Abstract
Keratin 8 and 18 (K8/K18) mutations are found in patients with cryptogenic cirrhosis, but the role of keratin mutations in noncryptogenic cirrhosis and the incidence of keratin mutations in the general population are not known. We screened for K8/K18 mutations in genomic DNA isolated from 314 liver explants of patients who primarily had noncryptogenic cirrhosis, and from 349 blood bank volunteers. Seven unique K8/K18 mutations were found in 11 independent patients with biliary atresia, hepatitis B/C, alcohol, primary biliary cirrhosis, and fulminant hepatitis. Seven of the 11 patients had mutations previously described in patients with cryptogenic cirrhosis: K8 Tyr-53 → His, K8 Gly-61 → Cys, and K18 His-127 → Leu. The four remaining patients had mutations at one K8 and three other K18 new sites. Of the 349 blood bank control samples, only one contained the Tyr-53 → His and one the Gly-61 → Cys K8 mutations (P < 0.004 when comparing cirrhosis versus control groups). Two additional mutations were found in both the liver disease and blood bank groups and, hence, likely represent polymorphisms. Livers with keratin mutations had cytoplasmic filamentous deposits that were less frequent in livers without the mutations (P = 0.03). Therefore, K8/K18 are likely susceptibility genes for developing cryptogenic and noncryptogenic forms of liver disease.
Keratin mutations are associated with several skin, oral, esophageal, ocular and cryptogenic liver diseases that reflect tissue-specific expression of the particular keratin involved (1–6). The resulting cellular and tissue defects are manifestations of the clearly defined function of keratins that provides cells with ability to cope with mechanical stresses. This keratin cytoprotective effect is evident in the blistering phenotype of several human keratin skin diseases such as epidermolysis bullosa simplex (1–6), and the phenotypes of animal models that lack or express a mutant keratin (7). Also, emerging evidence suggests that keratins protect cells from nonmechanical forms of injury via several mechanisms that may include keratin regulation of cell signaling cascades, regulation of the availability of other cellular proteins, and protein targeting to subcellular compartments (5, 6).
The function of keratins in protecting cells from mechanical stress is related to their unique properties and abundance as one of three major cytoskeletal protein families, which include intermediate filaments (IF), microfilaments and microtubules (8). Keratins (K) are members of the IF protein family, and are specifically expressed in epithelial cells and their appendages. They consist of >20 members (K1–K20) (5, 9), and are further classified into type I (K9–K20) and type II (K1–K8) keratins, which form obligate, noncovalent heteropolymers (9). Keratins serve as important cell-type-specific markers. For example, unique keratin complements distinguish different epithelial cell types and thereby reflect epithelial subtype-specific diseases that result from keratin-specific mutations. As such, keratinocytes express K5/K14 basally and K1/K10 suprabasally, and hepatocytes express K8/K18. K8/K18 are also found in other glandular cells including enterocytes, with variable complements of K19/K20/K7 depending on the cell type (8).
Most keratin diseases are autosomal-dominant with near complete penetrance. Exceptions appear to be K18 (10) and K8 (11) mutations in patients with cryptogenic cirrhosis. To date, six patients have been described with K8 (five patients) or K18 (one patient) mutations, from a group of 55 patients with cryptogenic cirrhosis (10, 11). Most patients with cryptogenic cirrhosis, including those with K8/K18 mutations, do not have a well-defined liver disease family history. Absence of a clear family history suggests that K8/K18 mutations predispose to, rather than cause, liver disease. The presence and frequency of keratin mutations in noncryptogenic liver disease is unknown, which prompted us to study the incidence of K8/K18 mutations in a variety of noncryptogenic liver diseases as compared with the general population. Our findings show a clear association of K8/K18 mutations with various forms of liver disease, and indicate that such mutations likely predispose to subsequent development of cirrhosis. We also describe a histologic feature that associates preferentially with cirrhotic livers that harbor K8/K18 mutations.
Methods
Patients.
We included for the analysis specimens of 467 explanted livers that were obtained from the liver transplantation units at Stanford University, the University of California San Francisco, and California Pacific Medical Center. Peripheral blood samples from 349 healthy volunteers were obtained from the Stanford Blood Bank and used for genomic DNA isolation. In addition and when available, blood samples were obtained from patients with the identified keratin mutations and/or from their children. Of the 467 liver samples, 153 were previously analyzed (11) and the remaining 314 liver explants are newly described and were used for genomic DNA isolation, immunofluorescence staining, and other biochemical tests. The patients' sex and racial/ethnic background were determined from patients' medical records. No information could be found on 15 patients due to lack of records or to retransplant. For the control samples, anonymous information regarding sex and race was provided by the Stanford Blood Bank. The diagnoses were based on the United Network for Organ Sharing transplant listing. Medical records of all patients with a keratin mutation were reviewed and the diagnosis was confirmed.
