Abstract
Noncytopathic RNA viruses persist in their natural hosts at various levels as highly mutating quasispecies. They exhibit only one known serotype. In most inbred DBA/2 mice infected with 2 × 104 or 2 × 106 plaque-forming units (pfu) of lymphocytic choriomeningitis virus (LCMV), the virus is transiently controlled below detectable levels measured with conventional assays (<1.7 pfu), but reemerges despite a common neutralizing Ab (nAb) response. Wild-type virus and cloned mutant viruses that had escaped polyclonal nAb responses in vivo induced nAb titers in new hosts that were usually cross-reactive; some sera were highly specific for certain mutants. The few mice that controlled LCMV infection for >170 days produced not only nAb against wild-type but also variably against many other mutants isolated from other mice with reemerging viremia. When DBA/2 mice were immunized and boosted with 200 pfu of a LCMV mutant, the neutralizing Ab response was limited to the immunizing “personal” clone. Thus, in contrast to classical serotype-defined cytopathic viruses (e.g., polio viruses) that induce strictly non-cross-reactive nAb titers, LCMV, a noncytopathic RNA virus, represents a dynamic multiplicity of personal serological submutants. Together, these mutants form a generally recognized “public” serotype. These findings may help to explain aspects of human infections and Ab responses against hepatitis B virus, hepatitis C virus, and HIV.
The key role of serotype-specific neutralizing Abs (nAbs) in controlling acute cytopathic virus infections is well accepted. In contrast, the role of nAbs in the control of poorly or noncytopathic virus infections [HIV or hepatitis C virus (HCV) in humans and lymphocytic choriomeningitis virus (LCMV) in mice], which develop quasispecies (1–6) in an individual host, is not well understood. In immunocompetent hosts, HIV, HCV, or LCMV are initially controlled by a strong cytotoxic T cell (CTL) response, whereas the nAb response usually appears late in the course of infection and sometimes remains low titered (7–9). This is in contrast to many of the acute cytopathic epidemic virus infections such as polio viruses I, II, and III, which are essentially controlled by very early and high titered nAb responses. Nevertheless, and despite their late appearance, there is good evidence that nAbs are important for the long-term virus control of hepatitis B virus (HBV), HCV, HIV, or LCMV. These low- or noncytopathic RNA viruses tend to persist in the host and also tend to develop nAb-escape mutants within an individual host. Such nAb-escape mutants have been observed not only in experimental mice infected with LCMV (10–12) but also during HCV (13–14) and HBV infections (15–18), or in animals and humans infected with simian immunodeficiency virus (SIV), or HIV, respectively (19–23). Here we evaluated LCMV-escaping nAb responses in nonmanipulated inbred mice to study virus and host parameters influencing the emergence of nAb-escape mutants in blood. The results correlate different levels of nAb with virus control and define low titered public and higher titered personal serotypes of noncytopathic RNA virus quasispecies.
Materials and Methods
Mice and Virus.
DBA/2 mice were obtained from BRL (Füllinsdorf, Switzerland) and Charles River Breeding Laboratories and were kept under specific pathogen-free conditions. The animal care was in accordance with institutional guidelines. LCMV strain WE (LCMV-WE) was originally obtained from F. Lehmann-Grube (Heinrich Pette Institute, Hamburg, Germany) and propagated on L929 cells. Mice were infected with 2 × 102, 2 × 104, or 2 × 106 plaque-forming units (pfu) of LCMV-WE or mutant virus intravenously. LCMV-neutralizing Ab escape mutants were isolated from the whole blood of DBA/2 mice (20, 90, or 110 days after infection), grown on MC57 cells for 48 h, and subsequently plaque-purified two times in vitro as described (11).
Neutralizing Activity.
Neutralizing activity against LCMV was measured in a focus reduction assay. The neutralizing titer was defined as the dilution causing half-maximal reduction of foci of LCMV when compared with the same amount of virus incubated with control sera from uninfected mice or medium only. A titer of <1 indicates neutralization comparable to the medium or naïve serum control.
T Cell-Mediated Cytotoxicity.
T cell-mediated cytotoxicity was performed as described (24).
Molecular Analysis.
