Abstract
The NF-κB-like transcription factor Relish plays a central role in the innate immune response of Drosophila. Unlike other NF-κB proteins, Relish is activated by endoproteolytic cleavage to generate a DNA-binding Rel homology domain and a stable IκB-like fragment. This signal-induced endoproteolysis requires the activity of several gene products, including the IκB kinase complex and the caspase Dredd. Here we used mutational analysis and protein microsequencing to demonstrate that a caspase target site, located in the linker region between the Rel and the IκB-like domain, is the site of signal-dependent cleavage. We also show physical interaction between Relish and Dredd, suggesting that Dredd indeed is the Relish endoprotease. In addition to the caspase target site, the C-terminal 107 aa of Relish are required for endoproteolysis and signal-dependent phosphorylation by the Drosophila IκB kinase β. Finally, an N-terminal serine-rich region in Relish and the PEST domain were found to negatively regulate Relish activation.
Innate immune responses rely on transcription factors of the Rel/NF-κB family. In unstimulated cells, Rel proteins reside in the cytoplasm complexed with an inhibitory IκB molecule. After an immune challenge, the inhibitor is phosphorylated by the IκB kinase (IKK) complex, ubiquitinated, and degraded by the 26S proteasome. The released Rel protein translocates to the nucleus where it activates target genes. The signaling cascades that activate Rel proteins are remarkably conserved between flies and human (1–3). Many proteins involved in the mammalian tumor necrosis factor receptor pathway have close homologs in the Drosophila immune deficiency (imd) pathway, which controls the immune-induced production of antimicrobial peptides. Recent genetic studies have established an order in which the participating genes may act in this signaling pathway (4–7). The central transcription factor in the imd signaling cascade is the NF-κB factor Relish (8, 9). With its composite structure, comprising a Rel homology domain and an IκB-like domain, Relish is similar to the mammalian NF-κB precursors p100 and p105.
But in striking contrast to its mammalian counterparts, the activation of Relish does not require proteasome-dependent degradation of the IκB-like region. Instead, Relish is processed by rapid, signal-dependent endoproteolysis, generating two stable fragments: REL-68, which contains the Rel homology domain and translocates to the nucleus, and REL-49, which includes the IκB-like region and remains cytoplasmic (10). Unexpectedly, a role for a caspase in Relish activation was indicated by the fact that mutants in Dredd, a Drosophila caspase gene, are deficient in Relish processing and antimicrobial peptide production (10–12). But whether Dredd acts directly on Relish has been an open question.
In addition to Dredd, we found that the Drosophila IKK complex regulates Relish processing. The IKK complex is activated by immune stimulation and Drosophila IKKβ can directly phosphorylate Relish in vitro (13). Moreover, mutants in ird5 (IKKβ) and kenny (IKKγ) have the same immune phenotype as Relish mutants (14, 15). It has not been clear though whether IKKβ-mediated phosphorylation of Relish occurs in response to an immune stimulus and whether it is required for Relish cleavage in vivo.
Here, we further investigated the roles of Dredd and IKKβ in Relish cleavage and characterized those sequences in Relish that are required for its endoproteolysis. We report the actual cleavage site and direct interactions between Dredd and Relish, which together provide strong evidence that Relish endoproteolysis is indeed carried out by the caspase Dredd.
Materials and Methods
Cell and Fly Culture.
The culture of flies and the Drosophila cell lines Schneider L2* and mbn-2 (16, 17) have been described (10). As a Drosophila WT strain we used Canton-S. The mutant fly strains are described elsewhere: imd (18), ird52 (14), key1 (15), and DreddB118 (12). FLAG-Relish-RGSH6 (FRH)/ΔS29-S45 transgenic flies will be described elsewhere. Third-instar larvae were challenged and protein extracts were prepared according to ref. 10. Transient transfections of cell cultures were carried out according to ref. 19.
Plasmids and Immunoreagents.
The double-tagged full-length FRH construct was generated by PCR from a cDNA clone (8) using Relish-specific primers that also contained the sequences for the FLAG epitope (5′-TGTCTAATCTAGACCAAAATGGACTATAAGGACGATGACGACAAAAACATGAATCAGTACTACGACC-3′) and the RGSH6 epitope (5′-TTAGACATCTAGATCAACTGTGATGGTGATGGTGATGCGATCCTCTAGTTGGGTTAACCAGTAGGGCG-3′). The PCR product was first inserted into the pCR 4Blunt-Topo vector (Invitrogen) and then subcloned as an XbaI fragment into the transfection vector pPacPL (20). N-terminal deletions were created by restriction enzyme digests as indicated in Fig. 1. C-terminal deletions were generated by PCR, deleting the indicated amino acids but restoring the RGSH6 tag. Internal deletions and the point mutation were PCR-generated by using site-directed mutagenesis (QuikChange, Stratagene). For the kinase assay, FLAG-tagged Relish constructs were inserted into the pCITE-2A vector (Novagen). All PCR products were verified by sequencing. Metallothionein promoter expression constructs of the c-myc-tagged δ* isoform of Dredd are described in refs. 21 and 22. The Relish anti-C antibody is described in ref. 10. Mouse anti-FLAG IgG and mouse anti-c-myc antibodies were obtained from Sigma; mouse anti-RGSH6 was from Qiagen (Chatsworth, CA). Secondary antibodies were goat anti-mouse/horseradish peroxidase (Amersham Pharmacia) and donkey anti-mouse/Cye3 conjugates (Jackson ImmunoResearch).
