA recombination apparatus (30). The gene, comprising 22 exons, was predicted to encode a 43-aa mitochondrial targeting presequence with mitochondrial targeting values of 0.916 (mitoprot), 0.943 (predotar, www.inra.fr/Internet/Produits/Predotar/), and 0.856 (targetp, www.cbs.dtu.dk/services/TargetP/). RNA gel blots showed that the transcript derived from this gene was 3.5 kb (data not shown), and the encoded protein was 1,118 aa in length, predicting a 124-kDa polypeptide.
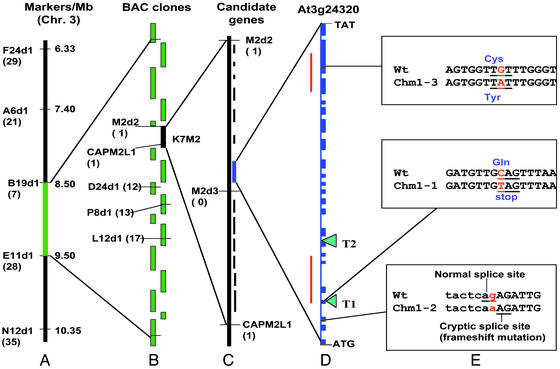
Figure 1.
Positional cloning of the CHM candidate locus. The use of molecular markers permitted the establishment of a genetic map (A) and identification of the intervening overlapping bacterial artificial chromosome clones for physical mapping (B). All physical mapping information was derived from the Arabidopsis Genome Initiative (50). High-resolution mapping with three markers permitted delimitation of the locus to an 80-kb interval contained within a single bacterial artificial chromosome clone (C). A gene candidate was identified within the interval based on predicted mitochondrial targeting features. The candidate CHM locus contains 22 exons (D) with two MutS-like conserved intervals denoted by red lines. Analysis of two ethyl methanesulfonate-derived mutants (chm1-1 and chm1-2) and one tissue culture-derived mutant (chm1-3), as well as two T-DNA insertion mutations (T1 and T2), provided definitive evidence of CHM identity (E). The numbers in parentheses in A correspond to the number of recombinants identified between the marker and the gene.
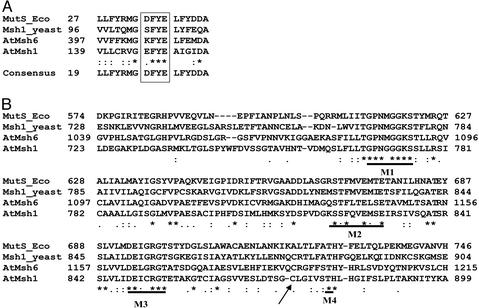
Figure 2.
Alignment of AtMSH1 with MutS and MutS homologs. The amino acid sequence alignment was performed by using CLUSTALW software and includes the MutS sequence from E. coli, MSH1 from Saccharomyces cerevisiae, and AtMSH6 and CHM (AtMSH1) from Arabidopsis. (A) Alignment of the region of the DNA binding domain that encompasses the conserved motif for mismatch recognition and DNA binding. (B) Alignment of a portion of the ATPase domain. The characteristic motifs for this domain are indicated by bold lines. M1, Walker motif; M2, ST motif; M3, DE motif (Walker B motif); M4, TH motif (31, 32). The asterisks indicate residues that are identical, and the arrow indicates the site of amino acid substitution in mutant chm1-3.
Confirmation of CHM Identity.