Total RNA of MC57 cells infected with either LCMV-WE wild-type mutant virus isolates for 48 h at an initial multiplicity of infection of 0.01 was extracted by using Trizol. RT-PCR was performed by using LCMV-glycoprotein (GP)1-specific primers R1 (5′-1037TCG TAG CAT GTC ACA GAA CTC TTC1014-3′) for the reverse transcription and the primer pairs 001/RC1 (001, 5′-1CGC ACC GGG GAT CCT AGG CTT21-3′; and RC1, 5′-965GAG CTC TGC AGC AAG GAT CAT CC942-3′) for Hot Start PCR amplification. PCR products were sequenced by an Applied Biosystems/Bio-Rad cycle sequencing kit, using the primers 001 and RC1.
Results
DBA/2 Mice Transiently Control a High Dose of LCMV and Produce Cross-Reactive nAbs.
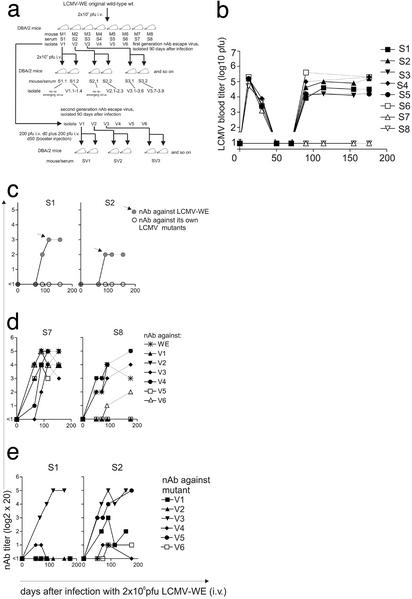
We analyzed DBA/2 mice, which are susceptible to LCMV, in part, probably because they have low CD8+ T cells compared with most other mouse strains (25). After infection with 2 × 106 pfu of LCMV-WE, DBA/2 mice (Fig. 1a) were able to clear the virus from the blood within 30–50 days (Fig. 1b). However, virus reemerged in the blood after 90 days in six of eight mice [serum (S)1–S6]. The detection limits by standard focus forming assay are <1.7 pfu/ml blood. Importantly, two mice (S7 and S8) were able to control LCMV for >170 days (Fig. 1b). All mice (S1-S8) developed nAbs against wild-type LCMV-WE with titers ranging from 1:40 to 1:320 (shown for mice 1 and 2 in Fig. 1c).
Figure 1.
(a) Schema of infection with LCMV-WE wild-type and the various nAb-escape mutants. (b) Transient or long-term control of LCMV-WE viremia in DBA/2 mice. Eight DBA/2 mice were i.v. infected with 2 × 106 pfu of LCMV-WE. LCMV titers were measured in the blood by focus forming assays. In six of eight mice (S1-S6) LCMV reappeared in the blood by day 90 after infection. Two mice (S7 and S8) controlled LCMV for >170 days. (c) nAb against LCMV-WE and first-generation nAb-escape mutant viruses V1 and V2. Sera from LCMV-WE-infected mice (S1 and S2) were tested for nAb against LCMV-WE and the mutant viruses V1 and V2. All mice mounted a nAb response against LCMV-WE, but no nAb titers could be detected against the mutants in the mice in which they had arisen (<1 represents a titer <1:20). Gray symbols represent neutralization response against the infecting virus (LCMV-WE). (d) Sera from mouse 7 and 8 (S7 and S8) were able to neutralize not only LCMV-WE but also many of the mutant viruses V1-V6. (e) Sera from mice infected with LCMV-WE (S1 and S2), in which LCMV mutant V1 and V2 had emerged, were analyzed for cross-neutralization against mutant viruses (V1–V6).
It has been shown earlier that LCMV persists in many, if not all, mice at very low levels (25).
Isolation of the reemerging virus 90 days after the infection of DBA/2 mice and subsequent subcloning, plus sequence analysis, revealed various single-point mutations within the LCMV envelope GP I. GP I forms the single neutralizing target on the intact virion (Table 1). No mutations were found in virus isolated from the blood on day 20 after infection when three subclones per mouse were sequenced (data not shown).
Table 1.
Sequence of GP I of LCMV-WE wild-type, first- and second-generation nAb-escape mutants
Virus was isolated from blood of infected DBA/2 mice and plaque-purified twice. First-generation nAb-escape mutant viruses were isolated from six mice infected with LCMV-WE (2 × 106 pfu) 90 days after infection and named V1–V6. Second-generation nAb-escape mutant viruses were isolated from seven mice 110 days after infection with 2 × 104 pfu of first-generation nAb mutants (V1–V6) and named V1.1–V1.3, V2.1–V2.3, etc. Several subclones derived from one mouse are shown. Subclones V3.1–V3.6 and subclones V3.7–V3.9 are from two different mice infected with mutant V3. Similarly subclones V4.1–4.3 and subclones V4.4–V4.6 are from two different mice infected with mutant V4. LCMV-WE are bold, first-generation nAb-escape mutants are underlined, and second-generation nAb-escape mutants are in italics.