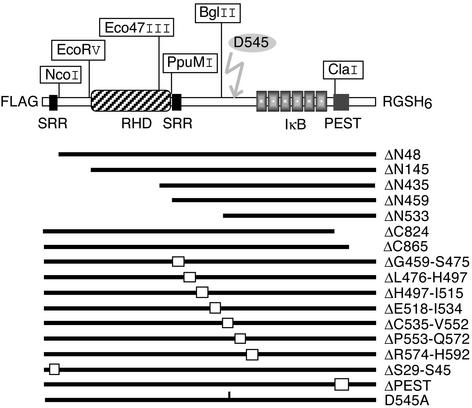
Figure 1.
Relish deletion constructs. Map of the double-tagged full-length FRH construct (Upper) and all of the mutations created from this basic construct. The FLAG epitope, IκB-like domain, PEST domain, Rel homology domain (RHD), RGSH6 epitope, the serine-rich region (SRR), and sites for restriction enzymes used in the cloning procedure are indicated. Internally deleted sequences appear as white boxes. A flash indicates the cleavage site.
Protein Extracts, Immunoblotting, and Gel-Shift Assay.
Transfected cells were challenged for 45 min and harvested. The PBS-washed cell pellets were lysed in either 3 vol of 20 mM Hepes (pH 7.9), 0.56 M KCl, 0.2 mM EDTA, 1.5 mM MgCl2, 2 mM DTT, and 25% glycerol for gel-shift assays or in 4–5 vol of immunoprecipitation-lysis buffer (10 mM Tris, pH 8/140 mM NaCl/1% Triton X-100) for all other purposes. Protease inhibitor mixture (Roche Molecular Biochemicals) and the phosphatase inhibitors sodium vanadate (1 mM) and sodium fluoride (50 mM) were added to lysis buffers. Protein concentrations were determined by Bradford assay (Bio-Rad). Both types of protein preparations were suitable for Western blotting and reporter enzyme assays. Immunoprecipitations followed a down-scaled protocol from ref. 23: 100 μg of protein extract was diluted 1:5 in immunoprecipitation-lysis buffer containing 1% BSA and incubated overnight at 4°C with 1 μl of the precipitating antibody. This mixture was loaded onto 30 μl of preblocked Protein G Sepharose (50% slurry, Amersham Pharmacia) and mixed end-over-end for 3–4 h at 4°C. The beads were then washed according to protocol, and protein complexes were eluted by warming the beads at 56°C for 1 h in 30 μl of 2× sample buffer for SDS/PAGE. 2-Mercaptoethanol (5%) was added to the eluate after separation from the beads and before boiling and gel electrophoresis. Immunoblotting was according to ref. 10 with minor modifications. Protein samples were separated on 10% SDS polyacrylamide gels and blotted. Membranes were incubated with primary antibody in RPMI medium 1640 (Invitrogen) and washed in PBS/0.05% Tween 20. Gel-shift assays were essentially carried out as described (10, 19).
Phosphatase Treatment.
Protein extracts were diluted 1:5 in 50 mM Tris⋅HCl (pH 7.5), 0.1 mM EDTA, 5 mM DTT, and 0.01% Brij 35 and treated with 40 units of λ protein phosphatase (New England Biolabs) per 25-μl reaction for 45 min at 30°C before SDS/PAGE.
Kinase Assay.
Versions of Relish were translated in vitro in reticulocyte lysates (Promega) and then immunoprecipitated by using anti-FLAG agarose (Sigma). Half of these immunoprecipitates were used in the control Western blot, the other half in an in vitro kinase reaction with recombinant Drosophila IKKβ (13).
Immunohistochemistry.
Immunohistochemistry was carried out as described (10).
Reporter Enzyme Assays.