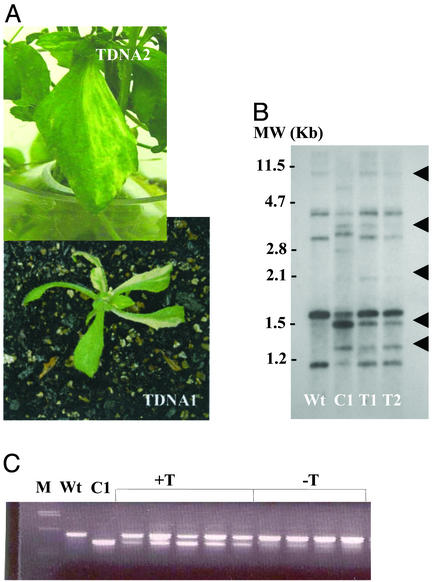
To test whether the identified locus might be CHM, we obtained two Arabidopsis lines (Col-0 ecotype) containing T-DNA insertion mutations within the candidate gene [GenBank accession nos. BH748178 (SALK041951) and BH908253 (SALK046763)]. The first insertion was located within the fourth exon and the second within the eighth intron. Analysis of the T-DNA mutants (T3 generation) revealed mild green-white leaf variegation, growing more intense in the selfed generation that followed (Fig. 3). Variegated plants carried a mitochondrial genome rearrangement similar to that observed in the mutants chm1-1 and chm1-2 (Fig. 3 B and C and data not shown). A population of 60 T4 plants segregating for one of the T-DNA (SALK041951) mutations (16 wild type, 31 hemizygous, 13 homozygous for the T-DNA) showed cosegregation of the T-DNA with the mitochondrial shifting phenotype. Of the 13 progeny homozygous for the T-DNA insertion, eight were variegated and the remaining five showed no obvious variegation phenotype. Incomplete penetrance of the variegation phenotype is characteristic of chm1-1 and chm1-2 mutants (25). All progeny homozygous for the T-DNA insertion mutation showed the mitochondrial shifting phenotype (data not shown). None of the segregants hemizygous for, or lacking, the T-DNA mutation showed evidence of variegation. The hemizygous plants showed no mitochondrial shifting. Similar cosegregation results were obtained for the second T-DNA (SALK046763) mutation as well (data not shown).
Figure 3.
The T-DNA insertion mutation phenotype. (A) Green-white variegation phenotype observed in two independent mutants derived by T-DNA insertion within the candidate CHM locus. T-DNA 1, SALK041951; T-DNA 2, SALK046763. (B) DNA gel blot hybridization analysis of mitochondrial genome configuration using the mitochondrial atp9-rpl16 junction sequence associated with substoichiometric shifting (16) as probe. Total genomic DNA was digested with BamHI, subjected to gel electrophoresis, blotted, and probed. Lane Wt designates wild-type ecotype Col-0, lane C1 designates mutant chm1-1, and lanes T1 and T2 designate two sister lines containing the T-DNA 1 insertion mutation. Arrowheads indicate DNA band pattern changes associated with substoichiometric shifting (24). (C) Cosegregation analysis of mitochondrial substoichiometric shifting with the T-DNA 1 insertion mutation. A three-primer PCR-based assay to detect substoichiometric shifting (16) was used to assay wild-type Col-0 (Wt), mutant chm1-1 (C1), and individual plants segregating for presence of the T-DNA insertion within the candidate CHM locus. All lanes labeled +T represent individual population segregants that are homozygous for the T-DNA insertion, whereas lanes labeled −T include two segregants that are hemizygous for the T-DNA insertion and two segregants that do not contain the T-DNA insertion. M designates the molecular weight marker lane.
To test further the possibility that the identified MutS-homologous sequence was CHM, we sequenced the chm1-1 and chm1-2 alleles of the gene. The chm1-1 line had a single nucleotide (C-T) substitution that gave rise to a premature stop codon within the fourth exon (Fig. 1E). The chm1-2 mutant had a single nucleotide (G-A) substitution at the intron–exon junction of exon 2 (Fig. 1E). This substitution resulted in a two-nucleotide slippage of the intron splice site, producing a frameshift and premature termination of translation five amino acids beyond the mutation site. Therefore, in both chm1-1 and chm1-2 mutant lines, the CHM candidate locus is predicted to give rise to highly truncated, inactive peptides.
