Abstract
CD45 (leukocyte common) antigen is a hemopoietic cell-specific tyrosine phosphatase essential for antigen receptor-mediated signaling in lymphocytes. The molecule undergoes complex alternative splicing in the extracellular domain, and different patterns of CD45 splicing are associated with distinct functions. Lack of CD45 leads to severe combined immunodeficiency, and alterations of CD45 splicing, because of a polymorphism in exon 4, have been associated with altered immune function. Here we describe a polymorphism in exon 6 (A138G) of the gene encoding CD45 that interferes with alternative splicing. The polymorphism results in an amino acid substitution of Thr-47 to Ala in exon 6, a potential O- and N-linked glycosylation site. This exon 6 A138G variant is present at a frequency of 23.7% in the Japanese population but is absent in Caucasoids. Peripheral blood T cells from individuals carrying the A138G variant show a significant decrease in the proportion of cells expressing the A, B, and C CD45 isoforms and a high frequency of CD45R0+ cells. These phenotypic alterations in the A138G carriers may lead to changes in ligand binding, homodimerization of CD45, and altered immune responses, suggesting the involvement of natural selection in controlling the A138G carrier frequency.
The leukocyte common antigen CD45 is an abundant tyrosine phosphatase expressed on all leukocytes (1). The phosphatase activity of CD45 is essential for lymphocyte antigen receptor signal transduction. CD45 can also function as a Janus kinase phosphatase, negatively regulating cytokine receptor signaling (2). Both CD45 knockout mice (3, 4) and humans lacking CD45 expression (5, 6) are severely immunodeficient, with very few peripheral T lymphocytes and impaired T and B cell responses. These studies provide evidence for the crucial role of CD45 in the proper functioning of the immune system.
Multiple CD45 isoforms can be generated by alternative splicing of exons 4 (A), 5 (B), and 6 (C) of the extracellular domain (7). CD45 alternative splicing is highly conserved between species and is tightly regulated. In humans, naive T cells express high-molecular-weight CD45 isoforms recognized by CD45RA mAbs. Activation of the cells results in a change of expression to low-molecular-weight isoforms detected by a CD45R0 mAb (8).
These two major subsets of T lymphocytes expressing CD45RA and CD45R0 have been termed naive and memory cells. A polymorphism (C77G) in exon 4 of CD45 causing abnormal CD45 splicing has been described in humans (9). Activated or memory lymphocytes in these individuals continue to express both high-CD45RA and low-molecular-weight CD45R0 isoforms, in contrast to the normal pattern of low-molecular-weight CD45R0 isoform expression. Recently, another point mutation in exon 4 of CD45 (C59A) causing aberrant splicing has been identified, but it appears to be relatively rare (10).
The C77G polymorphism and abnormal CD45 splicing have been further linked to the development of multiple sclerosis in German (11) and Italian (12) patient cohorts, although other studies do not support such an association (13, 14). We have shown an increased frequency of the C77G variant allele in HIV-1-infected individuals in the United Kingdom (15). All of these observations suggest that abnormal CD45 splicing is associated with altered immunological function, autoimmunity, and viral infections.
Here we report a polymorphism in exon 6 A138G in the gene encoding CD45 with a very high prevalence in Japanese and Korean populations. We analyzed the expression of CD45 isoforms in peripheral blood mononuclear cells (PBMC) of individuals homozygous and heterozygous for the A138G variant. Our results show that T cells in individuals carrying the A138G allele display altered cell-surface CD45 isoform expression because of changes in alternative splicing. Analysis of exon 6 A138G and exon 4 C77G variants in different populations showed striking differences in the frequency and distribution of these mutations, suggesting effects of natural selection.
Materials and Methods
Materials.
One hundred seventy-five Japanese genomic DNAs were collected from Osaka City University Medical School (Osaka), of which 49 were from patients with malignant gynecological cancer. PBMC were isolated by centrifugation on a Ficoll-Paque (Amersham Biosciences) density gradient and genomic DNA was extracted by using DNA blood Minikit (Qiagen, Tokyo). One hundred fifty-five of these samples are from individuals aged 25–65 years, and 20 are from individuals over 65. For the phenotypic analysis on PBMC (see below), the ages of the individuals studied are as follows: A138A common variant controls, 31, 28, 37, and 27; A138G heterozygotes, 27, 27, 35, and 33; G138G homozygotes, 71, 29, 49, and 30. Two hundred nine Ugandan samples were provided by J. Whitworth and A. Hill (Wellcome Trust Centre for Human Genetics, Oxford; ref. 16). One hundred eighty-one genomic DNAs from British individuals consisted of 96 samples obtained from the local Blood Bank of the U.K. National Blood Transfusion Service (London), and 85 were provided by the Cancer and Immunogenetics Laboratory (Cancer Research UK, Oxford). Seventy-two Orkney samples were provided by the Cancer and Immunogenetics Laboratory, 48 Korean samples were provided by J. C. Kim (College of Medicine and Asan Medical Centre, University of Uslan, Seoul, South Korea), and 74 Russian and 65 Tatar samples were provided by Ruslan Rusibakiev (Academy of Science, Tashkent, Uzbekistan). Ethical approval was obtained and the patients gave consent for the study.