Reporter plasmids for chloramphenicol transferase (CAT) and Cecropin A1-lacZ (24) were cotransfected along with the Relish constructs. Before lipopolysaccharide (LPS) addition, half of each culture was harvested to serve as the uninduced sample. To monitor the transfection efficiency, the amount of CAT was determined spectrophotometrically by ELISA (Roche Molecular Biochemicals). CAT correction was used to standardize all other analyses. β-Galactosidase activity was measured spectrophotometrically at 420 nm after substrate conversion: 10 μl of protein extract was added to 250 μl of substrate solution (0.8 mg/ml o-nitrophenyl-d-galactoside in 60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, modified from ref. 25). These values were first normalized for CAT expression and then compared with empty vector transfections. Finally, the values for the mutated forms of Relish were related to the one for the full-length protein.
Results
Sequences Required for Relish Endoproteolysis.
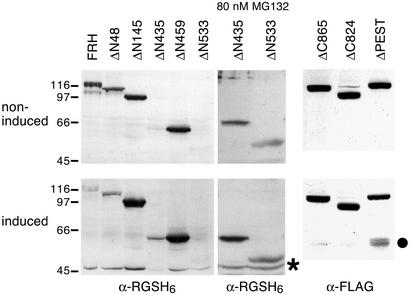
To identify regions of the Relish protein required for its signal-dependent cleavage, a series of deletion mutants was constructed (Fig. 1). We first analyzed truncations from the N-terminal or C-terminal ends. All constructs were expressed in cultured Drosophila cells and gave rise to proteins of the expected molecular weight. The responsiveness of WT and mutant Relish proteins to LPS treatment was tested, and Fig. 2 shows that all of the N-terminally truncated forms were endoproteolytically processed upon LPS treatment in the same way as the full-length FRH protein. The ΔN435 and ΔN533 proteins were unstable and could be detected only after application of the proteasome inhibitor MG132, which does not interfere with normal Relish cleavage (10). Even the highly truncated ΔN533 protein was processed into the slightly smaller REL-49 cleavage product. In contrast to the N-terminal deletions, truncations of the C terminus at position 865 or 824 greatly reduced LPS-induced cleavage. However, the ΔPEST protein, which lacks the domain between these two deletion end points, was cleaved normally. We conclude that the C-terminal 107 aa of Relish are necessary for signal-dependent endoproteolysis, whereas the entire N-terminal half is dispensable.
Figure 2.
Effects of terminal truncations on Relish processing. Western blots of protein extracts from mbn-2 cells transfected with the indicated Relish construct. The proteasome inhibitor MG132 (Calbiochem) was added to the cultures 1 h before LPS challenge. Antibodies used for detection are indicated at the bottom. Relish cleavage products are marked by an asterisk (REL-49) or a dot (REL-68).
Identification of the Relish Cleavage Site.
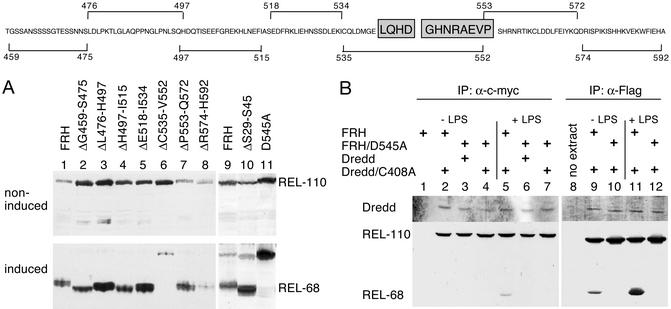
We also generated a series of internal deletions in the 130-residue linker between the Rel homology domain and the IκB-like domain, each removing ≈20 amino acid residues (Figs. 1 and 3). Lanes 1–8 in Fig. 3A show that all of the mutant proteins were processed normally in response to an immune stimulus, except for ΔC535-V552 (lane 6). Thus, residues 535–552 are required for recognition by the endoproteolytic machinery. Interestingly, this region contains a potential caspase target site, L-Q-H-D-G, in positions 542–546 that is similar to the consensus target site for group III caspases, L-E-x-D (26). The aspartate in the fourth position is known to be critical for recognition and cleavage by caspase proteases (27–29). Therefore, D545 was substituted by an alanine and the substitution mutant was tested for its ability to be cleaved in response to LPS. Fig. 3A, lane 11 shows that this mutant protein is completely resistant to signal-induced cleavage. Thus, this single aspartate residue within the caspase site is absolutely necessary for Relish endoproteolysis.
Figure 3.