Sequence analysis of the chm1-3 allele, derived from a tissue culture line by Martinez-Zapater et al. (24), revealed an amino acid (Cys-Tyr) substitution within the ATP binding domain (Fig. 1E). The mutant phenotype in this case may be due to the substitution of a bulkier amino acid within a site essential for protein function.
The CHM Candidate Has Features of a Mismatch Repair Component.
The MutS-homologous gene identified as a candidate for CHM displayed several features characteristic of a mismatch repair component. These features included an ATP-binding domain (amino acids 761–946) comprised of four well conserved motifs designated M1–M4 (ref. 31; Fig. 2B). In addition to ATPase function, this domain seems to be involved in dimerization of the protein (31, 32), although this has not yet been demonstrated for mitochondrial MutS homologs. A DNA binding domain (amino acids 129–206) was also identified (Figs. 1 and 2) to contain the aromatic doublet (FY) motif that is characteristic of this domain in MutS and MutS-like proteins (Fig. 2A). This doublet was shown to be essential for mismatch recognition and specific DNA binding activity (33, 34). We were unable to detect three other conserved domains characteristic of MutS. A connector domain (involved in interdomain interactions), a core domain, and a clamp domain (involved in nonspecific double-strand DNA binding) did not seem to be well conserved.
The CHM Candidate Protein Likely Localizes to Mitochondria.
To confirm that the MutS-like protein localized to the mitochondrion, we conducted RACE/PCR and discovered a transcript start site 578 residues upstream of the site predicted in the Munich Information Center for Protein Sequences (MIPS) database (ref. 35; http://mips.gsf.de/proj/thal/db/index.html) and in GenBank (accession no. AP000382). No start site was observed by RACE analysis at the point predicted by the MIPS database, and three clustered transcription start sites were detected at the upstream site (data not shown). The confirmed start site added 102 amino acids to the predicted protein product and permitted the identification of a mitochondrial targeting presequence that was omitted from the previous database entries. The sequence was annotated based on cDNA sequence analysis and is available as GenBank accession no. AY191303.
A transgene construction was developed that included the cauliflower mosaic virus 35S constitutive promoter linked to the first 213 nucleotides of the gene, encompassing the targeting presequence, and fused to the enhanced gfp reporter gene. This gene construction was introduced to Arabidopsis leaf cells by tungsten particle bombardment for transient expression assay. Confocal microscopy permitted the confirmation of transgene expression and gfp localization in mitochondria (Fig. 4). The potential for dual localization of the protein to plastids could not be definitively ascertained in these experiments.
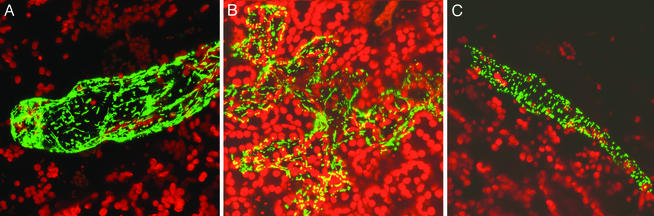
Figure 4.
Evidence for mitochondrial targeting capacity by the AtMSH1 protein. Particle bombardment experiments involved delivery of the AtMSH1 targeting presequence fused with enhanced gfp in association with the cauliflower mosaic virus 35S promoter. A and B show gfp localization as green within transformed epidermal cells. Mitochondria were identified by their characteristic movement and rapid interconversions from small, round to highly elongated shapes (A). Plastids located in the cells beneath emit red autofluorescence. Positive controls for mitochondrial (F1-ATPase γ-subunit provided by D. Stern, Boyce Thompson Institute, Ithaca, NY) (C) and chloroplast (Rubisco Pea/SSU/TPSS, provided by L. Alison, University of Nebraska) (data not shown) targeting were included with each experiment.