Histopathology and Statistical Analysis.
Pathology slides from the explanted livers with keratin mutations, and matched liver disease controls, were reviewed by a single pathologist (R.K.S.) who did not know which specimens harbored the keratin mutations. Slides from the explanted livers of all 17 patients with keratin mutations, as well as disease-matched controls, were examined and scored for the presence or absence of features including Mallory and acidophil bodies, cell size, ground glass cytoplasm, and dysplasia. Images of the hematoxylin and eosin stained liver sections were obtained by using a Nikon Eclipse E1000 microscope with a ×40 objective. Data analysis was conducted by using the Fisher's exact test performed with Statistical Analyzing System software (SAS, Cary, NC) (12).
Molecular Methods.
Genomic DNA was prepared by using a Dneasy tissue kit (Qiagen, Valencia, CA). Exonic regions were amplified by using previously described primers corresponding to amino acids: 67–131, 225–273, and 322–389 (for K18) and 50–107 and 341–401 (for K8) (11). Regions chosen for amplification included the epidermal keratin domains where most of the mutations have been identified (4). The PCR amplified products were analyzed by using Mutation Detection Enhancement gels (FMC Bioproducts, Rockland, ME), and any samples with a “shift” pattern suggestive of a mutation were sequenced in the forward and reverse directions to confirm the presence of a mutation.
Biochemical Methods.
Tissues were homogenized in PBS containing 1% n-dodecyl-N,N-dimethylglycine (Empigen BB, Calbiochem-Novabiochem, San Diego), 5 mM EDTA, and protease inhibitors. Homogenized samples were solubilized for 30 min, pelleted, and then used for immunoprecipitation of K8/K18 (13). Precipitates were analyzed by (i) SDS/PAGE under reducing or nonreducing conditions, followed by Coomassie staining (14), (ii) SDS/PAGE followed by immunoblotting, or (iii) two-dimensional gels by using isoelectric focusing (horizontal direction) and SDS/PAGE (vertical direction) followed by immunoblotting (15).
Mass Spectrometry Analysis.
Separated K8 and K18 bands were cut out from preparative gels, reduced with DTT, alkylated with iodoacetamide, and then digested with trypsin in 50 mM ammonium bicarbonate (pH 7.8) by using a standard in-gel-digestion procedure. Extracted K8 or K18 tryptic peptides were desalted by using a C18 ZipTip (Millipore, MA) then eluted with 50% acetonitrile/0.1% trifluoroacetic acid (TFA). A 1-μl aliquot of the eluant was mixed with equal volume of matrix solution (saturated α-cyano-4-hydroxycinnamic acid in 0.1% TFA-50% acetonitrile in water) and analyzed by a matrix-assisted laser desorption ionization time-of-flight mass spectrometer (Bruker Biflex III) equipped with a nitrogen 337-nm laser. The mass spectra were acquired in the reflectron mode. Internal mass calibration was performed with two trypsin autodigested fragments (842.5 and 2,211.1 Da). K8 tryptic peptides were also digested with CNBr and similarly analyzed.
Immunofluorescence Staining.
Snap-frozen liver explants were embedded in optimum cutting temperature compound, sectioned, then fixed in acetone (−20°C, 10 min). Sections were double-stained with antibodies directed to K8/K18 (13) or vimentin (NeoMarkers, Freemont, CA). Fluorescence images were obtained by using an MRC 1024ES confocal scanner (Bio-Rad) coupled to a Nikon Eclipse TE300 microscope.
Results
Identification of K8 and K18 Mutations.
We tested DNA extracted from liver explants or peripheral blood for the presence of K8 or K18 mutations. Two cohorts were examined (whose demographics are summarized in Table 4, which is published as supporting information on the PNAS web site, www.pnas.org): a group of 314 patients with a variety of liver diseases, and a control group of 349 blood bank donors. The ethnic background of the two cohorts was similar except for a higher preponderance of white patients in the control group. The etiology of the liver diseases is broad (Table 1), most of which is noncryptogenic (based on clinical criteria, 13 of the 314 patients were classified as having cryptogenic liver disease; Table 1). We included the control blood bank cohort to address which mutations identified in the liver disease cohort are likely to represent “true” mutations versus polymorphisms found in the general population. Eleven of the 314 liver disease patients had K8 or K18 heterozygous missense point mutations that were confirmed by DNA sequencing, which resulted in amino acid substitutions (Table 2). When combined with our previous cohort of 153 patients with liver disease (11), which had a significant number of patients with cryptogenic cirrhosis (i.e., 55), the total number of unrelated patients with K8/K18 mutations was 17 of 467 total patients (Table 2). In addition to these 17 mutations, two identified mutations (K8 I62V and K18 S229T) likely represent polymorphisms because they were significantly overrepresented in the control cohort (Table 2). Also, “silent” point mutations that did not result in any amino acid substitutions (K8 L71L/E376E; K18 Y330Y) were identified in the liver disease cohort but not in the control group (Table 2). These “silent” mutations were not studied further.