Known potential glycosylation sites.
Subsequent autologous neutralization assays (starting with 1/20 final dilutions) showed that sera S1–S6 were unable to neutralize virus isolated from each mouse on day 90. In contrast, we detected nAb titers against the original LCMV-WE (Fig. 1c S1 and S2; Fig. 4, which is published as supporting information on the PNAS web site, www.pnas.org, S3–S6) up to 170 days.
The first generation of nAb-escape-mutant viruses exhibited mutations in three regions of GP I: amino acid positions 122 and 128, around amino acid positions 181 and 182, and around amino acid positions 211 and 212, respectively. Interestingly, here we found identical mutations (122 Phe to Ser, 211 Ala to Thr, 182 Ser to Asn, and 212 Gly to Asp) or mutations within the same region (128 and 181) as documented (11–12) in nAb-escape mutants of infected C57BL/6 CD8−/− mice.
Due to the fact that LCMV T helper epitopes in H-2d mice are not known, we cannot exclude that some of the mutant viruses are CD4 T helper cell escape mutants, as described for H-2b-mice (26).
Surprisingly, those mice (S7 and S8), which had been able to control LCMV-WE below detectable levels (<1.7 pfu/ml blood) for >170 days had sera with a broad range of neutralizing activities. These sera neutralized the wild-type and all escape mutants more or less as well (Fig. 1d). This result indicates that long-term control of LCMV is probably associated with a nAb response against many mutants developing over time. We therefore examined whether sera from mice in which a nAb-escape mutant had arisen were also able to neutralize mutant viruses V1-V6, which had emerged in the other mice. The following patterns were observed: a specific nAb response could be found against V3 in all sera (Fig. 1e, S1 and S2 and Fig. 4, S3–S6) and a more complex pattern with the neutralization of V5 to 1:320, V1 to a mean of 1:80 versus V2, V4, and V6 with neutralization titers of 1:20–1:40 (Fig. 1e and Fig. 4, S3–S6). This finding perhaps suggests two overall groups of neutralization-specificity patterns: specific (personal) and more general (public).
Correlation of Virus Persistence and Cytotoxic T Cell Unresponsiveness.
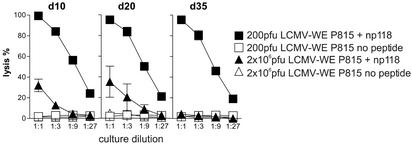
We monitored anti-LCMV CD8+ T cell responses of DBA/2 mice in parallel with nAb responses. When spleen cells obtained from acutely infected (2 × 106 pfu) DBA/2 mice on days 10, 20, and 35 were restimulated for 5 days with specific peptide, we still found CTL responses on days 10–20 as assessed by Cr51 release, but by day 35 the same restimulation yielded no measurable CTL responses (Fig. 2; ref. 24). In contrast, DBA/2 mice infected with 200 pfu of LCMV-WE yielded potent cytotoxic T cell responses at all tested time points (Fig. 2). Comparably efficient CTL deletion after infection with 2 × 106 pfu of LCMV-WE was also found with intracellular IFN-γ staining (data not shown). This discovery shows a strict correlation of CTL unresponsiveness and long-term LCMV viremia.
Figure 2.
Correlations between persistent viremia and the absence of specific CTLs. Cytotoxic CD8+ T cells were functionally tested by Cr51 release assay on P815 cells 10, 20, and 35 days after infection with 200 pfu or 2 × 106 pfu of LCMV-WE (i.v.) after 5 days in vitro restimulation with the np118 peptide.
NAb Responses Against First-Generation Escape Mutant.