Discussion
The predicted features of the candidate CHM-encoded protein suggest that the gene encodes the mitochondrial MSH1 counterpart in higher plants. MSH1 encodes a mitochondrial mismatch repair protein in yeast, although its counterpart in animals has not yet been identified. The CHM candidate sequence showed strongest homology with the Arabidopsis nuclear MSH6 sequence (Fig. 2), consistent with suggestions that nuclear mismatch repair components likely derived from a progenitor to MSH1 (36).
Although the predicted CHM candidate protein displayed several features suggesting its involvement in mismatch repair, lines containing mutations in the locus showed no evidence of mitochondrial point mutation accumulation. The primary effect within the mitochondrion seemed to be the reproducible substoichiometric shifting phenomenon. This assumption is based on the observation of identical mitochondrial DNA restriction fragments arising on substoichiometric shifting in all chm mutants when tested repeatedly (refs. 16 and 24 and this article). Moreover, no evidence of progressive decline in plant growth features has been observed over time. The chm1-1 and chm1-2 mutants, reported in the 1970s (25), appear identical to one another in phenotype and mitochondrial DNA configuration. Although detailed sequence analysis would be required to estimate the incidence of mismatch accumulation in the chm mutants, one would anticipate a random pattern of mitochondrial DNA polymorphism and progressive phenotypic decline in chm mutants were the mismatch accumulation rate enhanced.
Mutation of the MSH1 locus in yeast results in rapid accumulation of mitochondrial genomic rearrangements leading to disruption of mitochondrial function. Interestingly, a reproducible pattern of DNA restriction fragment polymorphism was reported in some of the petite mutants arising in yeast MSH1 mutant strains (37). This observation may be an indication that msh1-associated mitochondrial genomic rearrangements are similar in plants and fungi. Alignment between the yeast MSH1 protein and the Arabidopsis CHM (AtMSH1) candidate shows only 17% amino acid identity overall, with ≈28% identity within the predicted functional domains for ATP and DNA binding, but with well conserved motifs (Fig. 2). The yeast MSH1 protein has been shown to have both DNA mismatch binding and ATPase activity (38, 39). These properties have not been tested in the MSH1 candidate from Arabidopsis.
The protein seemed to be highly conserved among plant species. We have identified several ESTs that present high homology (>60% amino acid identity) with AtMSH1 in soybean, tomato, potato, rice, wheat, and barley (data not shown). Although we have not yet tested for DNA binding and ATPase activities of the plant protein, identification of the chm1-3 mutation as a cysteine-tyrosine substitution within the predicted ATP binding domain does suggest the importance of this region to protein function. Substitution of the bulkier tyrosine would likely create distortion in the region, affecting ATP binding or hydrolysis.
Mismatch repair components seem to be involved in not only the binding and excision of nucleotide mismatches during the replication process, but also suppression of ectopic recombination (40, 41). Investigation of the mitochondrial substoichiometric shifting phenomenon suggests two alternative models for the influence of MSH1. It is conceivable that the AtMSH1 gene has shared or relinquished its mismatch repair function, such that its primary role in the plant mitochondrial genome is to regulate nonhomologous recombination. Disruption of MSH1 could thus result in the enhancement of intramolecular ectopic recombination activity detected as apparent amplification of novel mitochondrial DNA forms. A possible weakness in this model arises in reports that several plant systems with mitochondrial DNA molecules susceptible to shifting seem to be derived from a DNA exchange that involved at least one molecular form no longer present in high copy number. Some also seemed to contain unique sequences. Therefore, the shifted molecules were thought to replicate autonomously (14, 20, 42).
If mitochondrial DNA molecules that undergo shifting are, in fact, replicated autonomously, an alternative model for molecule-specific substoichiometric shifting might apply. The Arabidopsis MSH1 product likely participates as a component of the DNA replication apparatus. Mitochondrial DNA molecules subject to copy number shifting may have originated by earlier ectopic recombination events during the evolution of the lineage. In this case, the resulting chimeric sites might serve to trigger a process of site-specific replication stalling by the MSH1 protein during vegetative growth.
