Abstract
As the central component of the human endotoxin sensor, Toll-like receptor 4 (TLR4) functions in the early detection and response to Gram-negative infection. We therefore examined a large collection of patients with meningococcal sepsis, comparing the frequency of rare TLR4 coding changes to those in an ethnically matched control population. TLR2 sequences were also acquired and compared. Total nucleotide variation at TLR4 and TLR2 loci was assayed by using a novel computational method. A total of 3.01 megabases of coding sequence was captured at these loci from white subjects with or without meningococcal disease. Authentic mutations were found and high-quality, bidirectional coverage was measured across the coding region by using mutationseeker, a program specifically designed to assay locus-specific genetic load. Using a method that obviates the confounding effect of linkage disequilibrium, we observed that rare heterozygous missense mutations of TLR4 contribute to the development of systemic meningococcal disease among white populations of the southern United Kingdom (P = 0.02; odds ratio 8.2). When results from all white populations were pooled, an overwhelmingly significant excess of such mutations was observed among individuals with disease (P = 2 × 10−6; odds ratio 27.0). The common white TLR4 variant (TLR4B), synonymous TLR4 substitutions, and variant TLR2 alleles were not significantly over-represented among patients with systemic meningococcal infections. No single variant of TLR4 was significantly over-represented in the meningococcal population. Collectively, however, rare TLR4 coding variants were markedly over-represented. Sensing via TLR4 probably contributes to the early containment of meningococcal infection, and sensing defects create increased risk of disease.
Premature death from infection (for the most part, from septic shock) is strongly heritable in human populations, more so than premature death from any other cause, including cardiovascular disease and neoplasia (1). Although a number of mutations are known to cause severe immunodeficiency states, milder and more prevalent immunodeficiency states presumably account for the overall heritability of infectious death, because most individuals who succumb to infection were previously considered to be healthy, and because the total number of people dying from infection far exceeds the number people with classical immunodeficiency syndromes. Little is known of the genes and the mutations that contribute to this heritability and hence are responsible for the preponderance of mortality caused by infectious diseases. A strict Mendelian pattern of inheritance does not apply to sepsis. Hence, the genetic components responsible for sepsis are weakly penetrant, markedly polygenic, or both. Logical considerations suggest that many mutations might contribute to a faulty immune response, and hence to sepsis, either incrementally or in a cooperative fashion. These mutations could, in principle, affect any aspect of the innate or acquired immune response.
The innate immune system is of central importance to the early containment of infection, in that it offers a response within minutes following invasion of the host. Hence, mutations that disrupt innate immune sensing of infectious organisms can permit sepsis to occur pursuant to the introduction of a small number of pathogenic microbes. Two of us (M.L.H. and M.L.) have previously shown, for example, that a mutation in mannose binding lectin, a plasma protein that binds to and activates complement on bacterial surfaces, is associated with childhood infection (2) and specifically with increased risk of meningococcal disease (3). Other genes have also fallen under suspicion. Among them are genes encoding the Toll-like receptors (TLRs), 10 of which are presently recognized in the human genome (4–6). TLR2 and TLR4 are the best understood in terms of ligand specificity.
In mice, mutations at the Lps locus abolish responses to bacterial lipopolysaccharide (LPS) (7–9). These mutations have been shown to enhance susceptibility to infection by Salmonella typhimurium (10, 11), Neisseria meningitidis (12), Francisella tularensis (13), Escherichia coli (14), Legionella pneumophila (15), and perhaps other Gram-negative organisms (16). Positional cloning revealed that Lps is identical to the TLR4 gene (Tlr4 in mice, or TLR4 in humans), which led two of us (I.S. and B.B.) to the conclusion that the TLR4 protein functions as the membrane-spanning component of the mammalian LPS receptor (17, 18). Moreover, genetic complementation studies (19, 20) have indicated that LPS enters into physical contact with TLR4 to elicit a signal, and that structural differences between the human and mouse TLR4 proteins are solely responsible for the observed interspecies difference in reactivity to specific LPS partial structures (lipid A vs. tetra-acyl lipid A).