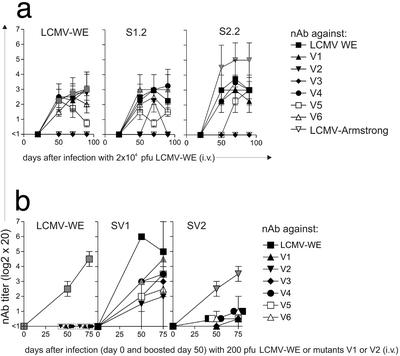
Because the LCMV-WE strain is a laboratory strain that had been subcloned three times and has been used at low multiplicity from frozen stocks, the following question arises: Are escape mutants “attenuated” or are they subject to similar immunological pressures in new hosts as the starting so-called wild-type isolate? To examine nAb responses against first-generation nAb-escape mutant and possible emerging second-generation escape mutants, we infected DBA/2 mice with 2 × 104 pfu (see Fig. 1a). This dose was chosen to avoid regrowing stocks. Mice infected with 2 × 104 pfu of nAb-escape mutants V3, V4, and V5 were able to control the virus at least until day 90, whereas only ≈50% of mice infected with the escape mutants V1, V2, and V6 transiently controlled LCMV below detectable levels. Interestingly, almost all mice that controlled LCMV at least until day 90 after infection mounted the best nAb response against the virus mutant with which they were infected (Fig. 3a V1 and V2; see Fig. 5, which is published as supporting information on the PNAS web site, V3–V6). All mutants, except V3, infecting the second host also induced a nAb response against the original LCMV-WE wild-type virus with titers ranging from 1:20 to 1:80 (Fig. 3a; Fig. 5, V3–V6). Surprisingly, the sera from these second hosts were able to neutralize not only the escape mutant with which they had been infected, but all sera also neutralized to variable extent the other nAb-escape mutants V1 and V2 and V4-V6 (Fig. 3a, V1 and V2; Fig. 5, V3–V6). Only a weakly neutralizing Ab response was observed against mutant V3 with the mutation at position 211 (Ala to Thr). This result is particularly interesting because all mice in which the first generation of nAb-escape mutants emerged had antisera that neutralized the V3 mutant to some extent (Fig. 1e). In addition, V3 failed to induce specific anti-LCMV-WE wild-type responses in a secondary host. This exceptional isolate needs to be studied in further detail.
Figure 3.
(a) NAb responses against wild-type virus and mutant viruses generated by mice infected with LCMV-WE and first-generation nAb-escape virus V1 or V2 (2 × 104 pfu). DBA/2 mice were infected with 2 × 104 pfu of LCMV-WE, mutants V1 or V2 (as indicated in the figures) and nAb against all mutant viruses, and LCMV-WE was measured in the sera at the indicated time points. Mean values with standard deviation of two independent assays are shown. (b) Induction of cross-reactive nAb is dose dependent (200 pfu). DBA/2 mice were infected with 200 pfu of LCMV-WE wild-type or first-generation nAb-escape mutant V1 or V2. Fifty days after infection, all mice were boosted with 200 pfu of the same virus with which they were initially infected. NAb in the sera (SV1 and SV2) was measured against the infecting virus and all other first-generation escape viruses (V1–V6). Mean values of two mice are shown. Day 0 represents the time point of primary infection. Gray symbols show neutralization response against the infecting virus.
The sera of mice infected with the mutant virus were also tested in a neutralization assay against the Armstrong strain of LCMV. All sera were below a titer of 1:20 (Fig. 3a, V1 and V2; Fig. 5, V3–V6). Vice versa, we were not able to detect neutralizing Ab against LCMV-WE in sera of LCMV-Armstrong-infected mice above a titer of 1:20 (not shown).
Low-Dose Infection Results in Virus Control and Low Cross-Reactive nAbs.
We evaluated whether the generation of widely cross-neutralizing Abs was associated with a prolonged viremia. Therefore, we infected DBA/2 mice with a low dose of 200 pfu of LCMV-WE or of the mutants V1–V6 (see Fig. 1a). Fifty days later, as expected mice were free of detectable viremia (except after infection with mutant V1; see below). Because nAb responses are usually low under these conditions, we boosted the mice by reinfection on day 50 with 200 pfu of the same virus isolate (e.g., V1 and V2 in Fig. 3b or V3–V6 in Fig 5). A nAb response against the immunizing virus isolate was clearly detectable 50 and 70 days later (a range of 1:80–1:320). Only low- or no cross-neutralization above a titer of 1:20 against the other mutant viruses was seen. Surprisingly, infection with mutant virus V1 resulted in a strong response against itself, against LCMV-WE wild-type, and, to a lesser extent, against other mutants (Fig. 3b, SV1 and SV2). However, DBA/2 mice were not able to clear the V1 mutant after infection with a low dose (200 pfu). This finding is particularly surprising, because DBA/2 mice infected with 2 × 104 pfu of mutant virus V1 were able to variably control V1. Some mice were able to clear the V1 mutant and, in others, new virus mutant reemerged (Fig. 1a). A similar paradox virus kinetic was already observed in H-2b CD8 knockout mice (12–13), where only high-dose (2 × 106 pfu) LCMV-WE infection resulted in transient clearance and virus reemergence, whereas low-dose infection (200 pfu) was always associated with high-level virus persistence. This finding needs to be studied further.