Table 1.
Liver disease etiologies in the new and combined patient cohorts
| Etiology of liver disease | New cohort no. (no. with keratin mutations) | New and old* cohorts no. (no. with keratin mutations) |
|---|---|---|
| Hepatitis C | 70 (2) | 80 (2) |
| Hepatitis C/alcohol | 28 (1) | 28 (1) |
| Hepatitis B | 43 (1) | 44 (1) |
| Biliary atresia | 47 (1) | 47 (1) |
| Alcohol | 13 (1) | 33 (1) |
| Cryptogenic | 13 (0) | 68 (6) |
| Primary biliary cirrhosis | 10 (1) | 12 (1) |
| Primary sclerosing cholangitis | 15 (0) | 15 (0) |
| Acute fulminant hepatitis | 14 (1) | 35 (1) |
| Neonatal hepatitis | 6 (0) | 9 (0) |
| Autoimmune hepatitis | 6 (0) | 33 (0) |
| Metabolic/genetic† | 18 | 23 |
| Cystic fibrosis | (1) | (1) |
| Metabolic/other | (1) | (1) |
| Primary liver cancer‡ | 5 (0) | 5 (0) |
| Drug-induced liver failure | 2 (0) | 9 (0) |
| Other§ | 20 | 22 |
| Hepatic artery thrombosis | (1) | (1) |
| Unknown | 4 (0) | 4 (0) |
| Total | 314 (11) | 467 (17) |
The old liver disease cohort was enriched for crytogenic cirrhosis (11).
Metabolic/genetic diagnoses included four Wilson's disease, three hemochromatosis, three α1-antitrypsin deficiency, two cystic fibrosis, three primary oxalosis, one Crigler-Najjar, one ornithine transcarbamoylase deficiency, one Nieman–Pick, one arginosuccinicaciduria, one tyrosinemia, one cirtrolemia, one glycogen storage, and one “metabolic disease” that is unclassified.
Patients with viral hepatitis and hepatoma were included under the appropriate viral hepatitis category. Primary liver cancers included one hepatoma, three hemangio-endotheliomas, and one hepatosarcoma.
The “Other” category of liver diseases included three Budd–Chiari, three hepatic artery thrombosis, two polycystic disease, one Caroli's disease, one Byler's disease, two chronic rejection, one primary graft nonfunction, one Klatskin tumor, one parenteral nutrition-induced, one carcinoid, one hepatitis B and C, one hepatitis B and alcohol, one multiple adenomas, one secondary biliary cirrhosis, one veno-occlusive disease, and one congenital hepatic fibrosis. At least 11 of the patients screened had received a prior orthotopic liver transplant.
Table 2.
K8/18 mutations in human liver diseases
| Mutations
|
No. of mutation carriers from
|
||||
|---|---|---|---|---|---|
| Amino acid | Nucleotide | 314 new cohort | 467 combined cohorts | 349 blood bank controls | |
| K8 | G52V | GGC → GTC | 1 | 1 | None |
| Y53H | TAT → CAT | 3 | 5 | 1 | |
| G61C | GGC → TGC | 3 | 6 | 1 | |
| I62V* | ATC → GTC | 1 | 1 | 7 | |
| L71L** | CTG → CTA | 1 | 1 | None | |
| E376E** | GAG → GAA | 1 | 2 | – | |
| K18 | T102A | ACC → GCC | 1 | 1 | None |
| H127L | CAT → CTT | 1 | 2 | None | |
| R260Q | CGG → CAG | 1 | 1 | None | |
| G339R | GGG → AGG | 1 | 1 | None | |
| S229T* | AGC → ACC | 2 | 2 | 4 | |
| Y330Y** | TAC → TAT | 1 | 1 | None | |
| K8 | 7/314 | 12/467 | 2/349 | ||
| K18 | 4/314 | 5/467 | None | ||
| K8/18 | 11/314 | 17/467 | 2/349 | ||
Single-letter standard abbreviations are used to represent amino acids and nucleotides. Sequences in boldface refer to mutations that pose a potential risk factor for subsequent development of cirrhosis, based on analysis of the liver disease cohort and the blood bank control group. Single asterisks (*) highlight amino acid substitutions (K8 I62V and K18 S229T) that are considered polymorphisms because they were found at similar, or higher, incidence in the control cohort as compared with the liver disease group. Double asterisks (**) highlight “silent” nucleotide mutations that do not result in any amino acid change. The total number of K8/K18 mutations shown represents true mutations and does not include “silent” mutations or polymorphisms.