The TLR2 protein confers a different but overlapping spectrum of microbial recognition. A knockout mutation of Tlr2 enhances susceptibility to Gram-positive infection in mice (21), and because TLR2 is known to recognize peptidoglycan, bacterial lipopeptides (22), which are constituents of both Gram-negative and -positive organisms, and certain exotic LPS species (23), it might be assumed that the recognition conferred by TLR2 is very broad.
In a recent survey of TLR4 polymorphism among different human populations, Smirnova et al. (24) showed that nonsynonymous variation (i.e., amino acid variation) is suppressed as compared with synonymous or intronic variation and, based on the pattern of mutation (a statistically significant excess of low-frequency coding variants), concluded that most mutations affecting TLR4 structure are mildly deleterious. This is particularly true of low-frequency coding variants, as opposed to the more common TLR4B allele (GenBank accession no. AF177766), which may have achieved relatively high frequency because it does not have a deleterious effect. The diminished fitness conferred by low-frequency mutations might, of course, reflect impairment of LPS sensing and consequent hypersusceptibility to Gram-negative infections. Although nothing is known of the LPS sensing potential of most of the low-frequency mutants, the TLR4B allele is reportedly a hypomorph (25), and the TLR4 locus is known to be haploinsufficient in mice so that a heterozygote effect would be anticipated for all hypomorphs.
In an effort to determine whether mutations affecting TLR4 or TLR2 structure render humans more susceptible to a specific Gram-negative bacterial infection and, further, to determine whether such mutations account for a substantial proportion of the Gram-negative sepsis that is observed in human populations, we have sequenced the complete coding region of both genes in DNA obtained from a large number of patients with meningococcal disease. These patients are known to usually succumb to disease in the absence of specific antibodies, exposing innate immunity as the principal means of defense, and suggesting that mutations affecting pathogen sensing might indeed cause enhanced susceptibility to infection.
Methods
Patients and Controls.
One hundred ninety-seven unrelated white patients with systemic meningococcal infections from the south of England, that attended the pediatric intensive care unit at St. Mary's hospital London between 1995 and 2000, were included in this study. The meningococcal patients were diagnosed, with the exclusion of other bacterial or viral causes, by the characteristic petechial rash, fever, and either meningitis or septicemia. Meningococcal meningitis is classified by evidence of meningism without shock or impaired perfusion, whereas meningococcal septicemia is diagnosed if there is no evidence of meningism but features of shock (prolonged capillary refill time, tachycardia, rapid base deficit, oliguria, and impaired oxygenation). In 146 patients diagnosis was confirmed by laboratory methods (detection of bacterial DNA, culture, or rising-specific antibody). Patients ranged in age from 1 month to 17.8 years (median 2.9 years) on admission. There were 108 boys. There were eight deaths (4%).
As a control group, 127 DNA samples were obtained from unrelated white subjects from the United Kingdom. The control samples came from persons who had no known history of serious disease, including infectious disease, and were unrelated contacts of the case. There were 64 boys. The study was approved by the St. Mary's Hospital Research Ethics Committee.
In an additional analysis, a total of 23 additional meningococcal samples were obtained from white meningococcal patients: 21 from the Netherlands and 2 from the United States. The Dutch patients ranged from 0.9 to 16.6 (median 3.8) years of age. The age of the two patients from the United States is unknown. As an additional control group, 256 control DNA samples were obtained from white subjects in the United States: either normal laboratory personnel without a history of serious illness or visitors to a health assessment clinic, also without history of serious illness. The age range of the individuals was 25–60 years, and the median age was 49.
Experimental Procedures
The single coding exon of TLR2 and the three coding exons of TLR4 were amplified from human DNA. The amplified DNA was purified from residual amplification primer by spin dialysis over Sepharose CL4B or by agarose gel electrophoresis. The samples were then sequenced by using internal primers to give complete coverage of the coding region and splice junctions of the TLR4 gene on both strands. For TLR4, if coverage by an initial set of 10 internal exon III primers was inadequate, a reserve set of 14 “secondary” primers was used to fill gaps. On average, 16 reads were required per sample to cover all three TLR4 exons, and 10 reads were required to cover the single TLR2 exon, using Applied Biosystems 373, 377, and 3100 sequencers and a Beckman Coulter CEQ sequencer. All primer sequences will be supplied on request.