Denaturing High-Performance Liquid Chromatography (DHPLC) and Sequencing.
Genomic DNA was amplified by PCR by using the following primers flanking the relevant exons: ex4 forward (5′-CATATTTATTTTGTCCTTCTCCCA-3′), ex4 reverse (5′-GTGCAGAAATGCAGGAAAT-3′), ex6 forward (5′-GGAGAAGTGCTTGAAGATT-3′), and ex6 reverse (5′-GTGCCAGATATTATTTGTAGG-3′), generating fragments of 384 and 372 bp, respectively. A two-stage, 34-cycle PCR was performed, with an initial 10-min denaturation at 95°C, then 14 cycles of 30 s at 95°C, 30 s at 61.5°C, and 30 s at 72°C, followed by 20 cycles of 30 s at 95°C, 30 s of annealing at 54°C for exon 4 and 58°C for exon 6, 30 s at 72°C, and a final 6-min extension at 72°C. PCRs were performed in a volume of 50 μl, containing 10 pmol of each primer, 200 μM dNTP, 2.5 mM MgCl2, and 0.5 units of AmpliTaq Gold (Perkin–Elmer) in 1× KCl Perkin–Elmer buffer II. Two microliters of the PCR product was resolved on 2% agarose to test product size, and the remaining product was denatured for 4 min at 95°C, followed by 42 cycles of 1 min at 95°C, dropping by 1.6°C per cycle to 28.8°C to hybridize. Products were run on the DHPLC machine (WAVE, Transgenomic, Crewe, U.K.). Purified PCR products were subjected to automated sequencing by using the same primers as for DHPLC.
Amplification Refractory Mutation System (ARMS) PCR.
To detect carriers of the exon 4 C77G and exon 6 A138G mutations, we used ARMS PCR with two separate reaction mixes containing one forward primer and one of the two reverse primers. For exon 4 the original forward primer was used (5′-CATATTTATTTTGTCCTTCTCCCA-3′), amplifying both the wild-type and variant alleles. The reverse primer, ex4 rev A (5′-GAAAGTTTCCACGAACGG-3′), amplified only the wild-type allele, whereas ex4 rev B (5′-GAAAGTTTCCACGAACGC-3′) amplified only the variant allele. Similarly, for exon 6 the original forward primer (5′-GGAGAAGTGCTTGAAGATT-3′) and ex6 rev A (5′-CGTATCAGTCTGGACTCCA-3′) amplified the wild-type allele, whereas ex6 rev B (5′-CGTATCAGTCTGGACTCCG-3′) amplified the mutant allele only. Annealing temperatures were 56°C for C77G and 60.5°C for A138G. ARMS PCR products were resolved by alkaline-mediated differential integration (17). Samples were quantitated on a Fluorostar plate reader (BMG Labtechnologies, Offenburg, Germany). A random subset of samples was checked on 2% agarose gel.
RT-PCR.
Total RNA was extracted from PBMC, before and after stimulation with phytohemagglutinin, by using Tri-Reagent (Sigma). First-strand cDNA synthesis was performed using random hexadeoxynucleotide primers and the First-Strand cDNA Synthesis kit (Amersham Biosciences). The CD45 cDNA was amplified by using primers spanning the alternatively spliced CD45 exons: ex2 forward 5′-CGAAGCTTGCTGTTTCTTAGGGACACG-3′ and ex7 reverse 5′-GTGAATTCCAGAAGGGCTCAGAGTGGT-3′. The PCR conditions for amplification of CD45 cDNA included a 4-min incubation at 94°C followed by 30 reaction cycles (1 min at 94°C, 1 min at 55°C, and 4 min at 72°C) and a final 16-min extension at 72°C. The PCR products were resolved on a Visigel Separation Matrix (Stratagene). Bands were quantitated by using quality one software (Bio-Rad).
Flow Cytometric Analysis.