The “true” mutations that were identified in 17 of 467 patients represent a mutation frequency of 3.6%, as compared with a mutation frequency of 0.6% found in 2 of 349 controls (P < 0.004; Tables 2 and 3). Given the demographics of patients with keratin mutations, there does not appear to be any obvious accumulation of keratin mutations in a particular sex or ethnic background (Table 3). Notably, 6 of 68 patients (8.8%) with cryptogenic cirrhosis had K8/K18 mutations (Table 1), whereas 11 of the remaining 399 patients (2.8%) with noncryptogenic cirrhosis had such mutations (P < 0.03 when comparing keratin mutations in cryptogenic versus noncryptogenic cirrhosis).
Table 3.
Diseases and ethnicities associated with different keratin mutations in liver disease patients and controls
| Keratin mutation* | Combined cohorts of liver disease patients
|
Blood bank controls, no. with mutation/ethnicity | |
|---|---|---|---|
| Liver disease | No. with mutation/ethnicity† | ||
| K8 G52V | Viral hepatitis | 1/White | None/– |
| K8 Y53H | Viral hepatitis, BA, CC | 5/3 Black, 1 White, 1 Hispanic | 1/Black |
| K8 G61C | Viral hepatitis, alcohol, CC, CF | 6/1 Black, 4 White, 1 Unknown | 1/White |
| K18 T102A | Acute fulminant hepatitis | 1/Hispanic | None/– |
| K18 H127L | Metabolic, CC | 2/White | None/– |
| K18 R260Q | PBC | 1/Hispanic | None/– |
| K18 G339R | Hepatic artery thrombosis | 1/Unknown | None/– |
| Prevalence of keratin mutations | 17/467 (3.6%)‡ | 2/349 (0.6%)‡ | |
BA, biliary atresia; CC, cryptogenic cirrhosis; CF, cystic fibrosis; PBC, primary biliary cirrhosis.
Two of the keratin mutations detected were in previously transplanted patients: one K8 G61C mutation was detected in a patient undergoing transplant for hepatitis C who had been transplanted 3 years prior for hepatitis C and alcohol, and one K18 G339R mutation was detected in a patient who had undergone transplant for hepatitis B and developed hepatic artery thrombosis resulting in a second transplant 7 months later.
The ethnicities and sexes of the 17 liver disease patients with keratin mutations were eight White (three female/five male), four Black (two female/two male), three Hispanic (two female/one male), and two unknown.
P value < 0.004 (proportion of patients with keratin mutations from the combined liver disease cohorts compared with blood bank controls).
Because most of the specimen that we analyzed consisted of liver explants, we also tested blood specimen for the germ-line presence and transmission of the identified keratin mutations, to exclude the possibility that the mutations we identified occurred during development of disease. Of the 17 independent patients with K8/K18 mutations, we were able to locate four patients and/or their offspring. All four of these patients and/or their offspring blood specimen had the identical heterozygous keratin mutation to that identified in the diseased explanted liver (Table 5, which is published as supporting information on the PNAS web site). Therefore, the K8/K18 mutations we identified are not a consequence of the liver disease but, rather, predispose their carriers to subsequent development of liver disease.
Expression of Mutant Keratin Proteins.
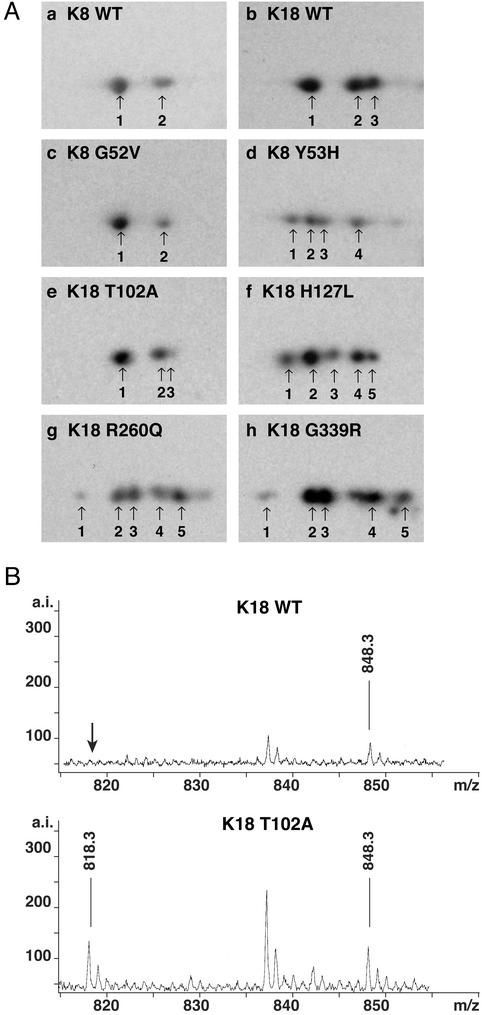
We examined, biochemically, the liver explant specimens from the 11 mutation carriers within the cohort of 314 patients to confirm the presence of the mutation at the protein level. These 11 patients had seven types of mutations: K8 G52V/Y53H/G61C and K18 T102A/H127L/R260Q/G339R (Table 2). The presence of the K8 G61C protein was confirmed as we did previously (11), by formation of K8 dimers (under nonreducing gel conditions) due to the newly introduced cysteine (normally absent in K8/K18, not shown). We also used two-dimensional gels to separate proteins based on their charge, and thereby confirmed the presence of the K8 Y53H and K18 H127L/R260Q/G339R species (Fig. 1A). These four K8/K18 mutations generated proteins with a different charge as compared with their wild-type counterparts, and resulted in new isoforms (four isoforms instead of two for mutant K8, and five isoforms instead of three for mutant K18; Fig. 1A).