Both models assume that the replicative form of the mitochondrial genome within meristematic (undifferentiated) tissues differs from that within vegetative (somatic) tissues. Hence, stoichiometric shifting events in vegetative tissues do not condition irreversible loss of the suppressed genetic information. Presumably, the complete mitochondrial genetic complement is retained within the transmitting (meristematic) tissues (19, 42).
During higher plant evolution, natural allelic variation for the MSH1 locus likely revealed the adaptive advantage that arises from sporadic copy number modulation of mitochondrial genomic variants. Some of these variants, when amplified, condition male sterility that could facilitate advantageous outcrossing activity in natural populations (19). It merits noting that the mitochondrial DNA metabolism apparatus also seems to be important to the incidence of mitochondrial heteroplasmy in humans. Reports of nuclear, autosomal mutations leading to human mitochondrial genetic disorders are numerous. Autosomal dominant progressive external ophthalmoplegia (AdPEO; ref. 43) is associated with two loci on chromosomes 10 and 4 (44, 45). Interestingly, two nonsense mutations in the adenine nucleotide translocator (ANT) gene were identified on chromosome 4, and mutations in a gene encoding a mitochondrial protein with homology to the primase/helicase of the phage T7 were identified on chromosome 10 (46). Moreover, cases of autosomal recessive progressive external ophthalmoplegia (ArPEO) have been associated with the nuclear gene for mitochondrial gamma DNA polymerase on chromosome 15 (47). Finally, cases of mitochondrial neurogastrointestinal encephalomyopathy (MNGIE; ref. 48), an autosomal recessive mitochondrial disorder that leads to mitochondrial deletions, have been associated with mutations in a gene on chromosome 22 encoding thymidine phosphorylase (49).
It remains to be determined whether the substoichiometric intermediates common to higher plant mitochondrial genomes are maintained by replicative or recombinational mechanisms. This question may be resolved by the identification of proteins with which the Arabidopsis MSH1 protein interacts, as well as mitochondrial DNA sequences to which it binds. Experiments are currently underway to down-regulate the MSH1-homologous genes of other plant species to assess mitochondrial genome influence. With this article we identify an intriguing variation on the role of eukaryotic MutS homologs that has contributed adaptive advantage by a strategy that very well may be unique to the plant kingdom.
Acknowledgments
We thank Ms. Sophia Alvarez for her efforts in determining MSH1 transcript size and the University of Nebraska Center for Biotechnology Microscopy Core Facility for assistance with confocal microscopy. This work was supported by grants from the National Science Foundation and the Department of Energy (to S.A.M.). R.V.A. was supported by a predoctoral fellowship provided by the Conselho Nacional de Pesquisas (Brazil). Funding for The Salk Institute Genomic Analysis Laboratory-indexed insertion mutant collection was provided by the National Science Foundation.
Abbreviation
- T-DNA
portion of the Ti (tumor-inducing) plasmid that is transferred to plant cells
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY191303).