PBMC were surface-stained with the following mAbs against human CD45 isoforms: CD45R0-phycoerythrin (PE) (clone UCHL1, PharMingen), CD45R0-fluorescein isothiocyanate (FITC) (clone UCHL1, PharMingen), CD45RB-FITC (clone PD7/26, DAKO), CD45RB-PE (clone MT4, PharMingen), CD45RA-FITC (clone HI10, PharMingen), CD45RA-PE (clone 4KB5, DAKO), and allophycocyanin-conjugated CD3 (PharMingen). For CD45RC (clone YTH80.103, BioSource International, Camarillo, CA) analysis, a second layer of affinity-purified F(ab)′2 goat anti-rat FITC or PE (Caltag-MedSystems, Silverstone, U.K.) was used. Isotype-matched mAbs were used as controls. Ten thousand events per sample were collected on a FACSCalibur flow cytometer (Becton Dickinson) and analyzed by using winmdi software (http://facs.scripps.edu/software.html). For stimulation studies, PBMC were stimulated for 12 days with 1 μg/ml PHA-P (Sigma).
Results
Identification of a Point Mutation in Exon 6 of CD45 in Japanese and Korean Populations.
To examine the CD45 locus for polymorphisms, we used denaturing high-performance liquid chromatography to detect mutations in the alternatively spliced exons 4, 5, and 6 of CD45, followed by sequencing of the target individuals.
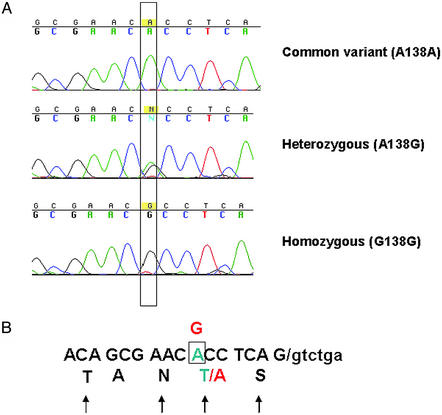
An A → G transversion at position 138 in exon 6 was found in Japanese samples. This A → G substitution located 7 bp before the splice donor site at the 3′ end of exon 6, and it results in a threonine-to-alanine semiconservative amino acid change at position 47 of the CD45RC exon 6 (Fig. 1). Thr-47 is a potential O-linked glycosylation site (18), but it is also adjacent to an asparagine and forms part of a consensus flanking sequence for an N-linked site. Therefore, a substitution of this Thr may lead to changes in the glycosylation of the extracellular domain of the molecule.
Figure 1.
Identification of A138G polymorphism in exon 6 of CD45. (A) An A → G transversion in position 138 of exon 6 was identified. Shown are examples of a common variant homozygote allele, a heterozygote, and a homozygote. The changed bases are boxed. (B) Schematic structure of exon 6, showing the relative position of the A138G mutation, which is 7 bp from the 3′ end of exon 6. The sequence of exon 6 is shown in a one-letter amino acid code, and the potential glycosylation sites are indicated by arrows. The mutation leads to the amino acid change 47T → A in the coded CD45RC domain, shown in red.
We used amplification refractory mutation system PCR to detect the presence of the A138G variant (Table 1), and we found 65 individuals of 175 Japanese samples that carried the variant allele, of which 9 were homozygotes for the G allele (allele frequency, 23.7%). The number of homozygotes was as expected according to the Hardy–Weinberg law. Note that the frequency of the A138G variant among the 49 Japanese patients with gynecological cancer was within the normal range (17 heterozygotes and 2 homozygotes), and the presence of the variant did not correlate with any distinctive clinical manifestation. The high frequency of this allele in the Japanese population was further confirmed by resequencing all individuals indicated as carrying the allele. We also found 7 heterozygotes of 48 Korean samples (allele frequency, 7.3%). No homozygotes have so far been found in this or other populations. The A138G variant was not detected in 209 Ugandan samples. We found 1 heterozygote of 181 United Kingdom samples and 1 of 72 Orkney samples. We also analyzed samples from Asia and found 6 A138G heterozygotes in 65 Tatars (from Kazan and the Crimea) but none in 74 Russians from Tashkent.
Table 1.
Frequency of CD45 exon 4 C77G and exon 6 A138G alleles in different populations
| Population | Total no. | Exon 6 (A138G)
|
Exon 4 (C77G)
|
||
|---|---|---|---|---|---|
| A138G | Allele frequency, % | C77G | Allele frequency, % | ||
| Japanese | 175 | 65 (9) | 23.7 | 0 | 0 |
| Korean | 48 | 7 | 7.3 | 0 | 0 |
| United Kingdom | 181 | 1 | 0.003 | 4 | 1.1 |
| Orkney | 72 | 1 | 0.7 | 5 | 3.5 |
| Tatar | 65 | 6 | 4.6 | 3 | 2.3 |
| Russian/Tashkent | 74 | 0 | 0 | 3 | 2 |
| Ugandan | 209 | 0 | 0 | 0 | 0 |
Homozygotes are shown in parentheses.