Determination of the Relish cleavage site and interaction with Dredd. (Upper) Outline of the linker sequence. Borders of the deletion constructs used in A are indicated, and the caspase site and the sequence obtained from Edman degradation are boxed (see Fig. 7). (A) Analysis of internal deletion constructs and the point mutation D545A by immunoblotting. (B) Physical interaction between Dredd and Relish. Western blots showing coimmunoprecipitates of Relish with Dredd (Left) and vice versa (Right). Mbn-2 cell cultures were transfected with the indicated combination of expression plasmids for FLAG-tagged Relish and c-myc-tagged Dredd. Control reactions contained either lysates from only FRH-expressing cells (lane 1) or only buffer (lane 8). To avoid detection of the precipitating antibody, an alkaline phosphatase-conjugated anti-c-myc antibody and colorimetric detection were used to visualize Dredd. LPS induction was for 10 min. Forms of Relish were detected by using the anti-FLAG antibody.
Processing by a caspase at this site should result in peptide bond cleavage between D545 and the following glycine. To test this prediction, the N-terminal sequence of the REL-49 cleavage product was determined. A stably transfected cell line expressing the FRH protein was established, cells were challenged with LPS, and RGSH6-tagged REL-49 was enriched by nickel affinity chromatography and analyzed by Edman degradation (Fig. 7, which is published as supporting information on the PNAS web site, www.pnas.org). The obtained N-terminal sequence, G-H-N-R-A-E-V-P, is unique in the database and identical to residues 546–553 in Relish. We conclude that the LPS-induced endoproteolysis of Relish occurs between D545 and G546.
Thus, only the caspase site at 542–545 and the C terminus are required for Relish cleavage. In the human p100 and p105 proteins a glycine-rich sequence C-terminal to the nuclear translocation signal is important for the constitutive processing to p52 and p50, respectively. This region was proposed to function as a “stop signal,” preventing the complete degradation of the precursors by the proteasome (30, 31). Relish has a serine-rich region in a corresponding position and a similar one near the N terminus. However, the deletion of these motifs in the ΔG459-S475 and ΔS29-S45 mutants had no effect on Relish cleavage as determined by Western blot analysis (Fig. 3A, lanes 2 and 10). These results illustrate the difference between the signal-induced activation of Relish and the proteasome-dependent processing of p100 and p105.
Interactions Between Dredd and Relish.
The immune phenotype of Dredd mutant flies and their inability to process Relish showed that this caspase is involved in NF-κB activation (10–12). Our identification of the Relish cleavage site as a bona fide caspase target site further supports the proposal that Relish is actually cleaved by a caspase. To test whether Dredd interacts directly with Relish, we carried out coimmunoprecipitation experiments. Because Relish processing occurs rapidly, we sought to strengthen the interaction by using loss-of-function versions of both proteins. FLAG-tagged Relish (WT or noncleavable form) was expressed together with c-myc-tagged Dredd (WT or caspase-inactive) in different combinations in mbn-2 cells. Protein complexes were then precipitated by using either an anti-c-myc or the anti-FLAG antibody. As shown in Fig. 3B, Relish was coprecipitated with Dredd and vice versa. The interaction occurred with similar strength regardless of the enzymatic activity of the caspase or the form of Relish and was not LPS-induced. Note, that not only REL-110 but also small amounts of REL-68 bound to the caspase (Fig. 3B, lanes 5, 9, and 11). It is possible that Dredd acts as a dimer, and the combination of endogenous Dredd with the C408A mutant still produces cleavage product. We also observed that coexpression of WT Dredd and WT Relish resulted in reduced amounts of Relish (Fig. 8, which is published as supporting information on the PNAS web site).
Signal-Dependent Phosphorylation.
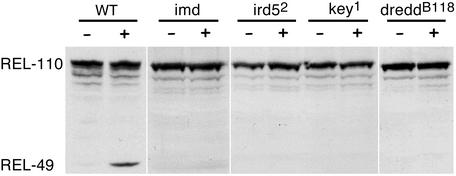
Among all of the genes known to act in the imd pathway, only the loss-of-function phenotypes of PGRP-LC and Dredd have so far been connected with a defect in Relish processing (10, 32). We extended these analyses with mutants in the ird5 (IKKβ) and kenny (IKKγ) genes along with those from imd mutants, which all display the same deficiency in Relish cleavage (Fig. 4). This result is consistent with our previous data on the importance of the Drosophila IKK complex for Relish cleavage in cell culture (13) and the published genetic findings (14, 15).
Figure 4.
Inhibition of Relish endoproteolysis in immunodeficient fly strains. Immunoblotting of protein extracts from untreated (−) and challenged (+) WT or mutant larvae. Appearance of REL-49 as visualized by the anti-C antibody indicates signal-induced endoproteolysis.