Figure 1.
Protein expression of mutant K8 and K18 in explanted livers. (A) K8/K18 immunoprecipitates were obtained from 1% Empigen solubilized normal liver or livers with keratin mutation. The immunoprecipitates were separated by isoelectic focusing followed by SDS/PAGE, then immunoblotting with anti-K8/K18 antibodies. Note that K8 and K18 in normal liver consists of two or three isoforms depending on their phosphorylation levels (a and b). In contrast, some of the mutant keratins contain four (K8) or five (K18) isoforms caused by coexpression of the wild-type and mutant keratin with subsequent generation of altered charged species that have a slightly different mutation-induced isoelectric focusing point (d and f–h). (B) K8/K18 immunoprecipitates were prepared from normal liver or liver with the K18 T102A mutation, then analyzed by SDS/PAGE. The K18 bands were cut out, digested with trypsin, then analyzed with a matrix-assisted laser desorption ionization time-of-flight mass spectrometer. Note that a peak position at 818.3 was detected only in liver specimen with the K18 T102A mutation but not in normal liver. The mass difference of 30 between the wild-type and T102A K18 tryptic peptides (848.3 vs. 818.3) corresponds to the HO-C-H species (two hydrogen, one oxygen atom and one carbon atom with a mass of 30 Da) that are present in threonine (the wild-type residue) but not in alanine (the mutant residue).
Two-dimensional gel analysis was, however, not informative for mutations that do not significantly alter the isoelectric point (Fig. 1A, K8 G52V and K18 T102A variants). For these mutations, we compared the mass spectrometric profiles of protease-generated fragments of wild-type and mutant keratins and tested for the presence of peptides that have a mutation-altered mass. As shown in Fig. 1B, presence of the K18 T102A protein was confirmed by detection of its alanine-102-containing peptide with a predicted mass of 818.3 Da (in addition to the wild-type threonine-102-containing peptide with a predicted mass of 848.3 Da). A similar analysis was attempted for K8 G52V but no peptide that corresponds to a valine-to-glycine substitution was detected, likely due to inability to recover it from the isolation column (data not shown).
Effect of Keratin Mutations on Keratin Filament Organization and Liver Histology.
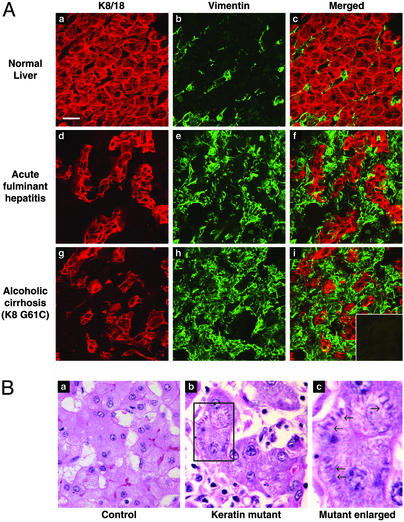
We compared keratin filament organization in liver explants of patients with and without keratin mutations. We did not observe any generalized keratin mutation-specific organization defects as tested by immunofluorescence staining (Fig. 2A). The diseased livers (with or without keratin mutations) had reorganization of the keratins filaments with the most prominent feature being thickening and partial collapse (Fig. 2A d and g) as compared with normal liver keratin staining (Fig. 2Aa). The diseased livers had variable but significant vimentin-positive staining (Fig. 2A e and h), which was used as a fibroblast/stellate cell marker, as compared with normal liver (Fig. 2Ab). Vimentin staining did not correlate with the presence of keratin mutations, and did not involve hepatocytes (Fig. 2A c, f, and i). Analysis of additional liver samples with proper attention to sample handling will be needed to better assess any potential keratin mutation-induced effects on keratin organization.
Figure 2.
Keratin filament organization in human liver explants, and histologic findings of livers harboring the keratin mutations. (A) Human livers were sectioned, fixed in acetone, and double-stained with rabbit anti-K8/18 (red) or mouse anti-vimentin (green) antibodies. (i Inset) Control double staining using red and green fluorochrome-conjugated goat anti-rabbit and goat anti-mouse antibodies without adding any primary antibodies. For each tissue, the “Merged” image corresponds to K8/K18 plus vimentin staining. All images were obtained by using the same magnification. (Bar in a = 20 μm.) (B) Hematoxylin and eosin staining of explanted liver from two patients with acute fulminant hepatitis. a is from a patient without a keratin mutation, whereas b is from a patient with the K18 T102A mutation. The region outlined by a box in b is magnified in c to illustrate the cytoplasmic filamentous deposits noted primarily in livers of patients with keratin mutations.