In general, there seems to be an association between low-dose (200 pfu; rapid virus clearance) or high-dose (2 × 106 pfu; prolonged viremia) infection and nAb responses: low-dose infections induce CTL-mediated virus control, lower and shorter viremia, lower Ab titers, and less cross-reactivities.
Reemergence of Second-Generation nAb-Escape Mutants.
In all DBA/2 mice infected with 2 × 104 pfu of the first generation of nAb-escape mutant viruses (except for V5) LCMV reappeared after 90–120 days. Therefore, we reisolated and cloned reemerging second-generation nAb-escape virus (see Fig. 1a and Table 1). The second generation of escape virus was named V1.1, V1.2, etc. nAb-escape mutants that arose in V1-infected mice and V2.1, V2.2, etc. in V2-infected mice and so on (see scheme in Fig. 1a). Interestingly several new mutations accumulated, again focusing on the three known regions (amino acid positions 114–133, amino acid positions 170–182, and amino acid positions 210–220). In some cases, mutants reverted at some positions to the original amino acid of WE, but mutations at other sites came up near the original mutations. For example, the second-generation nAb-escape virus of the V6 mutant reverted to the mutation at position 122 (Ser back to Phe; see box Table 1) but had a mutation at position 128 (Ala to Ser) and V4 (V4.4–V4.6) mutant reverted the mutation at position 181 (Thr back to Ile; see box in Table 1), but had a mutation at position 182 (Ser to Gly; V4.4–V4.5). Two escape viruses had mutations at known potential glycosylation sites: V1.2 at position 114 (Asp to Tyr) and V1.1 at position 124 (Asn to Thr). We also sequenced the known LCMV strains Armstrong 53b, Docile, and the so-called aggressive WE (data not shown). We could not observe an obvious tendency for mutant viruses to mutate toward the sequences of other LCMV strains such as Armstrong, Docile, or aggressive WE (data not shown).
nAb Responses Against Second-Generation Escape Mutants.
In all cases, sera from mice in which virus reemerged were not able to neutralize the new mutant, which is indicative of a neutralization escape virus (data not shown, but comparable to the first-generation escape virus shown in Fig. 1). We next examined cross-neutralization by using sera from LCMV wild-type-infected mice and sera from mutant LCMV-infected (V1–V6) mice. Cross-neutralization could still be observed. The neutralizing titers of sera from mice infected with first-generation nAb-escape virus was variable, but, in general, a tendency for higher neutralizing titers against several second-generation nAb-escape mutants was noted when they were compared with sera from LCMV-WE wild-type virus-infected mice. The data are summarized in Table 2 (the highest titers at any tested time point are shown).
Table 2.
nAb against second-generation escape mutants
| Mice | Virus
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| V1.1 | V1.2 | V2.2 | V2.3 | V3.2 | V3.7 | V3.9 | V4.1 | V4.5 | V6.3 | |
| V1 infected; cleared | <1 | <1 | 2–3 | ≥4 | 2–3 | 2–3 | 2–3 | <1 | 2–3 | 2–3 |
| V1 infected; V1.1–4 emerged | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| V2 infected; cleared | ≥4 | ≥4 | ≥4 | <1 | <1 | 2–3 | <1 | 2–3 | <1 | ≥4 |
| V2 infected; V2.1–3 emerged | <1 | 2–3 | <1 | <1 | <1 | <1 | <1 | <1 | ≥4 | <1 |
| V3 infected; V3.1–6 emerged | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| V3 infected; V3.7–9 emerged | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| V4 infected; V4.1–3 emerged | ≥4 | ≥4 | 2–3 | 2–3 | ≥4 | ≥4 | 2–3 | <1 | ≥4 | ≥4 |
| V4 infected; V4.4–6 emerged | <1 | <1 | <1 | <1 | <1 | <1 | <1 | 2–3 | <1 | <1 |
| V5 infected; cleared | 2–3 | <1 | <1 | <1 | ≥4 | ≥4 | 2–3 | <1 | ≥4 | ≥4 |
| V6 infected; cleared | <1 | 2–3 | <1 | <1 | <1 | ≥4 | <1 | 2–3 | 2–3 | <1 |
| V6 infected; V6.1–3 emerged | 2–3 | <1 | <1 | <1 | <1 | 2–3 | 2–3 | <1 | <1 | <1 |
| M1 | <1 | <1 | <1 | <1 | <1 | <1 | 2–3 | 2–3 | <1 | ≥4 |
| M2 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| M3 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| M4 | <1 | 2–3 | <1 | <1 | <1 | ≥4 | <1 | <1 | <1 | <1 |
| M5 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| M6 | 2–3 | 2–3 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| M7 | ≥4 | ≥4 | 2–3 | ≥4 | 2–3 | 2–3 | 2–3 | 2–3 | ≥4 | ≥4 |
| M8 | 2–3 | 2–3 | 2–3 | 2–3 | 2–3 | ≥4 | 2–3 | ≥4 | <1 | ≥4 |
Highest titers at any tested time point are shown. Titers are differentiated between nonneutralizing (<1), low neutralizing (2–3; 1/40–1/80), and high neutralizing (≥4; ≥1/160) activity.