All reads captured from the template were given an alpha-numeric designation specific for the individual from which the template was obtained. Collections of reads from up to 50 individuals at a time were read and mass-assembled by using the programs phred and phrap (written by Brent Ewing and Phil Green, respectively, of the University of Washington Genome Center, Seattle, and sequentially executed by using the phredphrap perl script). The program polyphred (Version 4.0) was used to tag mutations at maximum sensitivity (setting 6).
The program mutationseeker was written to give a precise, unbiased estimate of bidirectional coverage in millions of base pairs of aligned DNA sequence, and to assay locus-specific genetic load. The program first surveys reads acquired from each individual in the assembly, excluding those regions that fail to meet a user-specified level of quality. It then demarcates those regions that are covered bidirectionally to the specified level of quality. Within these regions of bidirectional coverage, it finds bases that were tagged in both forward and reverse direction by polyphred and, finally, exonerates those tagged positions in which one or more other reads of a specified quality conflict with the mutation. For the purposes of this study, the inclusion criterion was a mean phred score of 10 or greater over a window 30 nt. The exoneration threshold was set at 20 (i.e., a mean phred score of 20 at positions two bases to the left and two bases to the right of a putative mutation was required to exclude the mutation). mutationseeker surveys ≈500,000 bp of DNA per hour, applying highly objective criteria to the identification of mutations. It is more rapid and more sensitive than a human observer, and most importantly, produces a “denominator” with which to assess how much sequence of a specified quality was actually examined, either across an entire coding region or at a specific point within the coding region.
The viewing program consed [Version 11; David Gordon, University of Washington Genome Center (26)] was used to examine all hits produced by mutationseeker. Wherever a true hit was identified, all reads in the column above and below the mutation were manually checked for mutations as well.
The term chromosome equivalents (CE) is used to denote partial coverage of coding regions. Two CE correspond to the coverage of 2,355 bp of nonredundant coding sequence from TLR2 or 2,520 bp of nonredundant coding sequence from TLR4.
Statistical Analysis.
The differences in mutation frequencies were analyzed by using Fisher's exact test or a 2 × 2 χ2 test (two-tailed) as stated.
Results
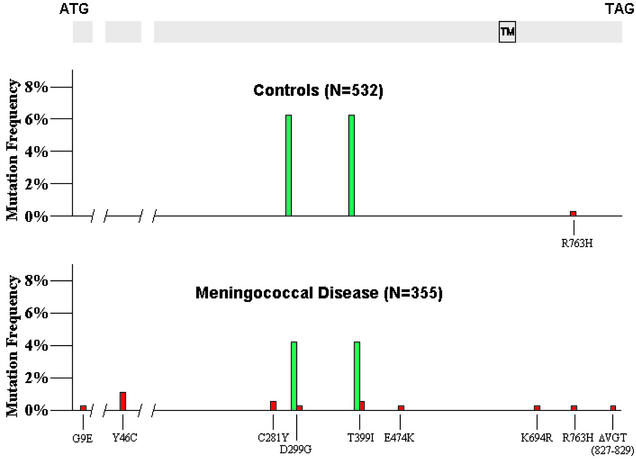
An obvious discrepancy between disease and control populations was evident at the TLR4 locus. Among 321 CE derived from the 197 patients with meningococcal disease in the United Kingdom, 11 rare missense mutations of TLR4 were observed. Only one rare missense mutation was identified in 238 control CE from the 127 United Kingdom controls (Fisher's exact test, P = 0.03; Tables 1 and 2, Fig. 1). In the disease group, 9 of the 11 rare mutations were located in the ectodomain, whereas 2 were located in the smaller but more strongly conserved (27) cytoplasmic domain (Fig. 1). Among the rare mutation group there were two deaths (18%), compared with six deaths in the rest of the group (3.2%; Fisher's exact test, P = 0.07), suggesting a trend toward more severe disease. The single variant observed among the control chromosomes was in the cytoplasmic domain. The TLR4B allele was slightly, but not significantly, less common among meningococcal disease patients as compared with the controls (10 of 321 CE for the meningococcal group vs. 13 of 238 CE for the control group; P = 0.1).
Table 1.