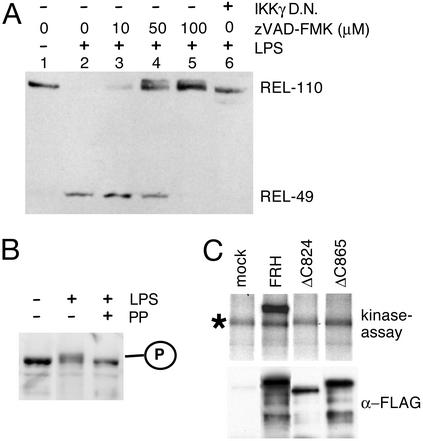
Consistent with the role of Dredd in the imd pathway, we found that the caspase inhibitor zVAD-fmk blocks Relish processing in a dose-dependent manner in Drosophila cell culture (Fig. 5A). Under these conditions an increase in the apparent molecular weight of REL-110 was observed upon stimulation with LPS. This modification could be reversed by phosphatase treatment (Fig. 5B), a result that we also obtained with the noncleavable D545A mutant after LPS induction (Fig. 9, which is published as supporting information on the PNAS web site). Thus, Relish is phosphorylated in a signal-dependent manner before proteolytic cleavage. The phosphorylation is likely to be mediated by the IKK complex, which can directly phosphorylate Relish in vitro (13). Inhibition of this kinase activity by overexpressing a dominant negative form of IKKγ prevented Relish cleavage and did not result in the accumulation of the modified Relish species (Fig. 5A, lane 6).
Figure 5.
Signal-dependent phosphorylation of Relish. (A) Signal-dependent modification of endogenous Relish demonstrated by immunoblotting of protein extracts from Schneider L2* cells that were treated with the caspase inhibitor zVAD-FMK (Calbiochem) 20 min before challenge. In lane 6 Relish processing was inhibited by expression of a dominant negative form of Drosophila IKKγ (13). (B) Phosphatase treatment of LPS-activated endogenous REL-110 from Schneider L2* cells treated with caspase inhibitor. PP, λ protein phosphatase. (C) In vitro kinase assay showing IKKβ-mediated phosphorylation of full-length Relish, but not of the C-terminally truncated forms. Synthesis and immunoprecipitation of the proteins in question was controlled by immunoblotting (Lower). IKKβ autophosphorylation is indicated by *. In B and C Relish was detected with the anti-C antibody.
As the ΔC865 and ΔC824 mutants were not cleaved, the deleted regions could be involved in phosphorylation. Therefore, we tested both proteins in an in vitro kinase assay with Drosophila IKKβ. Fig. 5C shows that neither the ΔC865 nor the ΔC824 truncation was phosphorylated whereas the full-length protein was. Deletion of the PEST domain resulted only in a slight reduction of phosphorylation (not shown). From these data we conclude that the 107 C-terminal residues of Relish are required both for phosphorylation by IKKβ and subsequent cleavage of Relish. It remains to be shown whether the phosphorylation actually occurs within this region. Interestingly, phosphorylation of human p105 by IKKβ requires a docking site in the PEST region and a phospho-acceptor site further C terminally (33, 34). A similar separation of functional sites might exist in Relish.
Effects on Rel Factor Function.
After the signal-induced cleavage of endogenous Relish, the N-terminal Rel homology domain is translocated to the nucleus, binds to the promoters of target genes, and activates their transcription (10). We tested whether these activities were affected in the Relish deletion mutants.
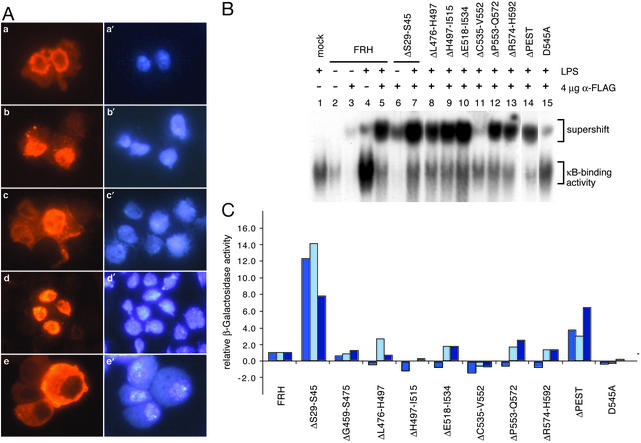
First, the subcellular localization of the mutated proteins was analyzed in cultured cells. Using the anti-FLAG antibody, the full-length protein and all of the mutant precursor proteins were detected in the cytoplasm of unstimulated cells (Fig. 6A a and a′). After stimulation with LPS, the staining was concentrated in the nucleus for all cleavable forms of Relish, as shown for the full-length protein and ΔS29-S45 in Fig. 6A b and d. Only the two noncleavable mutants, ΔC535-V552 and D545A, remained in the cytoplasm after stimulation, as shown for D545A in Fig. 6Ae. These results are consistent with the conclusion that Relish cleavage is a prerequisite for efficient nuclear translocation (10). Surprisingly, we also observed constitutive nuclear staining in cells expressing ΔS29-S45 (Fig. 6Ac) and ΔPEST (not shown).