References
- 1.Golden T R, Melov S. Mech Aging Dev. 2001;122:1577–1589. doi: 10.1016/s0047-6374(01)00288-3. [DOI] [PubMed] [Google Scholar]
- 2.Naviaux R K. Eur J Pediatr. 2000;159:S219–S226. doi: 10.1007/pl00014407. [DOI] [PubMed] [Google Scholar]
- 3.Ravagnan L, Roumier T, Kroemer G. J Cell Physiol. 2002;192:131–137. doi: 10.1002/jcp.10111. [DOI] [PubMed] [Google Scholar]
- 4.Epstein C B, Waddle J A, Hale W, Dave V, Thornton J, Macatee T L, Garner H R, Butow R A. Mol Biol Cell. 2001;12:297–308. doi: 10.1091/mbc.12.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Small I, Wintz H, Akashi K, Mireau H. Plant Mol Biol. 1998;38:265–277. [PubMed] [Google Scholar]
- 6.Peeters N, Small I. Biochim Biophys Acta. 2001;1541:54–63. doi: 10.1016/s0167-4889(01)00146-x. [DOI] [PubMed] [Google Scholar]
- 7.Knoop V, Brennicke A. Crit Rev Plant Sci. 2002;21:111–126. [Google Scholar]
- 8.Oldenburg D J, Bendich A J. J Mol Biol. 2001;310:549–562. doi: 10.1006/jmbi.2001.4783. [DOI] [PubMed] [Google Scholar]
- 9.Backert S, Nielsen B L, Borner T. Trends Plant Sci. 1997;2:477–483. [Google Scholar]
- 10.Maleszka R, Skelly P J, Clark-Walker G D. EMBO J. 1991;10:3923–3929. doi: 10.1002/j.1460-2075.1991.tb04962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lecrenier N, Foury F. Gene. 2000;246:37–48. doi: 10.1016/s0378-1119(00)00107-4. [DOI] [PubMed] [Google Scholar]
- 12.Backert S, Borner T. Curr Genet. 2000;37:304–314. doi: 10.1007/s002940050532. [DOI] [PubMed] [Google Scholar]
- 13.Fauron C, Casper M, Gao Y, Moore B. Trends Genet. 1995;11:228–235. doi: 10.1016/s0168-9525(00)89056-3. [DOI] [PubMed] [Google Scholar]
- 14.Andre C, Levy A, Walbot V. Trends Genet. 1992;8:128–132. doi: 10.1016/0168-9525(92)90370-J. [DOI] [PubMed] [Google Scholar]
- 15.Mackenzie S, McIntosh L. Plant Cell. 1999;11:571–585. doi: 10.1105/tpc.11.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakamoto W, Kondo H, Murata M, Motoyoshi F. Plant Cell. 1996;8:1377–1390. doi: 10.1105/tpc.8.8.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Small I D, Isaac P G, Leaver C J. EMBO J. 1987;6:865–869. doi: 10.1002/j.1460-2075.1987.tb04832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janska H, Sarria R, Woloszynska M, Arrieta-Montiel M, Mackenzie S. Plant Cell. 1998;10:1163–1180. doi: 10.1105/tpc.10.7.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arrieta-Montiel M, Lyznik A, Woloszynska M, Janska H, Tohme J, Mackenzie S. Genetics. 2001;158:851–864. doi: 10.1093/genetics/158.2.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanazawa A, Tsutsumi N, Hirai A. Genetics. 1994;138:865–870. doi: 10.1093/genetics/138.3.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith R L, Chowdhury M K U. Theor Appl Genet. 1991;81:793–798. doi: 10.1007/BF00224992. [DOI] [PubMed] [Google Scholar]
- 22.Small I D, Suffolk R, Leaver C J. Cell. 1989;58:69–76. doi: 10.1016/0092-8674(89)90403-0. [DOI] [PubMed] [Google Scholar]
- 23.Mackenzie S, Chase C D. Plant Cell. 1990;2:905–912. doi: 10.1105/tpc.2.9.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez-Zapater J, Gil P, Capel J, Somerville C. Plant Cell. 1992;4:889–899. doi: 10.1105/tpc.4.8.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Redei G P. Mutat Res. 1973;18:149–162. [Google Scholar]