Summary of mutations identified within the bounds of the coding region of all three exons of the TLR4 gene
| TLR4 | Meningococcal disease
|
Normal controls
|
P*
|
|||
|---|---|---|---|---|---|---|
| United Kingdom | All white populations | United Kingdom | All white populations | United Kingdom | All white populations | |
| Rare missense | 11 (8 alleles) | 14 (10 alleles) | 1 | 1 | 0.018 | 0.000002 |
| (TLR4B) | 10 | 11 | 13 | 30 | 0.162 | 0.0593 |
| Synonymous | 7 | 9 | 6 | 12 | 0.396 | 0.4819 |
| Diploid coverage | 810.0 kb | 893.7 kb | 599.9 kb | 1341.0 kb | ||
| (321.4 CE) | (354.6 CE) | (238 CE) | (532 CE) | |||
| Number of subjects | 197 | 220 | 127 | 283 | ||
Relative locations of individual missense mutations are presented in Fig. 1, and details concerning all mutations are given in Table 4.
P value refers to the difference between meningococcal and control groups from the United Kingdom or to the difference between meningococcal and control groups from the total white population (United Kingdom and other). χ2 test, one-tailed. Calculations are performed with respect to diploid coverage values, i.e., the number of coding substitutions per number of nucleotides of coding sequence examined.
Table 2.
Characteristics of exonic mutations of TLR4 observed in United Kingdom white populations with systemic meningococcal disease (n = 197 individuals and 321 CE examined) and controls (n = 127 individuals and 238 CE examined)
| Number of chromosomes | Nucleotide change* | Exon | Amino acid change | Receptor domain† | Allele frequency‡ | P§ |
|---|---|---|---|---|---|---|
| Meningococcal disease | ||||||
| 1 | 4350 G → A | 1 | G9E | Ectoplasmic | 0.0040 | 0.53 |
| 4 | 8457 A → G | 2 | Y46C | Ectoplasmic | 0.0168 | 0.064 |
| 1 | 12293 T → C | 3 | None | — | 0.0030 | 0.59 |
| 3 | 12413 C → A | 3 | None | — | 0.0091 | 0.65 |
| 1 | 12820 G → A | 3 | C281Y | Ectoplasmic | 0.0031 | 0.56 |
| 11¶ | 12874 A → G and 13174 C → T | 3 | D299G and T399I | Ectoplasmic | 0.0344 | 0.20 |
| 1 | 13040 A → G | 3 | None | — | 0.0031 | 0.58 |
| 2 | 13174 C → T | 3 | T399I | Ectoplasmic | 0.0063 | 0.32 |
| 1 | 13398 G → A | 3 | E474K | Ectoplasmic | 0.0031 | 0.57 |
| 1 | 13937 G → A | 3 | None | — | 0.0031 | 0.11 |
| 1 | 13982 T → G | 3 | None | — | 0.0031 | 0.57 |
| 1 | 14059 A → G | 3 | K694R | Cytoplasmic | 0.0031 | 0.57 |
| 1 | 14266 G → A | 3 | R763H | Cytoplasmic | 0.0031 | 0.68 |
| Controls | ||||||
| 2 | 12413 C → A | 3 | None | — | 0.0083 | |
| 13 | 12874 A → G | 3 | D299G | Ectoplasmic | 0.0529 | |
| 13174 C → T | T399I | |||||
| 4 | 13937 G → A | 3 | None | — | 0.0161 | |
| 1 | 14266 G → A | 3 | R763H | Cytoplasmic | 0.0041 | |
Assigned with reference to genomic sequence of human TLR4 (GenBank accession no. AF177765).
The human TLR4 protein is 839 aa in length. Residues 1–631 are designated here as the ectodomain, residues 632–662 are assumed to span the plasma membrane, and residues 663–839 are considered to comprise the cytoplasmic domain.
Allele frequencies refer to the number of observations divided by the number of loci that could be examined at the site in question. All coding mutations occurred in isolation from one another; i.e., no compound heterozygotes were observed.
P value refers to the likelihood (two-sided) that the excess incidence of each allele would be observed in the meningococcal population by chance.
Among the 10 individuals in whom the mutation was observed, one was a homozygote for the common variant allele; hence a total of 11 chromosomes with the mutation were detected.
Figure 1.