We also asked whether any histologic features identified by light microscopy could distinguish cirrhotic livers of patients with and without keratin mutations. Features such as Mallory's hyaline, acidophil bodies, enlarged hepatocytes, ground glass cytoplasm and dysplasia were found in keratin-mutant and non-keratin-mutant livers (data not shown). However, close inspection of the liver specimen(s) showed a unique accumulation in some hepatocytes of cytoplasmic filamentous arrays (Fig. 2B). When coding of the slides (with mutant or nonmutant keratin) was opened, the findings indicated that the filamentous deposits were found in 10 of 17 patients with keratin mutations but in only 3 of 16 disease-matched controls (P = 0.03). When patients with only primary hepatocellular diseases (cryptogenic, viral hepatitis, alcohol, acute fulminant hepatitis) were included in the analysis, 10 of 11 explants with a keratin mutation contained the cytoplasmic filamentous deposits as compared with 3 of 13 disease-matched controls (P = 0.001). The nature of the filamentous deposits remains to be determined, but they do not appear to correspond to aggregated keratins because they were not recognized by antikeratin antibodies (which may reflect epitope masking, not shown).
Discussion
Mutations in keratins and other IF family members, including lamins, desmin, glial fibrillary acidic protein (GFAP) and neurofilaments, are well-established causes of a wide range of tissue-specific human diseases (4, 16–22). The list of newly identified diseases associated with IF proteins continues to grow, including the latest association of K8 with cryptogenic cirrhosis (11), GFAP with Alexander disease (20) and the neurofilament-L chain with Charcot-Marie-Tooth type-2 (21, 22). One distinguishing feature of K8/K18 mutations, as compared with epidermal keratin mutations involving K5/K14/K1/K10, is that the epidermal keratin diseases are typically autosomal dominant with ≈100% penetrance. In contrast, K8/K18 mutations appear to be risk factors with variable penetrance, rather than direct causes of disease (5, 6). In support of this, the location of the characterized K8/K18 mutations does not involve conserved pan-keratin domain mutation hot spots that have been identified in epidermal keratins. Apparent absence of such mutations suggests that they may be lethal, given that K8/K18 are among the earliest expressed keratins during embryogenesis (23). Also, three independent patients with K8 G61C, K18 H127L, or K18 R260Q mutations had germ-line transmission of the mutations to their children (Table 5). The ages of the offspring with the keratin mutations range from 31–52, but none of these carriers have apparent liver disease, based on clinical history and serologic testing. These observations support a “multi-hit” hypothesis, whereby one major “hit” is carrying a relevant K8/K18 mutation with subsequent “hits” including underlying liver disease or exposure to injurious factors such as toxins or viruses. Clinical and natural history studies will be needed to define the relative risk of subsequent development of liver disease, or the relative increase in progression of an underlying liver disease, in those who carry specific K8/K18 mutations.
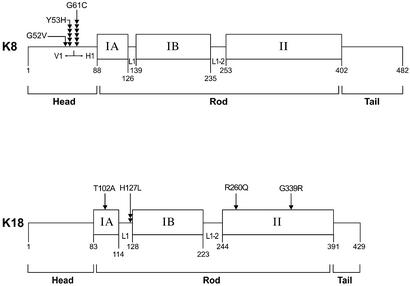
The significant number of patients described in this study, with new and previously described K8/K18 mutations, provide several insights into keratin-associated liver diseases. For example, only one K18 H127L mutation was previously described in a patient with cryptogenic cirrhosis (10), but the results herein corroborate this mutation in a different liver disease patient and add three other new K18 mutation sites (Fig. 3). It appears that K18 H127L and K8 Y53H/G61C are emerging as mutation hot spots. These three mutations were found in 2, 5, and 6 of the 17 patients with K8/K18 mutations, respectively (Fig. 3), and collectively make up ≈75% of the K8/K18 mutations identified to date. Analysis of additional liver disease patients will help determine whether these mutation hot spots maintain their frequency. Also, search for additional keratin mutation carriers in a broad range of cryptogenic and noncryptogenic liver diseases is warranted. For example, although the frequency of keratin mutations in noncryptogenic liver diseases is only 2.8% (11 of 399 patients), the frequency of keratin mutations in cryptogenic liver disease(s) is 8.8% (6 of 68 patients) (Table 1). This raises the possibility that some diseases that are linked with cryptogenic cirrhosis, such as nonalcoholic steatohepatitis (24–26), may be associated with keratin mutations. Furthermore, our calculation of keratin mutation frequencies in association with cirrhosis are probably underestimated, given that our search was restricted to regions that harbor epidermal disease keratin mutations. As such, the regions we analyzed correspond to 25% and 42% of K8 and K18 exonic sequences, respectively.