- 26.Jander G, Norris S R, Rounsley S D, Bush D F, Levin I M, Last R L. Plant Physiol. 2002;129:440–450. doi: 10.1104/pp.003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Chory J. In: Arabidopsis Protocols: Methods in Molecular Biology. Martinez-Zapater J M, Salinas J, editors. Totowa, NJ: Humana; 1998. pp. 55–60. [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 29.Claros M G, Vincens P. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 30.Marti T M, Kunz C, Fleck O. J Cell Physiol. 2002;191:28–41. doi: 10.1002/jcp.10077. [DOI] [PubMed] [Google Scholar]
- 31.Obmolova G, Ban C, Hsieh P, Yang W. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 32.Lamers M H, Perrakis A, Enzlin J H, Winterwerp H H, de Wind N, Sixma T K. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 33.Tachiki H, Kato R, Masui R, Hasegawa K, Itakura H, Fukuyama K, Kuramitsu S. Nucleic Acids Res. 1998;26:4153–4159. doi: 10.1093/nar/26.18.4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malkov V A, Biswas I, Camerini-Otero R D, Hsieh P. J Biol Chem. 1997;272:23811–23817. doi: 10.1074/jbc.272.38.23811. [DOI] [PubMed] [Google Scholar]
- 35.Schoof H, Zaccaria P, Gundlach H, Lemcke K, Rudd S, Kolesov G, Arnold R, Mewes H W, Mayer K F. Nucleic Acids Res. 2002;30:91–93. doi: 10.1093/nar/30.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Culligan K M, Meyer-Gauen G, Lyons-Weiler J, Hays J B. Nucleic Acids Res. 2000;28:463–471. doi: 10.1093/nar/28.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reenan R A G, Kolodner R D. Genetics. 1992;132:975–985. doi: 10.1093/genetics/132.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chi N-W, Kolodner R D. J Biol Chem. 1994;269:29984–29992. [PubMed] [Google Scholar]
- 39.Chi N-W, Kolodner R D. J Biol Chem. 1994;269:29993–29997. [PubMed] [Google Scholar]
- 40.Harfe B D, Jinks-Robertson S. Annu Rev Genet. 2000;34:359–399. doi: 10.1146/annurev.genet.34.1.359. [DOI] [PubMed] [Google Scholar]
- 41.Chen W, Jinks-Robertson S. Genetics. 1999;151:1299–1313. doi: 10.1093/genetics/151.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Janska H, Mackenzie S A. Genetics. 1993;135:869–879. doi: 10.1093/genetics/135.3.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeviani M, Servidei S, Gellera C, Bertini E, DiMauro S, DiDonato S. Nature. 1989;339:309–311. doi: 10.1038/339309a0. [DOI] [PubMed] [Google Scholar]
- 44.Suomalainen A, Kaukonen J, Amati P, Timonen R, Haltia M, Weissenbach J, Zeviani M, Somer H, Peltonen L. Nat Genet. 1995;9:146–151. doi: 10.1038/ng0295-146. [DOI] [PubMed] [Google Scholar]
- 45.Kaukonen J, Zeviani M, Comi G P, Piscaglia M G, Peltonen L, Suomalainen A. Am J Hum Genet. 1999;65:256–261. doi: 10.1086/302445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spelbrink J N, Li F Y, Tiranti V, Nikali K, Yuan Q P, Tariq M, Wanrooij S, Garrido N, Comi G, Morandi L, et al. Nat Genet. 2001;28:223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- 47.Van Goethem G, Dermaut B, Lofgren A, Martin J J, Van Broeckhoven C. Nat Genet. 2001;28:211–212. doi: 10.1038/90034. [DOI] [PubMed] [Google Scholar]
- 48.Hirano M, Silvestri G, Blake D M, Lombes A, Minetti C, Bonilla E, Hays A P, Lovelace R E, Butler I, Bertorini T E, et al. Neurology. 1994;44:721–727. doi: 10.1212/wnl.44.4.721. [DOI] [PubMed] [Google Scholar]
- 49.Nishino I, Spinazzola A, Hirano M. Science. 1999;283:689–692. doi: 10.1126/science.283.5402.689. [DOI] [PubMed] [Google Scholar]
- 50.The Arabidopsis Initiative. Nature. 2000;408:796–801. [Google Scholar]