We further compared the distribution of exon 6 A138G and exon 4 C77G variants, the latter being the only described common polymorphism in CD45 causing abnormal CD45 splicing (Table 1). The C77G variant was absent in samples from an African population (Ugandan), as has been shown (16), and was not detected among the Far Eastern Japanese and Korean populations. Interestingly, the exon 4 C77G variant was found at a higher frequency (3.5%) in the United Kingdom Orkney islands than elsewhere, but, as might be expected, no C77G homozygotes were found in the samples studied.
CD45 Isoform Expression on PBMC of Individuals with the Exon 6 A138G Variant.
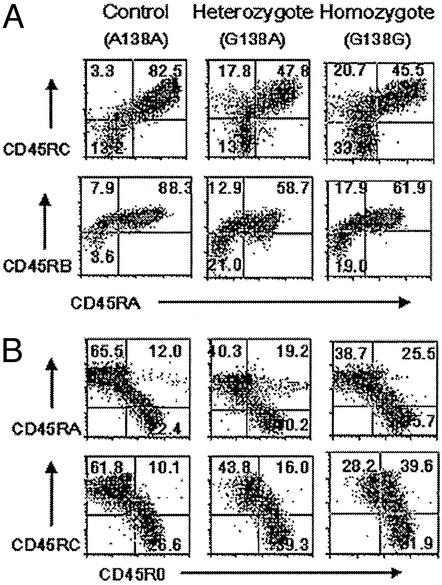
We next examined whether the A138G polymorphism affects CD45 isoform expression on the cell surface. Cryopreserved PBMC from four healthy A138G heterozygotes, four G138G homozygotes, and four common variant A138A homozygous controls were analyzed by flow cytometry. CD45RA, CD45RB, CD45RC, and CD45R0 antibodies were used to determine the expression of CD45 isoforms on these samples. There was a marked decrease in the proportion of cells expressing CD45RA and CD45RC or CD45RA and CD45RB isoforms in A138G-positive individuals, with homozygotes and heterozygotes showing similar changes (mean of 41.5% and 49.6% for CD45RA+CD45RC+ and 56.3% and 55.9% for CD45RA+CD45RB+; Table 2) from the common variant controls (73.7% and 71.3% for CD45RA+CD45RC+ and CD45RA+CD45RB+). This suggests a dominant effect on splicing of the A138G variant. There was a corresponding increase in CD45RA−CD45RC− or CD45RA−CD45RB− cells. Representative profiles are shown in Fig. 2A.
Table 2.
CD45 isoform expression on CD3+ cells from A138G and control individuals
| Cell subset | Control (A138A) | Heterozygote (A138G) | Homozygote (G138G) |
|---|---|---|---|
| CD45RA+ CD45RC+ | 73.7 ± 12.0 | 49.6 ± 1.9 | 41.5 ± 5.5 |
| CD45RA− CD45RC− | 18.7 ± 8.4 | 34.5 ± 5.5 | 36.0 ± 3.7 |
| CD45RA+ CD45RB+ | 71.3 ± 14.1 | 55.9 ± 12.5 | 56.3 ± 5.5 |
| CD45RA− CD45RB− | 10.1 ± 7.8 | 20.7 ± 7.6 | 21.8 ± 2.1 |
| CD45RC+ CD45R0+ | 11.2 ± 5.4 | 13.4 ± 6.3 | 40.6 ± 5.7 |
| CD45RC− CD45R0+ | 20.1 ± 6.6 | 32.0 ± 6.4 | 36.1 ± 5.0 |
| CD45RA+ CD45R0+ | 29.8 ± 22.8 | 56.5 ± 12.5 | 49.8 ± 23.3 |
| CD45RA− CD45R0+ | 20.5 ± 7.8 | 31.8 ± 6.0 | 31.4 ± 3.6 |
Means and standard deviations of data expressed as the percentage of CD3+ T cells from four homozygote (G138G), four heterozygotes (A138G), and four homozygotes for the common variant (A138A) control individuals.
Figure 2.