TLR4 mutations observed in white patients with meningococcal sepsis and in normal controls. The common TLR4B allele is shown in green and occurs at a similar frequency in both groups. All rare coding mutations are shown in red. Fourteen rare mutations are observed in the meningococcal sample, representing 10 separate alleles. One rare mutation is observed in the control group (P = 0.000002). Note that the isolated mutations at residue positions 299 and 399 represent rare alleles in the white population, although they are each component parts of the common TLR4B allele. Exons are drawn in proportion to the length of coding sequence that they contain, but intron lengths are not shown in proportion to actual length. TM, location of the transmembrane domain; N, the number of chromosome equivalents analyzed.
Because a significant excess of rare coding variants were found at the TLR4 locus in white patients from the United Kingdom, additional meningococcal sepsis templates were collected from white subjects in The Netherlands and the United States, along with additional control templates. These samples were processed in the same way as the United Kingdom templates. Among 33 additional meningococcal CE, three more rare variants were found, two of which represented previously uncharacterized mutant alleles (a second instance of G → A at position 12820; C281Y, as well as single instances of A → G at position 12874; D299G, and del[14453–14461]; del[VGT, 827–829]). Among a total of 294 additional control CE, no rare variants were found. These data suggest that the findings in the United Kingdom population are likely to be true of white populations in general. By combining the white populations, the significance of the association between low-frequency mutations and meningococcal disease becomes overwhelming (14 mutations identified in 897.7 kb of coding sequence from 230 patients with meningococcal disease vs. 1 mutation identified in 1,341 kb of coding sequence from 421 controls; χ2 = 22.6, P = 0.000002; odds ratio = 27).
At the TLR2 locus (Tables 3 and 4), a variant allele represented in 9.5% of the control population (8 of 168 chromosomes examined) and specifying an amino acid substitution within the cytoplasmic domain of the TLR2 protein (P631H) was less common among patients with meningococcal disease (observed in 2 of 192 chromosomes examined; P = 0.05). A second cytoplasmic domain variant (R753Q) was observed in 5 of 152 control chromosomes and 6 of 192 chromosomes from meningococcal sepsis patients (P = 1.0). A rather common synonymous substitution (C → T at position 1752) was slightly over-represented among patients with meningococcal disease (P = 0.02). However, because C → T at position 1752 is clearly not in linkage disequilibrium with an adjacent polymorphism and is unlikely to have a functional effect, the relative excess among patients is likely to be due to chance. There was no significant excess of missense alleles (no enhancement of locus-specific genetic load at TLR2) in the meningococcal population, and the trend was, overall, to the contrary.
Table 3.
Summary of mutations identified within the bounds of the coding region of the TLR2 gene
| TLR2 | Meningococcal disease | Normal controls | P* |
|---|---|---|---|
| Missense | 8 | 13 | 0.132 |
| Synonymous | 136 | 105 | 0.972 |
| Diploid coverage† | 440.9 kb (187.2 CE) | 336.0 kb (142.7 CE) | |
| Number of subjects | 102 | 104 |
Details concerning all mutations are given in Table 4.
P value refers to the difference between meningococcal and normal groups collected in the United Kingdom. χ2 test, two-tailed. Calculations are performed with respect to diploid coverage values, i.e., the number of coding substitutions per number of nucleotides of coding sequence examined.
Total coding sequence acquired on both strands, given as kb or CE.
Table 4.
Characteristics of TLR2 mutations observed in white subjects with systemic meningococcal disease (n = 102 individuals and 187.2 CE examined) and normal controls (n = 104 individuals and 142.7 CE examined)
| Number of chromosomes | Nucleotide (genomic) | Amino acid | Receptor domain* | Allele frequency† | P‡ |
|---|---|---|---|---|---|
| Meningococcal disease | |||||
| 105§ | 726 C → T | None | — | 0.5198 | 0.533 |
| 1 | 768 G → C | None | — | 0.0050 | 1.000 |
| 15 | 1479 T → C | None | — | 0.0773 | 0.388 |
| 13 | 1752 C → T | None | — | 0.0663 | 0.023 |
| 2 | 1818 G → A | None | — | 0.0102 | 0.500 |
| 2 | 2021 C → A | P631H | Cytoplasmic | 0.0104 | 0.050 |
| 6 | 2387 G → A | R753Q | Cytoplasmic | 0.0313 | 1.000 |
| Controls | |||||
| 94 | 726 C → T | None | — | 0.5529 | |
| 8 | 1479 T → C | None | — | 0.0506 | |
| 3 | 1752 C → T | None | — | 0.0174 | |
| 8 | 2021 C → A | P631H | Cytoplasmic | 0.0476 | |
| 5 | 2387 G → A | R753Q | Cytoplasmic | 0.0329 | |
The human TLR2 protein is 784 aa in length. Residues 1–588 are designated here as the ectodomain, residues 589–615 are assumed to span the plasma membrane, and residues 616–784 are considered to comprise the cytoplasmic domain.