Figure 6.
Effects of mutations in Relish on its function as a transcription factor. (A) Subcellular localization of mutated forms of Relish before (a and c) and 10 min after (b, d, and e) an immune stimulus. Relish proteins were detected with the anti-FLAG antibody and a Cye3-conjugated secondary antibody (red, a–e). The nuclei were counterstained with 4′,6-diamidino-2-phenylindole (blue, a′–e′). (a and b) Nuclear translocation of the FRH protein is shown and is representative for all other internal deletions except for those mentioned below. (c and d) Constitutive nuclear staining for the ΔS29-S45 protein is shown. (e) The lack of translocation for the D545A protein is shown. The same result was obtained for ΔC535-V552 (not shown). (B) κB-binding activity of mutated Relish proteins as tested by gel-shift assay using the same protein extracts as in Fig. 3A and a CecA1 κB oligonucleotide. (C) Relative transcriptional activity of mutated forms of Relish as determined by β-galactosidase activity. Results from uninfected cells of three independent transfection series are shown.
Second, electrophoretic mobility-shift assays were carried out with the Relish mutants by using a Cecropin A1 κB site as probe and whole-cell extracts (Fig. 6B). After stimulation with LPS, the oligonucleotide was bound by activated Relish. This was the case both for the endogenous and the overexpressed protein (Fig. 6B, lanes 1 and 4). A supershift assay with anti-FLAG antibody was used to specifically detect the DNA-binding capacity of the transfected Relish proteins (Fig. 6B, lanes 5–15). Except for the ΔC535-V552 and D545A mutations (Fig. 6B, lanes 11 and 15), all other Relish mutants exhibited a signal-dependent supershift, demonstrating that their respective REL-68 products can bind a κB site. The ΔS29-S45 protein also showed elevated κB binding in the absence of an LPS stimulus (Fig. 6B, lane 6) and a weaker constitutive binding was observed with ΔPEST (not shown).
Third, the transactivation capacity of the mutant Relish proteins was monitored after cotransfection with a Cecropin-lacZ reporter. Fig. 6C shows that in unstimulated cells full-length Relish and many of the mutants gave a modest activation of the reporter, ≈2-fold higher than empty vector transfections. The addition of LPS gave only a minor increase (data not shown). As expected, the two noncleavable mutants, ΔC535-V552 and D545A, showed reduced activity. ΔH497-I515 also appeared to be inactive. Consistent with their constitutive activity in the first two functional assays, ΔS29-S45 had ≈10-fold and ΔPEST ≈5-fold higher activity relative to full-length FRH. This result was also confirmed in a Northern blot, showing that the expression of the endogenous Diptericin gene was up-regulated in cells transfected with ΔS29-S45 and by ΔS29-S45 transgenic flies that have constitutively elevated levels of Diptericin RNA (not shown). The ΔS29-S45 and ΔPEST proteins were neither constitutively cleaved (Figs. 2 and 3A) nor were they more stable (data not shown). However, as shown above, both exhibited increased nuclear localization and κB binding without LPS stimulus. We cannot fully explain these observations but it seems likely that the N-terminal serine-rich region and the PEST domain are both important to prevent the Relish precursor from entering the nucleus (see below).
Discussion
The data presented here demonstrate that the signal-dependent cleavage of Relish occurs at a caspase target site. The residues immediately adjacent to the cleavage site fit the caspase consensus and the critical aspartate within this site, at position 545, is required for cleavage. These data strongly argue for a caspase as the Relish endoprotease. Although Dredd, by homology to the human caspases-8 and -10, is thought to be an initiator rather than an effector caspase, it is the prime candidate for the Relish endoprotease. Dredd mutants are unable to process Relish. Here, we demonstrated that Dredd and Relish interact physically. Furthermore, we did not find any of the other six known Drosophila caspases to be involved in Relish activation when we used RNA interference in cell culture (data not shown). However, we have so far failed to reconstitute cleavage in vitro with purified Dredd and IKKβ-phosphorylated Relish (P. Chen, N.S., and J. Abrams, unpublished data).
We noted that Dredd and Relish are bound to each other before an immune stimulus, suggesting the existence of a preassembled Dredd/Relish complex that is awaiting the incoming signal. This signal is most likely identical with phosphorylation by IKKβ. This set-up fits well with the speed of Relish processing, which occurs within seconds after LPS stimulation (10).