Expression of CD45 isoforms in human peripheral T cells. (A) PBMC were stained with isoform-specific antibodies against CD45RA and CD45RB or CD45RA and CD45RC together with anti-CD3. Analysis was performed on gated CD3+ cells. A138G individuals show a decrease in the proportion of cells expressing both CD45RA and CD45RC or CD45RA and CD45RB isoforms. (B) Expression of CD45R0, CD45RA, and CD45RC on CD3 cells from A138G and control individuals. PBMC were stained with anti-CD3 together with isoform-specific CD45R0 and CD45RA or CD45R0 and CD45RC antibodies. In the G138G variant, an increase in the proportion of CD45R0+ cells is seen. Examples are representative of similar analyses of four A138G homozygous, heterozygous, and control individuals. The ages of the individuals in the flow cytometric profiles shown are 31 (common variant control A138A), 35 (A138G heterozygote), and 29 (G138G homozygote) years.
A138G individuals, whether heterozygous or homozygous, also have higher proportion of cells expressing CD45R0 in either the presence or absence of CD45RA and in the absence of CD45RC (Table 2 and Fig. 2B). Thus, the effect of the variant on CD45R0 expression in the presence and absence of CD45RA appears dominant. In contrast, only A138G homozygotes had a higher proportion of cells expressing CD45R0 in association with CD45RC than either heterozygotes or controls (40.6% versus 13.4% in heterozygotes and 11.2% in controls), suggesting that in this context the A138G variant acts recessively.
After 12 days of stimulation with phytohemagglutinin, all of the CD3+ cells of the A138G homo- or heterozygous individuals showed very similar phenotypes to common variant control individuals with predominant expression of CD45R0 and CD45RB isoforms (data not shown).
No differences were observed in isoform expression on CD3-negative cells (data not shown).
Taken together, these data suggest that the exon 6 A138G carriers have fewer T cells expressing isoforms containing exons 4, 5, and 6 (naive phenotype cells) and have more activated CD45R0+ cells than the common variant CD45 controls. However, among A138G individuals, only homozygotes have more CD45R0+ cells that coexpress CD45RC.
Effect on CD45 Splicing.
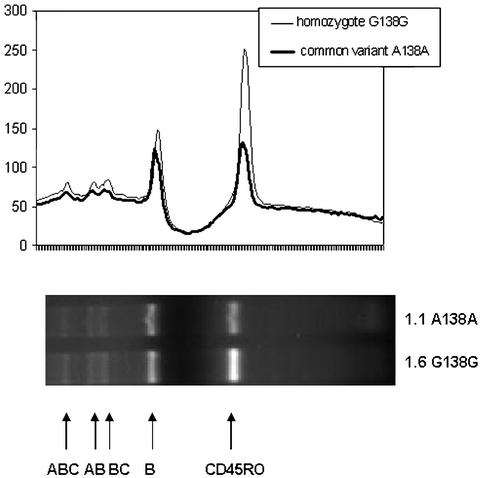
Because of the dramatic changes in the proportions of T cells expressing CD45 isoforms in A138G carriers, we next wanted to establish whether the exon 6 A138G variant interferes with CD45 splicing. RT-PCR analysis was performed on PBMC before and after stimulation with phytohemagglutinin. No qualitative differences in the expression of CD45 isoforms were observed among the G138G homozygotes, the heterozygous A138G individuals, and the A138A controls at either time point (Fig. 3). However, quantification of the intensity of the bands showed a significant difference in the level of CD45R0 transcripts, clearly demonstrating that splicing toward CD45R0 was increased in the mutated A138G gene when compared with the common variant.
Figure 3.
CD45 RNA expression in PBMC from homozygous G138G and homozygous A138A individuals. Total RNA was extracted from unstimulated PBMC. After reverse transcription the resulting cDNA was amplified with primers spanning exons 2–7 of the CD45 gene. PBMC from both homozygotes for G138G and common variant A138A allele individuals contained mRNA for the CD45R0 (197 bp), CD45RB (337 bp), CD45RBC (480 bp), CD45RAB (534 bp), and CD45RABC (677 bp) isoforms. Bands in each lane were quantitated, and densitograms are shown on top of the gel corresponding to the respective isoform. The ratio between the intensity of the CD45R0 and CD45RB bands is shown at the bottom of the gel. Data of three representative analyses of three G138G homozygotes and three control samples for the common variant A138A allele are shown.
These results suggest that the effect of this polymorphism is quantitative rather than qualitative, with A138G carriers expressing more CD45R0 transcript compared with the controls.