Allele frequencies refer to the number of observations divided by the number of loci that could be examined at the site in question. All coding mutations occurred in isolation from one another; i.e., no compound heterozygotes were observed.
P value refers to the likelihood (two-sided; Fisher's exact test) that the difference between the incidence of each variant allele in the meningococcal and control populations would occur by chance.
The ratio of T/T:C/T:C/C individuals among the meningococcal population was (32:30:23), whereas the ratio of T/T:C/T:C/C individuals among the control population is (25:55:21). Neither distribution represents a significant departure from predictions of the Hardy–Weinberg equilibrium.
There was no significant difference between the patient and control groups with respect to the frequency of synonymous mutations at either locus, whether these mutations were considered individually or collectively and whether they were common or rare (Tables 1 and 3).
Discussion
Although the meningococcus is an oropharyngeal commensal in a large fraction of the population, and most individuals are believed to harbor meningococcus for at least part of their lives, only a small fraction of the population (≈1 per 40,000–100,000) ever develops systemic meningococcal disease. When it occurs, systemic meningococcal disease is usually observed in previously healthy children and young adults without underlying medical or surgical conditions. It may be assumed that the disease begins following inoculation of a small number of organisms, which, under normal circumstances, would be eliminated through an innate immune mechanism that possibly leads to the generation of specific antibodies
LPS is readily shed from the meningococcus in vivo, and immunopathological studies in children with meningococcal sepsis have produced clear and consistent data suggesting an important role for endotoxin (28). In this sense, hyperreactivity to LPS might prove damaging during meningococcal infection. Equally, however, during the first minutes or hours following inoculation of the organism, relative hyporesponsiveness to LPS might prevent the mobilization of an effective innate immune response and set the stage for overwhelming sepsis.
The pattern of mutation at the human TLR4 locus (a relative excess of rare amino acid replacements) indicates that TLR4 has been subject to weak purifying selection (24). Hence, no structural variants of TLR4 have achieved a particularly high frequency among humans.
The observation that severe hypomorph mutations of Tlr4 cause mice to be selectively immunocompromised, enhancing susceptibility to Gram-negative infections, suggests that comparable mutations might yield enhanced susceptibility to Gram-negative infection in humans. This, in turn, would be manifested in the observation of a higher frequency of TLR4 coding mutations among patients with Gram-negative infection.
We have now presented data consistent with this expectation. Among white populations, allelic variation at TLR4 is exceedingly rare. Excluding a single common variant allele, only one coding variant was observed among 532 CE (1.34 megabases of DNA sequence) analyzed. Among patients with systemic meningococcal disease, an increased frequency of rare mutations at the TLR4 locus is clearly observed. Moreover, only rare mutations of TLR4, and only missense mutations, appear to have been concentrated within the meningococcal population.
At the TLR4 locus, the relatively common TLR4B variant, which has achieved an allele frequency of ≈6% in white populations, is absolved of suspicion as a causative factor in meningococcal sepsis, notwithstanding its reported codominant hypomorphic character (25) and its purported involvement in other infectious diseases (29). This observation is in agreement with the analysis of Read and colleagues (30). On the other hand, if we consider that 14 rare TLR4 coding mutations observed among 355 CE surveyed from white patients with meningococcal disease were functional and that only one rare mutation was observed among 532 CE surveyed from white controls, it may be surmised that ≈7.5 ± 3.7% of meningococcal sepsis cases in the white population can be directly attributed to coding mutations at this locus. An added contribution may be imparted by mutations that affect TLR4 promoter function or splicing, although this contribution would be more difficult to detect.