Taken together, we could not observe a correlation between the site of mutation of the LCMV GP1 of a virus and its capacities to induce nAb against a particular nAb-escape mutant virus. Also, there was no overall tendency toward lower nAb responses against later virus escape generations.
Discussion
Two general findings emerged in this study: (i) Mice successfully controlling viremia up to day 170 (the latest time point tested) develop nAb against many or sometimes against almost all possible first- and second-generation nAb-escape mutants, i.e., they make nAb against a great variety of escape viruses (ii). The escaping virus mutant represents a negative personal serotype in an individual mouse, in which nAb against this specific virus mutant failed to be produced. Both observations may help to understand findings with other poorly, variably, or noncytopathic viruses such as Dengue, HIV, and HCV in humans. During HIV infection in humans, no clear correlations between genetic subtypes, antigenic serotypes, and neutralizing serotypes have been found so far (27). It would be interesting to find out whether, in HIV infection, cross-reactive or specific nAb responses are mounted reflecting an evolutionary process driven by the generation of many quasispecies within one host, as suggested here for LCMV in mice.
Interestingly, infection of naïve mice with subcloned virus escape mutants not only induced nAb against wild-type (which had not been experienced by these mice) but also mounted nAb against many of the other escape mutants (first- and second-generation nAb-escape virus). The neutralization in vitro of the original wild-type virus (at usually high titer when compared with autologous neutralization) by sera from nAb-escape mutant-infected mice is reminiscent of “original antigenic sin” against influenza virus, where a “junior” virus induced nAbs against a senior virus (28). The difference is that the retrospectively asymmetric serotype specificity is selected at the population level for influenza virus and on the individual host level against LCMV.
It remains unclear how cross-neutralization of the various nAb-escape mutants by sera from mice infected with other nAb-escape mutants correlate with binding qualities (i.e., affinities and avidities). Unfortunately, binding qualities cannot be reliably measured yet, because purified LCMV GP suitable for such measurements is not available in sufficient purity and quantities.
The present findings may help to explain some aspects of Dengue virus infections. The presence of nAb against one Dengue virus sometimes enhances disease to a subsequent infection with a different Dengue virus serotype (29–30). In fact, Dengue-specific antisera cross-neutralize to 1:40 or 1:80 (public specificity) other Dengue serotypes variably and often asymmetrically. Such sera may well exhibit personal serotypes that may stay hidden, similar to LCMV, in a low titer of public reactivity. Our results may suggest, for Dengue, that rapid virus control favors narrow private serotype-specific responses and may not accumulate nAb reactivities against the other three major serotypes. Slow virus control may then correlate with the reverse, i.e., sera may contain more public cross-reactive Abs (31–33). In the case of Dengue virus, the distribution of virus mutants are limited by the cycle between the human host and the replication in mosquitoes. Nevertheless, the induced cross-reactive nAb will persist for a prolonged period in the infected host and may therefore interact with a second infection with a different Dengue serotype.