Discussion
Here we describe a polymorphism in exon 6 A138G of the gene encoding CD45 (PTPRC) that results in a semiconservative amino acid substitution, Thr47Ala, in the extracellular domain of the CD45 molecule. This variant allele is present, with a relatively high frequency in Korean (7.3%) and Japanese (23.7%) populations, with 5.1% homozygous individuals for the G allele among the Japanese. Although a thorough phenotypic and functional analysis has not yet been performed on A138G individuals, our results so far indicate that the carriers of the A138G mutation have a higher proportion of CD45R0+ T cells and a decrease in naive phenotype T cells expressing A, B, and C isoforms.
The altered CD45 isoform expression is most likely caused by changes in alternative splicing, as shown by the increased levels of CD45R0 transcripts detected by RT-PCR in the A138G carriers. These findings are in agreement with earlier studies (19) showing that mutations of nucleotides 134–144 at the most 3′ end of exon 6 resulted in mRNA that did not include exon 6 sequences. The exon 6 A138G mutation described here exerts a more subtle quantitative effect and does not induce complete splicing out of exon 6. It is plausible that, in a way similar to the model proposed by Tsai et al. (19), the A138G substitution reduces the overall similarity of the splice site to the consensus sequence, resulting in a less efficient recognition by the spliceosome. Alternatively, the exon 6 A138G change may induce alterations in exon splicing by disrupting regulatory elements within the exon itself (20). For example, the C77G polymorphism in exon 4 functions by disrupting an exon splicing silencer that normally represses the use of the 5′ splice site of exon 4 (21). Further studies using minigenes containing the mutation will be required to determine the precise mechanism for the altered CD45 expression in the A138G variant.
An alternative and more speculative explanation for the observed phenotypic differences of the PBMC of A138G carriers might be that the variant results in the expression of a structurally altered CD45 molecule. Thus, the A138G polymorphism results in the substitution of Thr47Ala, a potential glycosylation site for both O- and N-linked sugars, and may therefore change the reactivity with carbohydrate-dependent epitopes of anti-CD45 mAbs (22). Changes in the glycosylation not only might change the interactions with anti-CD45 antibodies, but may have important implications for the function of the CD45, because the crucial contribution of carbohydrates to the regulation of CD45 isoform function has been documented. Several lectin-like molecules have been shown to bind to carbohydrates of CD45, among other ligands. These include CD22 (23), galectins (24–26), mannose receptor (27), and serum mannan-binding protein (28, 29). The CD45 ectodomain has also been suggested to influence CD45 engagement in cis interactions with T cell antigen receptor, CD4, and CD8 (30–32), but no direct binding between the CD45 ectodomain and another protein has been shown so far. Another proposed role for the CD45 extracellular domain is the regulation of dimerization, and there is evidence that CD45 forms dimers on the cell surface (33, 34). These studies suggest that the structural differences caused by the A138G variant could affect the interactions of CD45 with potential ligands in cis and trans as well as dimerization between CD45 isoforms, and might have functional consequences for the immune response.
It is interesting that the exon 4 C77G and exon 6 A138G variants have quite different geographical distributions. This may suggest that the variants arose independently after the emigration of ancestral humans from Africa. The high frequency of the A138G variant in Japan suggests that it arose in the Far East, and its low frequency elsewhere would confirm this. The maintenance of these CD45 variants in different human populations, given their functional consequences, could be influenced by natural selection, e.g., with respect to differential resistance to infections. Further functional and disease-association studies may thus provide more convincing evidence for a selective effect, particularly of the A138G variant.
In Caucasoids the one polymorphism with an obvious phenotypic effect is the previously described C77G mutation in exon 4, which prevents normal splicing from high-molecular-weight (CD45RA) to low-molecular-weight (CD45R0) isoforms. We have previously shown that the frequency of the C77G variant allele in Northern Europe and North America is in the region of 0.85–1.6% and that it is absent in Africans (16). The data presented here confirm the previously observed frequency in the United Kingdom (on a different set of United Kingdom samples) and the lack of this variant in African Ugandan populations (16), and indicate a similar lack among the Far Eastern Japanese and Korean populations. It would be interesting to analyze whether the relatively high allele frequency in Orcadians is associated with the high incidence of multiple sclerosis in the Orkney Islands (35, 36).
In summary, our results suggest that individuals with the A138G variant have an increased proportion of T cells with an activated/effector phenotype, as determined by the increased proportion of CD45R0+ cells and reduced number of cells expressing the CD45 A, B, and C isoforms. The functional consequences of the mutation for immune responses remain to be elucidated. However, the altered CD45 expression may contribute to changes in interaction with potential ligands or homo- or heterodimerization of the CD45 isoforms. Xu and Weiss (34) recently proposed a model suggesting that expression of the CD45R0 isoform in activated cells shifts the equilibrium of cell-surface CD45 toward dimers and acts as a negative regulator, contributing to the cessation of the immune response. Increased expression of the CD45R0 isoforms caused by the A138G polymorphism would promote this negative regulation, resulting in a less vigorous immune response, which may reduce the risk of autoimmune disease in A138G carriers. Alternatively, the higher proportion of CD45R0+ cells may indicate that these individuals have a larger memory population and can make vigorous recall responses to pathogens. The high frequency of this allele in Japan and Korea may indicate that it confers a survival advantage and that functional and disease association studies are warranted.