For the most part, association studies linking variants at specific loci to meningococcal disease susceptibility or severity have dealt with polymorphisms that are represented at high frequency and reside either within the coding region or promoter of genes concerned with immune function. It is, in general, difficult to exclude linkage disequilibrium as the cause of associations that are observed. In the present instance, linkage disequilibrium cannot explain the observations that have been made, because numerous independent mutations contribute to the association with meningococcal disease.
Systemic meningococcal disease is relatively rare. However, it may be accepted that many Gram-negative organisms are detected primarily as a result of the LPS that they produce, and that mutations affecting TLR structure might affect the timely detection of these organisms just as they seem to affect the detection of the meningococcus. Moreover, there are 10 human TLRs (4–6), each of which is believed to make a distinct contribution to the detection of microbial infection. Accessory proteins such as CD14 (31, 32), MD-2 (33, 34), and MyD88 (35) are also believed to be required for at least some of the signals that alert the host to infection. Many other proteins yet unknown may also contribute to microbial sensing. Collectively, genes encoding proteins of the afferent pathways used by the innate immune system may represent a major repository of the heritability observed by Sorenson et al. (1). Here, we have established that rare codominant mutations affecting TLR4 structure contribute to disease susceptibility. Indeed, many human diseases may owe their heritability to low-frequency codominant mutation. When multiple inherited and environmental factors influence the development of disease, estimation of locus-specific genetic load, and perhaps no other method, can reliably implicate a specific gene in disease pathogenesis.
Acknowledgments
This work was funded by National Institutes of Health Grant R01-GM60031 and the Meningitis Research Foundation.
Abbreviations
- CE
chromosome equivalents
- LPS
lipopolysaccharide
- TLR
Toll-like receptor
References
- 1.Sorensen T I, Nielsen G G, Andersen P K, Teasdale T W. N Engl J Med. 1988;318:727–732. doi: 10.1056/NEJM198803243181202. [DOI] [PubMed] [Google Scholar]
- 2.Summerfield J A, Sumiya M, Levin M, Turner M W. Br Med J. 1997;314:1229–1232. doi: 10.1136/bmj.314.7089.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hibberd M L, Sumiya M, Summerfield J A, Booy R, Levin M. Lancet. 1999;353:1049–1053. doi: 10.1016/s0140-6736(98)08350-0. [DOI] [PubMed] [Google Scholar]
- 4.Du X, Poltorak A, Wei Y, Beutler B. Eur Cytokine Network. 2000;11:362–371. [PubMed] [Google Scholar]
- 5.Chuang T H, Ulevitch R J. Eur Cytokine Network. 2000;11:372–378. [PubMed] [Google Scholar]
- 6.Chuang T, Ulevitch R J. Biochim Biophys Acta. 2001;1518:157–161. doi: 10.1016/s0167-4781(00)00289-x. [DOI] [PubMed] [Google Scholar]
- 7.Heppner G, Weiss D W. J Bacteriol. 1965;90:696–703. doi: 10.1128/jb.90.3.696-703.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watson J, Riblet R, Taylor B A. J Immunol. 1977;118:2088–2093. [PubMed] [Google Scholar]
- 9.Watson J, Kelly K, Largen M, Taylor B A. J Immunol. 1978;120:422–424. [PubMed] [Google Scholar]
- 10.O'Brien A D, Rosenstreich D L, Scher I, Campbell G H, MacDermott R P, Formal S B. J Immunol. 1980;124:20–24. [PubMed] [Google Scholar]
- 11.Rosenstreich D L, Weinblatt A C, O'Brien A D. CRC Crit Rev Immunol. 1982;3:263–330. [PubMed] [Google Scholar]