Figure 3.
Distribution of K8 and K18 mutations within the keratin protein backbone. Keratins, like all other intermediate filament proteins, have the prototype structure of a central and relatively conserved α-helical domain that is flanked by nonhelical N-terminal head and C-terminal tail domains. The rod domain is in turn divided into subdomains IA, IB, and II, which are interspaced by the linker regions (L1 and L1-2). The H1 and V1 subdomains of the K8 head domain, and the amino acid positions of the K8 and K18 domains/subdomains are also indicated. Each arrowhead represents an independently identified mutation at the indicated residue. Note that the most common mutations are K8 G61C (6 of 17), K8 Y53H (5 of 17), and K18 H127L (2 of 17), which collectively make up ≈75% of the mutations identified to date. Mutation location, based on codon numbers (not shown), would correspond to involved amino acid number plus one.
Although the mechanisms by which keratin mutations predispose to cirrhosis remain to be defined, already known and emerging keratin functions are likely to be involved (5, 6). For example, multiple transgenic mouse model studies showed that K8/K18 serve the essential function of protecting hepatocytes from a variety of stresses including agents that cause acute (e.g., acetaminophen) or chronic (e.g., griseofulvin) injury, and agents that induce apoptosis (e.g., Fas antibody) (6, 27, 28, 30). K8/K18 may also be involved in protein targeting to the apical compartment of polarized epithelia (29), interacting with apoptotic machinery proteins (6, 27, 28, 30, 31), cell signaling, and regulating the availability of abundant cellular proteins (5, 6). Hence, keratin mutations may potentially act at a number of functional cellular nodes. One surrogate marker of keratin function is cytoplasmic filament organization (5, 6), which was shown to be abnormally altered, only after stress exposure, in the K8 Y53H/G61C mutations (11). Our observation of preferential cytoplasmic filamentous deposits in cirrhotic livers of patients with keratin mutations is likely to be relevant and is reminiscent of Rosenthal fibers that are seen in association with Alexander Disease (20). The nature and pathogenesis of these deposits and their association with keratin-related liver disease remain to be investigated, but they are morphologically distinct from Mallory body-type deposits (32).
Supplementary Material
Acknowledgments
We are very grateful to Drs. Lauren Gerson and Ningguo Feng for performing the statistical analysis; to Tigist Belaye for assistance with patient liver samples and information; to Kris Morrow for preparing the figures; and to Linda Jacob and Blanca Pineda for assistance in manuscript preparation. We are also very thankful to the patients, their family members, and the blood bank volunteers who made their liver explants and blood samples available to allow us to carry out this study. This work was supported by a Department of Veterans Affairs Merit Award and National Institutes of Health Grant DK47918 (to M.B.O.), National Institutes of Health Training Grant DK07056 postdoctoral support (to J.M.D.), and National Institutes of Health Digestive Disease Center Grant DK56339. N.O.-K. is supported by a Veterans Administration Research Enhancement Award Program Award.
Abbreviations
- IF
intermediate filaments
- K
keratin
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Fuchs E, Coulombe P A. Cell. 1992;69:899–902. doi: 10.1016/0092-8674(92)90607-e. [DOI] [PubMed] [Google Scholar]
- 2.Steinert P M, Bale S J. Trends Genet. 1993;9:280–284. doi: 10.1016/0168-9525(93)90014-9. [DOI] [PubMed] [Google Scholar]
- 3.McLean W H I, Lane E B. Curr Opin Cell Biol. 1995;7:118–125. doi: 10.1016/0955-0674(95)80053-0. [DOI] [PubMed] [Google Scholar]
- 4.Irvine A D, McLean W H I. Br J Dermatol. 1999;140:815–828. doi: 10.1046/j.1365-2133.1999.02810.x. [DOI] [PubMed] [Google Scholar]
- 5.Coulombe P A, Omary M B. Curr Opin Cell Biol. 2002;14:110–122. doi: 10.1016/s0955-0674(01)00301-5. [DOI] [PubMed] [Google Scholar]