Acknowledgments
We thank Dr. David Tough for critical reading of the manuscript and Dr. Diana Wallace for helpful discussions and suggestions.
Abbreviation
- PBMC
peripheral blood mononuclear cells
References
- 1.Penninger J M, Irie-Sasaki J, Sasaki T, Oliveira-Dos-Santos A J. Nat Immunol. 2001;2:389–396. doi: 10.1038/87687. [DOI] [PubMed] [Google Scholar]
- 2.Irie-Sasaki J, Sasaki T, Matsumoto W, Opavsky A, Cheng M, Welstead G, Griffiths E, Krawczyk C, Richardson C D, Aitken K, et al. Nature. 2001;409:349–354. doi: 10.1038/35053086. [DOI] [PubMed] [Google Scholar]
- 3.Kishihara K, Penninger J, Wallace V A, Kundig T M, Kawai K, Wakeham A, Timms E, Pfeffer K, Ohashi P S, Thomas M L, et al. Cell. 1993;74:143–156. doi: 10.1016/0092-8674(93)90302-7. [DOI] [PubMed] [Google Scholar]
- 4.Byth K F, Conroy L A, Howlett S, Smith A J, May J, Alexander D R, Holmes N. J Exp Med. 1996;183:1707–1718. doi: 10.1084/jem.183.4.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kung C, Pingel J T, Heikinheimo M, Klemola T, Varkila K, Yoo L I, Vuopala K, Poyhonen M, Uhari M, Rogers M, et al. Nat Med. 2000;6:343–345. doi: 10.1038/73208. [DOI] [PubMed] [Google Scholar]
- 6.Tchilian E Z, Wallace D L, Wells R S, Flower D R, Morgan G, Beverley P C. J Immunol. 2001;166:1308–1313. doi: 10.4049/jimmunol.166.2.1308. [DOI] [PubMed] [Google Scholar]
- 7.Streuli M, Hall L R, Saga Y, Schlossman S F, Saito H. J Exp Med. 1987;166:1548–1566. doi: 10.1084/jem.166.5.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akbar A N, Terry L, Timms A, Beverley P C, Janossy G. J Immunol. 1988;140:2171–2178. [PubMed] [Google Scholar]
- 9.Schwinzer R, Wonigeit K. J Exp Med. 1990;171:1803–1808. doi: 10.1084/jem.171.5.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobsen M, Hoffmann S, Cepok S, Stei S, Ziegler A, Sommer N, Hemmer B. Immunogenetics. 2002;54:158–163. doi: 10.1007/s00251-002-0455-7. [DOI] [PubMed] [Google Scholar]
- 11.Jacobsen M, Schweer D, Ziegler A, Gaber R, Schock S, Schwinzer R, Wonigeit K, Lindert R B, Kantarci O, Schaefer-Klein J, et al. Nat Genet. 2000;26:495–499. doi: 10.1038/82659. [DOI] [PubMed] [Google Scholar]
- 12.Ballerini C, Rosati E, Salvetti M, Ristori G, Cannoni S, Biagioli T, Massacesi L, Sorbi S, Vergelli M. Neurosci Lett. 2002;328:325–327. doi: 10.1016/s0304-3940(02)00565-7. [DOI] [PubMed] [Google Scholar]
- 13.Vorechovsky I, Kralovicova J, Tchilian E, Masterman T, Zhang Z, Ferry B, Misbah S, Chapel H, Webster D, Hellgren D, et al. Nat Genet. 2001;29:22–23. doi: 10.1038/ng723. [DOI] [PubMed] [Google Scholar]
- 14.Barcellos L F, Caillier S, Dragone L, Elder M, Vittinghoff E, Bucher P, Lincoln R R, Pericak-Vance M, Haines J L, Weiss A, et al. Nat Genet. 2001;29:23–24. doi: 10.1038/ng722. [DOI] [PubMed] [Google Scholar]
- 15.Tchilian E Z, Wallace D L, Dawes R, Imami N, Burton C, Gotch F, Beverley P C. AIDS. 2001;15:1892–1894. doi: 10.1097/00002030-200109280-00024. [DOI] [PubMed] [Google Scholar]
- 16.Tchilian E Z, Dawes R, Ramaley P A, Whitworth J A, Yuldasheva N, Wells S, Watera C, French N, Gilks C F, Kunachiwa W, et al. Immunogenetics. 2002;53:980–983. doi: 10.1007/s00251-001-0410-z. [DOI] [PubMed] [Google Scholar]