We identified additional regions in Relish that control its activation. The N-terminal serine-rich region and the PEST domain seem to negatively regulate Relish activation. One attractive model for Relish activation is that the precursor is held in a closed conformation, via an interaction between the serine-rich region and the PEST domain. This closed conformation would prevent nuclear translocation, inappropriate cleavage, and DNA binding by concealing the nuclear localization signal and the poorly structured linker with the caspase target site. Upon stimulation, Relish is phosphorylated in a reaction that requires IKKβ and the C-terminal 107 residues of Relish. This modification results in an open conformation in which the nuclear localization signal and the caspase target site would become accessible.
The direct involvement of a caspase in Relish endoproteolysis represents a novel mechanism of NF-κB activation and caspase function. Interestingly, a similar mechanism may also exist in mammalian systems. For example, Chun et al. (35) recently reported a caspase-8 loss-of-function mutation in human patients that is connected with defective activation of lymphocytes, a process that is known to require NF-κB. This new function of caspase-8 is independent of death receptor signaling and apoptosis induction. Another parallel between Relish processing and NF-κB activation in mammals is given by the so-called noncanonical NF-κB pathway, which requires NF-κB inducing kinase and IKK for the signal-dependent processing of p100 (36, 37).
Supplementary Material
Acknowledgments
We thank Yiran Lu, Kathryn V. Anderson, Bruno Lemaitre, and Dominique Ferrandon for mutant fly strains and Po Chen and John Abrams for Dredd plasmids. This project was supported by grants from the Göran Gustafsson Foundation for Scientific Research (to D.H.), the Swedish Research Council and the Swedish Foundation for Strategic Research (to D.H. and Y.E.), the Swedish Cancer Society (to S.S and Y.E.), and the Helen Hay Whitney Foundation (to N.S.), and by National Institutes of Health Grants GM29379 and GM59919 (to T.M.).
Abbreviations
- IKK
IκB kinase
- LPS
lipopolysaccharide
- FRH
FLAG-Relish-RGSH6
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Silverman N, Maniatis T. Genes Dev. 2001;15:2321–2342. doi: 10.1101/gad.909001. [DOI] [PubMed] [Google Scholar]
- 2.Khush R S, Leulier F, Lemaitre B. Trends Immunol. 2001;22:260–264. doi: 10.1016/s1471-4906(01)01887-7. [DOI] [PubMed] [Google Scholar]
- 3.Hultmark D. Curr Opin Immunol. 2003;15:1–8. doi: 10.1016/s0952-7915(02)00005-5. [DOI] [PubMed] [Google Scholar]
- 4.Georgel P, Naitza S, Kappler C, Ferrandon D, Zachary D, Swimmer C, Kopczynski C, Duyk G, Reichhart J-M, Hoffmann J A. Dev Cell. 2001;1:503–514. doi: 10.1016/s1534-5807(01)00059-4. [DOI] [PubMed] [Google Scholar]
- 5.Vidal S, Khush R S, Leulier F, Tzou P, Nakamura M, Lemaitre B. Genes Dev. 2001;15:1900–1912. doi: 10.1101/gad.203301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann J A, Ferrandon D, Royet J. Nature. 2002;416:640–644. doi: 10.1038/nature734. [DOI] [PubMed] [Google Scholar]
- 7.Leulier F, Vidal S, Saigo K, Ueda R, Lemaitre B. Curr Biol. 2002;12:996–1000. doi: 10.1016/s0960-9822(02)00873-4. [DOI] [PubMed] [Google Scholar]
- 8.Dushay M S, Åsling B, Hultmark D. Proc Natl Acad Sci USA. 1996;93:10343–10347. doi: 10.1073/pnas.93.19.10343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hedengren M, Åsling B, Dushay M S, Ando I, Ekengren S, Wihlborg M, Hultmark D. Mol Cell. 1999;4:827–837. doi: 10.1016/s1097-2765(00)80392-5. [DOI] [PubMed] [Google Scholar]
- 10.Stöven S, Ando I, Kadalayil L, Engström Y, Hultmark D. EMBO Rep. 2000;1:347–352. doi: 10.1093/embo-reports/kvd072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elrod-Erickson M, Mishra S, Schneider D. Curr Biol. 2000;10:781–784. doi: 10.1016/s0960-9822(00)00569-8. [DOI] [PubMed] [Google Scholar]