The role of the nAb must be viewed in the context of virus control by CD8+ T and CD4+ T cells, which also exert evolutionary pressures on the persisting non- or poorly cytopathic virus and also select both CD8+ and CD4+ T cell escape mutants (26, 34–36). New nAb-escape viruses seem to reemerge all of the time, and, eventually, they will exhaust CD8+ T and CD4+ T cells. The process of CD4 T cell exhaustion impairs the generation of new nAb against new mutants (12, 26). Whether very long-term sequential selection may eventually result in an attenuated virus-host relationship if the virus remains noncytopathic remains a proposal that has to be examined. For LCMV-attenuated mutants, this means that nAb-escapees induce less immunopathology because they persist more readily and thereby causes more efficient T cell deletion or selection of T cell escape mutants. Overall, this could result in less immunopathological T cell responses and more efficient virus transfer by vertical (LCMV) or horizontal infection. LCMV or HIV 2 and HCV may have achieved this goal nearly optimally, whereas HIV 1 may not be there yet (36).
Supplementary Material
Acknowledgments
This work was supported by European Union Hepatitis Grant 23.8452, Swiss National Foundation for Science Grant 24.6362, and Kanton of Zurich Grant 24.6853.
Abbreviations
- pfu
plaque-forming unit
- LCMV
lymphocytic choriomeningitis virus
- LCMV-WE
LCMV strain WE
- HCV
hepatitis C virus
- nAb
neutralizing Ab
- CTL
cytotoxic T cell
- Sn
serum n
- GP
glycoprotein
References
- 1.Domingo E. J Virol. 2002;76:463–465. doi: 10.1128/JVI.76.1.463-465.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holmes E C, Moya A. J Virol. 2002;76:460–462. doi: 10.1128/JVI.76.1.460-462.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Domingo E, Holland J J. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 4.Farci P, Shimoda A, Coiana A, Diaz G, Peddis G, Melpolder J C, Strazzera A, Chien D Y, Munoz S J, Balestrieri A, et al. Science. 2000;288:339–344. doi: 10.1126/science.288.5464.339. [DOI] [PubMed] [Google Scholar]
- 5.Kalams S A, Johnson R P, Dynan M J, Hartman K E, Harrer T, Harrer E, Trocha A K, Blattner W A, Buchbinder S P, Walker D D. J Exp Med. 1996;183:1669–1679. doi: 10.1084/jem.183.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang W K, Lin S R, Lee C M, King C C, Chang S C. J Virol. 2002;76:4662–4665. doi: 10.1128/JVI.76.9.4662-4665.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kagi D, Ledermann B, Bürki K, Seiler P, Odermatt B, Olsen K J, Podack E R, Zinkernagel R M, Hengartner H. Nature. 1994;369:31–37. doi: 10.1038/369031a0. [DOI] [PubMed] [Google Scholar]
- 8.Schmitz J E, Kuroda M J, Santra S, Sasseville V G, Simon M A, Lifton M A, Racz P, Tenner-Racz K, Dalesandro M, Scallon B J, et al. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 9.Lechner F, Wong D K, Dunbar P R, Chapman R, Chung R T, Dohrenwend P, Robbins G, Phillips R, Klenerman P, Walker B D. J Exp Med. 2000;191:1499–1512. doi: 10.1084/jem.191.9.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seiler P, Senn B M, Brundler M A, Zinkernagel R M, Hengartner H, Kalinke U. J Immunol. 1999;162:4536–4541. [PubMed] [Google Scholar]
- 11.Ciurea A, Klenerman P, Hunziker L, Horvath E, Senn B M, Ochsenbein A F, Hengartner H, Zinkernagel R M. Proc Natl Acad Sci USA. 2000;97:2749–2754. doi: 10.1073/pnas.040558797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ciurea A, Hunziker L, Klenerman P, Hengartner H, Zinkernagel R M. J Exp Med. 2001;193:297–306. doi: 10.1084/jem.193.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weiner A J, Geysen H M, Christopherson C, Hall J E, Mason T J, Saracco G, Bonino F, Crawford K, Marion C D, Crawford K A. Proc Natl Acad Sci USA. 1992;89:3468–3472. doi: 10.1073/pnas.89.8.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimizu Y K, Hijikata M, Iwamoto A, Alter H J, Purcell R H, Yoshikura H. J Virol. 1994;68:1494–1500. doi: 10.1128/jvi.68.3.1494-1500.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogura Y, Kurosaki M, Asahina Y, Enomoto N, Marumo F, Sato C. J Infect Dis. 1999;180:1444–1451. doi: 10.1086/315094. [DOI] [PubMed] [Google Scholar]