- 17.Bartlett S, Straub J, Tonks S, Wells R S, Bodmer J G, Bodmer W F. Proc Natl Acad Sci USA. 2001;98:2694–2697. doi: 10.1073/pnas.041619998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van den Steen P, Rudd P M, Dwek R A, Opdenakker G. Crit Rev Biochem Mol Biol. 1998;33:151–208. doi: 10.1080/10409239891204198. [DOI] [PubMed] [Google Scholar]
- 19.Tsai A Y, Streuli M, Saito H. Mol Cell Biol. 1989;9:4550–4555. doi: 10.1128/mcb.9.10.4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith C W, Valcarcel J. Trends Biochem Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 21.Lynch K W, Weiss A. J Biol Chem. 2001;276:24341–24347. doi: 10.1074/jbc.M102175200. [DOI] [PubMed] [Google Scholar]
- 22.Pulido R, Schlossman S F, Saito H, Streuli M. J Exp Med. 1994;179:1035–1040. doi: 10.1084/jem.179.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stamenkovic I, Sgroi D, Aruffo A, Sy M S, Anderson T. Cell. 1991;66:1133–1144. doi: 10.1016/0092-8674(91)90036-x. [DOI] [PubMed] [Google Scholar]
- 24.Perillo N L, Pace K E, Seilhamer J J, Baum L G. Nature. 1995;378:736–739. doi: 10.1038/378736a0. [DOI] [PubMed] [Google Scholar]
- 25.Walzel H, Schulz U, Neels P, Brock J. Immunol Lett. 1999;67:193–202. doi: 10.1016/s0165-2478(99)00012-7. [DOI] [PubMed] [Google Scholar]
- 26.Symons A, Cooper D N, Barclay A N. Glycobiology. 2000;10:559–563. doi: 10.1093/glycob/10.6.559. [DOI] [PubMed] [Google Scholar]
- 27.Martinez-Pomares L, Crocker P R, Da Silva R, Holmes N, Colominas C, Rudd P, Dwek R, Gordon S. J Biol Chem. 1999;274:35211–35218. doi: 10.1074/jbc.274.49.35211. [DOI] [PubMed] [Google Scholar]
- 28.Uemura K, Yokota Y, Kozutsumi Y, Kawasaki T. J Biol Chem. 1996;271:4581–4584. doi: 10.1074/jbc.271.9.4581. [DOI] [PubMed] [Google Scholar]
- 29.Baldwin T A, Ostergaard H L. J Immunol. 2001;167:3829–3835. doi: 10.4049/jimmunol.167.7.3829. [DOI] [PubMed] [Google Scholar]
- 30.Alexander D R. In: Lymphocyte Signalling: Mechanisms, Subversion, and Manipulation. Harnett M M, Rigley K P, editors. New York: Wiley; 1997. p. 107. [Google Scholar]
- 31.Leitenberg D, Boutin Y, Lu D D, Bottomly K. Immunity. 1999;10:701–711. doi: 10.1016/s1074-7613(00)80069-2. [DOI] [PubMed] [Google Scholar]
- 32.Dornan S, Sebestyen Z, Gamble J, Nagy P, Bodnar A, Alldridge L, Doe S, Holmes N, Goff L K, Beverley P, et al. J Biol Chem. 2002;277:1912–1918. doi: 10.1074/jbc.M108386200. [DOI] [PubMed] [Google Scholar]
- 33.Majeti R, Xu Z, Parslow T G, Olson J L, Daikh D I, Killeen N, Weiss A. Cell. 2000;103:1059–1070. doi: 10.1016/s0092-8674(00)00209-9. [DOI] [PubMed] [Google Scholar]
- 34.Xu Z, Weiss A. Nat Immunol. 2002;3:764–771. doi: 10.1038/ni822. [DOI] [PubMed] [Google Scholar]
- 35.Roberts D F. Genet Epidemiol. 1991;8:147–151. doi: 10.1002/gepi.1370080302. [DOI] [PubMed] [Google Scholar]
- 36.Rothwell P M, Charlton D. J Neurol Neurosurg Psychiatry. 1998;64:730–735. doi: 10.1136/jnnp.64.6.730. [DOI] [PMC free article] [PubMed] [Google Scholar]