- 12.Leulier F, Rodriguez A, Khush R S, Abrams J M, Lemaitre B. EMBO Rep. 2000;1:353–358. doi: 10.1093/embo-reports/kvd073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silverman N, Zhou R, Stöven S, Pandey N, Hultmark D, Maniatis T. Genes Dev. 2000;14:2461–2471. doi: 10.1101/gad.817800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu Y, Wu L P, Anderson K V. Genes Dev. 2001;15:104–110. doi: 10.1101/gad.856901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rutschmann S, Jung A C, Zhou R, Silverman N, Hoffmann J A, Ferrandon D. Nat Immunol. 2000;1:342–347. doi: 10.1038/79801. [DOI] [PubMed] [Google Scholar]
- 16.Gateff E, Gissmann L, Shrestha R, Plus N, Pfister H, Schröder J, zur Hausen H. In: Invertebrate Systems in Vitro. Kurstak E, Maramorosch K, Dübendorfer A, editors. Amsterdam: Elsevier/North–Holland; 1980. pp. 517–533. [Google Scholar]
- 17.Samakovlis C, Åsling B, Boman H G, Gateff E, Hultmark D. Biochem Biophys Res Commun. 1992;188:1169–1175. doi: 10.1016/0006-291x(92)91354-s. [DOI] [PubMed] [Google Scholar]
- 18.Lemaitre B, Kromermetzger E, Michaut L, Nicholas E, Meister M, Georgel P, Reichhart J M, Hoffmann J A. Proc Natl Acad Sci USA. 1995;92:9465–9469. doi: 10.1073/pnas.92.21.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petersen U-M, Björklund G, Ip Y T, Engström Y. EMBO J. 1995;14:3146–3158. doi: 10.1002/j.1460-2075.1995.tb07317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urness L, Thummel C. Drosophila Inf Serv. 1993;72:193. [Google Scholar]
- 21.Chen P, Rodriguez A, Erskine R, Thach T, Abrams J M. Dev Biol. 1998;201:202–216. doi: 10.1006/dbio.1998.9000. [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez A, Oliver H, Zou H, Chen P, Wang X, Abrams J M. Nat Cell Biol. 1999;1:272–279. doi: 10.1038/12984. [DOI] [PubMed] [Google Scholar]
- 23.Edwards D N, Towb P, Wasserman S A. Development (Cambridge, UK) 1997;124:3855–3864. doi: 10.1242/dev.124.19.3855. [DOI] [PubMed] [Google Scholar]
- 24.Engström Y, Kadalayil L, Sun S-C, Samakovlis C, Hultmark D, Faye I. J Mol Biol. 1993;232:327–333. doi: 10.1006/jmbi.1993.1392. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 26.Thornberry N A, Rano T A, Peterson E P, Rasper D M, Timkey T, Garcia-Calvo M, Houtzager V M, Nordstrom P A, Roy S, Vaillancourt J P, et al. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 27.Blanchard H, Kodandapani L, Mittl P R, Marco S D, Krebs J F, Wu J C, Tomaselli K J, Grütter M G. Struct Fold Des. 1999;7:1125–1133. doi: 10.1016/s0969-2126(99)80179-8. [DOI] [PubMed] [Google Scholar]
- 28.Watt W, Koeplinger K A, Mildner A M, Heinrikson R L, Tomasselli A G, Watenpaugh K D. Struct Fold Des. 1999;7:1135–1143. doi: 10.1016/s0969-2126(99)80180-4. [DOI] [PubMed] [Google Scholar]
- 29.Nicholson D W. Cell Death Diff. 1999;6:1028–1042. doi: 10.1038/sj.cdd.4400598. [DOI] [PubMed] [Google Scholar]
- 30.Betts J C, Nabel G J. Mol Cell Biol. 1996;16:6363–6371. doi: 10.1128/mcb.16.11.6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin L, Ghosh S. Mol Cell Biol. 1996;16:2248–2254. doi: 10.1128/mcb.16.5.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choe K-M, Werner T, Stöven S, Hultmark D, Anderson K V. Science. 2002;296:359–362. doi: 10.1126/science.1070216. [DOI] [PubMed] [Google Scholar]
- 33.Heissmeyer V, Krappmann D, Hatada E N, Scheidereit C. Mol Cell Biol. 2001;21:1024–1035. doi: 10.1128/MCB.21.4.1024-1035.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salmeron A, Janzen J, Soneji Y, Bump N, Kamens J, Allen H, Ley S C. J Biol Chem. 2001;276:22215–22222. doi: 10.1074/jbc.M101754200. [DOI] [PubMed] [Google Scholar]
- 35.Chun H J, Zheng L, Ahmad M, Wang J, Speirs C K, Siegel R M, Dale J K, Puck J, Davis J, Hall C G, et al. Nature. 2002;419:395–399. doi: 10.1038/nature01063. [DOI] [PubMed] [Google Scholar]
- 36.Xiao G, Harhaj E W, Sun S-C. Mol Cell. 2001;7:401–409. doi: 10.1016/s1097-2765(01)00187-3. [DOI] [PubMed] [Google Scholar]
- 37.Senftleben U, Cao Y, Xiao G, Greten F R, Krähn G, Bonizzi G, Chen Y, Hu Y, Fong A, Sun S-C, Karin M. Science. 2001;293:1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.