- 6.Omary M B, Ku N-O, Toivola D M. Hepatology. 2002;35:251–257. doi: 10.1053/jhep.2002.31165. [DOI] [PubMed] [Google Scholar]
- 7.Magin T M, Hesse M, Schroder R. Protoplasma. 2000;211:140–150. [Google Scholar]
- 8.Ku N-O, Zhou X, Toivola D M, Omary M B. Am J Physiol. 1999;277:G1108–G1137. doi: 10.1152/ajpgi.1999.277.6.G1108. [DOI] [PubMed] [Google Scholar]
- 9.Moll R, Franke W W, Schiller D L, Geiger B, Krepler R. Cell. 1982;31:11–24. doi: 10.1016/0092-8674(82)90400-7. [DOI] [PubMed] [Google Scholar]
- 10.Ku N-O, Wright T L, Terrault N A, Gish R, Omary M B. J Clin Invest. 1997;99:19–23. doi: 10.1172/JCI119127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ku N O, Gish R, Wright T L, Omary M B. N Engl J Med. 2001;344:1580–1587. doi: 10.1056/NEJM200105243442103. [DOI] [PubMed] [Google Scholar]
- 12.SAS Institute. JMP Statistics and Graphics Guide. Cary, NC: SAS Institute; 1995. , Version 3.1. [Google Scholar]
- 13.Ku N-O, Michie S, Oshima R G, Omary M B. J Cell Biol. 1995;131:1303–1314. doi: 10.1083/jcb.131.5.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laemmli U K. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 15.Liao J, Ku N-O, Omary M B. Electrophoresis. 1996;17:1671–1676. doi: 10.1002/elps.1150171104. [DOI] [PubMed] [Google Scholar]
- 16.Mounkes L C, Burke B, Stewart C L. Trends Cardiovasc Med. 2001;11:280–285. doi: 10.1016/s1050-1738(01)00126-8. [DOI] [PubMed] [Google Scholar]
- 17.Novelli G, Muchir A, Sangiuolo F, Helbling-Leclerc A, D'Apice M R, Massart C, Capon F, Sbraccia P, Federici M, Lauro R, et al. Am J Hum Genet. 2002;71:426–431. doi: 10.1086/341908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlsson L, Thornell L E. Acta Physiol Scand. 2001;171:341–348. doi: 10.1046/j.1365-201x.2001.00837.x. [DOI] [PubMed] [Google Scholar]
- 19.Goudeau B, Dagvadorj A, Rodrigues-Lima F, Nedellec P, Casteras-Simon M, Perret E, Langlois S, Goldfarb L, Vicart P. Hum Mutat. 2001;18:388–396. doi: 10.1002/humu.1210. [DOI] [PubMed] [Google Scholar]
- 20.Brenner M, Johnson A B, Boespflug-Tanguy O, Rodriguez D, Goldman J E, Messing A. Nat Genet. 2001;27:117–120. doi: 10.1038/83679. [DOI] [PubMed] [Google Scholar]
- 21.Mersiyanova I V, Perepelov A V, Polyakov A V, Sitnikov V F, Dadali E L, Oparin R B, Petrin A N, Evgrafov O V. Am J Hum Genet. 2000;67:37–46. doi: 10.1086/302962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Jonghe P, Mersivanova I, Nelis E, Del Favero J, Martin J J, Van Broeckhoven C, Evgrafov O, Timmerman V. Ann Neurol. 2001;49:245–249. doi: 10.1002/1531-8249(20010201)49:2<245::aid-ana45>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 23.Thorey I S, Meneses J J, Neznanov N, Kulesh D A, Pedersen R A, Oshima R G. Dev Biol. 1993;160:519–534. doi: 10.1006/dbio.1993.1326. [DOI] [PubMed] [Google Scholar]
- 24.Falck-Ytter Y, Younossi Z M, Marchesini G, McCullough A J. Semin Liver Dis. 2001;21:17–26. doi: 10.1055/s-2001-12926. [DOI] [PubMed] [Google Scholar]
- 25.Reid A E. Gastroenterology. 2001;121:710–723. doi: 10.1053/gast.2001.27126. [DOI] [PubMed] [Google Scholar]
- 26.Harrison S A, Diehl A M. Semin Gastrointest Dis. 2002;13:3–16. [PubMed] [Google Scholar]
- 27.Marceau N, Loranger A, Gilbert S, Daigle N, Champetier S. Biochem Cell Biol. 2001;79:543–555. [PubMed] [Google Scholar]
- 28.Oshima R G. Cell Death Differ. 2002;9:486–492. doi: 10.1038/sj.cdd.4400988. [DOI] [PubMed] [Google Scholar]
- 29.Ameen N A, Figueroa Y, Salas P J. J Cell Sci. 2001;114:563–575. doi: 10.1242/jcs.114.3.563. [DOI] [PubMed] [Google Scholar]
- 30.Ku N-O, Soetikno R M, Omary M B. Hepatology. 2003;37:1006–1014. doi: 10.1053/jhep.2003.50181. [DOI] [PubMed] [Google Scholar]
- 31.Inada H, Izawa I, Nishizawa M, Fujita E, Kiyono T, Takahashi T, Momoi T, Inagaki M. J Cell Biol. 2001;155:415–426. doi: 10.1083/jcb.200103078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Denk H, Stumptner C, Zatloukal K. J Hepatol. 2000;32:689–702. doi: 10.1016/s0168-8278(00)80233-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.