- 12.Woods J P, Frelinger J A, Warrack G, Cannon J G. Infect Immun. 1988;56:1950–1955. doi: 10.1128/iai.56.8.1950-1955.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macela A, Stulik J, Hernychova L, Kroca M, Krocova Z, Kovarova H. FEMS Immunol Med Microbiol. 1996;13:235–238. doi: 10.1111/j.1574-695X.1996.tb00243.x. [DOI] [PubMed] [Google Scholar]
- 14.Hagberg L, Hull R, Hull S, McGhee J R, Michalek S M, Svanborg Eden C. Infect Immun. 1984;46:839–844. doi: 10.1128/iai.46.3.839-844.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gebran S J, Yamamoto Y, Newton C, Tomioka M, Widen R, Klein T W, Friedman H. J Leukocyte Biol. 1995;57:80–87. doi: 10.1002/jlb.57.1.80. [DOI] [PubMed] [Google Scholar]
- 16.Mitchell C R, Kempton J B, Scott-Tyler B, Trune D R. Otolaryngol Head Neck Surg. 1997;117:459–464. doi: 10.1016/S0194-59989770014-7. [DOI] [PubMed] [Google Scholar]
- 17.Poltorak A, Smirnova I, He X L, Liu M Y, Van Huffel C, McNally O, Birdwell D, Alejos E, Silva M, Du X, et al. Blood Cells Mol Dis. 1998;24:340–355. doi: 10.1006/bcmd.1998.0201. [DOI] [PubMed] [Google Scholar]
- 18.Poltorak A, He X, Smirnova I, Liu M-Y, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, et al. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 19.Muzio M, Ni J, Feng P, Dixit V M. Science. 1997;278:1612–1615. [Google Scholar]
- 20.Poltorak A, Ricciardi-Castagnoli P, Citterio A, Beutler B. Proc Natl Acad Sci USA. 2000;97:2163–2167. doi: 10.1073/pnas.040565397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takeuchi O, Hoshino K, Akira S. J Immunol. 2000;165:5392–5396. doi: 10.4049/jimmunol.165.10.5392. [DOI] [PubMed] [Google Scholar]
- 22.Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 23.Werts C, Tapping R I, Mathison J C, Chuang T H, Kravchenko V, Saint G I, Haake D A, Godowski P J, Hayashi F, Ozinsky A, et al. Nat Immunol. 2001;2:346–352. doi: 10.1038/86354. [DOI] [PubMed] [Google Scholar]
- 24.Smirnova I, Hamblin M, McBride C, Beutler B, Di Rienzo A. Genetics. 2001;158:1657–1664. doi: 10.1093/genetics/158.4.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arbour N C, Lorenz E, Schutte B C, Zabner J, Kline J N, Jones M, Frees K, Watt J I, Schwartz D A. Nat Genet. 2000;25:187–192. doi: 10.1038/76048. [DOI] [PubMed] [Google Scholar]
- 26.Gordon D, Abajian C, Green P. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 27.Smirnova I, Poltorak A, Chan E K L, McBride C, Beutler B. Genome Biol. 2000;1:1–10. doi: 10.1186/gb-2000-1-1-research002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brandtzaeg P, Ovstebo R, Kierulf P. Prog Clin Biol Res. 1995;392:219–233. [PubMed] [Google Scholar]
- 29.Lorenz E, Mira J P, Frees K L, Schwartz D A. Arch Intern Med. 2002;162:1028–1032. doi: 10.1001/archinte.162.9.1028. [DOI] [PubMed] [Google Scholar]
- 30.Read R C, Pullin J, Gregory S, Borrow R, Kaczmarski E B, Di Giovine F S, Dower S K, Cannings C, Wilson A G. J Infect Dis. 2001;184:640–642. doi: 10.1086/322798. [DOI] [PubMed] [Google Scholar]
- 31.Wright S D, Ramos R A, Tobias P S, Ulevitch R J, Mathison J C. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 32.Haziot A, Ferrero E, Kontgen F, Hijiya N, Yamamoto S, Silver J, Stewart C L, Goyert S M. Immunity. 1996;4:407–414. doi: 10.1016/s1074-7613(00)80254-x. [DOI] [PubMed] [Google Scholar]
- 33.Shimazu R, Akashi S, Ogata H, Nagai Y, Fukudome K, Miyake K, Kimoto M. J Exp Med. 1999;189:1777–1782. doi: 10.1084/jem.189.11.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagai Y, Akashi S, Nagafuku M, Ogata M, Iwakura Y, Akira S, Kitamura T, Kosugi A, Kimoto M, Miyake K. Nat Immunol. 2002;3:667–672. doi: 10.1038/ni809. [DOI] [PubMed] [Google Scholar]
- 35.Kawai T, Adachi O, Ogawa T, Takeda K, Akira S. Immunity. 1999;11:115–122. doi: 10.1016/s1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]