- 16.Carman W F, Zanetti A R, Karayiannis P, Waters J, Manzillo G, Tanzi E, Zuckerman A J, Thomas H C. Lancet. 1990;336:325–329. doi: 10.1016/0140-6736(90)91874-a. [DOI] [PubMed] [Google Scholar]
- 17.Hsu H Y, Chang M H, Ni Y H, Lin H H, Wang S M, Chen D S. Hepatology. 1997;26:786–791. doi: 10.1002/hep.510260336. [DOI] [PubMed] [Google Scholar]
- 18.Protzer-Knolle U, Naumann U, Bartenschlager R, Berg T, Hopf U, Meyer zum Buschenfelde K H, Neuhaus P, Gerken G. Hepatology. 1998;27:254–263. doi: 10.1002/hep.510270138. [DOI] [PubMed] [Google Scholar]
- 19.Albert J, Abrahamsson B, Nagy K, Aurelius E, Gaines H, Nystrom G, Fenyo E M. AIDS. 1990;4:107–112. doi: 10.1097/00002030-199002000-00002. [DOI] [PubMed] [Google Scholar]
- 20.Tremblay M, Wainberg M A. J Infect Dis. 1990;162:735–737. doi: 10.1093/infdis/162.3.735. [DOI] [PubMed] [Google Scholar]
- 21.Arendrup M, Nielsen C, Hansen J E, Pedersen C, Mathiesen L, Nielsen J O. J Acquired Immune Defic Syndr. 1992;5:303–307. [PubMed] [Google Scholar]
- 22.Burns D P, Collignon C, Desrosiers R C. J Virol. 1993;67:4104–4113. doi: 10.1128/jvi.67.7.4104-4113.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bradney A P, Scheer S, Crawford J M, Buchbinder S P, Montefiori D C. J Infect Dis. 1999;179:1264–1267. doi: 10.1086/314711. [DOI] [PubMed] [Google Scholar]
- 24.Moskophidis D, Lechner F, Hengartner H, Zinkernagel R M. J Immunol. 1994;152:4976–4983. [PubMed] [Google Scholar]
- 25.Ciurea A, Klenerman P, Hunziker L, Horvath E, Odermatt B, Ochsenbein A F, Hengartner H, Zinkernagel R M. Proc Natl Acad Sci USA. 1999;96:11964–11969. doi: 10.1073/pnas.96.21.11964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ciurea A, Hunziker L, Martinic M M, Oxenius A, Hengartner H, Zinkernagel R M. Nat Med. 2001;7:795–800. doi: 10.1038/89915. [DOI] [PubMed] [Google Scholar]
- 27.Moore J P, Parren P W, Burton D R. J Virol. 2001;75:5721–5729. doi: 10.1128/JVI.75.13.5721-5729.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fazekas de St. Groth B, Webster R G. J Exp Med. 1966;124:331–345. doi: 10.1084/jem.124.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Halstead S. Science. 1988;239:476–481. doi: 10.1126/science.3277268. [DOI] [PubMed] [Google Scholar]
- 30.Morens D M. Clin Infect Dis. 1994;19:500–512. doi: 10.1093/clinids/19.3.500. [DOI] [PubMed] [Google Scholar]
- 31.Guzman M G, Kouri G, Halstead S B. Lancet. 2000;355:1902–1903. doi: 10.1016/S0140-6736(00)02303-5. [DOI] [PubMed] [Google Scholar]
- 32.Halstead S B, Venkateshan C N, Gentry M K, Larsen L K. J Immunol. 1984;132:1529–1532. [PubMed] [Google Scholar]
- 33.Vaughn D W, Green S, Kalayanarooj S, Innis B L, Nimmannitya S, Suntayakorn S, Endy T P, Raengsakulrach B, Rothman A L, Ennis F A, Nisalak A. J Infect Dis. 2000;181:2–9. doi: 10.1086/315215. [DOI] [PubMed] [Google Scholar]
- 34.Pircher H P, Moskophidis D, Rohrer U, Bürki K, Hengartner H, Zinkernagel R M. Nature. 1990;346:629–633. doi: 10.1038/346629a0. [DOI] [PubMed] [Google Scholar]
- 35.Phillips R E, Rowland-Jones S, Nixon D F, Gotch F M, Edwards J P, Ogunlesi A O, McMichael A J. Nature. 1991;354:453–459. doi: 10.1038/354453a0. [DOI] [PubMed] [Google Scholar]
- 36.Goulder P J, Brander C, Tang Y, Tremblay C, Colbert R A, Addo M M, Rosenberg E S, Nguyen T, Allen R, Trocha A, et al. Nature. 2001;412:334–338. doi: 10.1038/35085576. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.