Abstract
Adeno-associated viruses (AAVs) are single-stranded DNA viruses that are endemic in human populations without known clinical sequelae and are being evaluated as vectors for human gene therapy. To better understand the biology of this virus, we examined a number of nonhuman primate species for the presence of previously uncharacterized AAVs and characterized their structure and distribution. AAV genomes were widely disseminated throughout multiple tissues of a variety of nonhuman primate species. Surprising diversity of sequence, primarily localized to hypervariable regions of the capsid protein, was detected. This diversity of sequence is caused, in part, by homologous recombination of co-infecting parental viruses that modify the serologic reactivity and tropism of the virus. This is an example of rapid molecular evolution of a DNA virus in a way that was formerly thought to be restricted to RNA viruses.
Adeno-associated viruses (AAVs) belong to the Parvoviridae family, which is characterized as small animal viruses with linear single-stranded DNA genomes that replicate in the presence of helper virus such as adenovirus (1). AAVs are being evaluated as vectors for human gene therapy (2). The initial characterization of this group of viruses was based on serologic crossreactivity by using complement fixation and neutralizing assays (3). Six distinct serotypes of AAV have been described, of which five were initially isolated as contaminants of adenovirus preparations (4–6). Sequence analysis of selected AAV isolates revealed divergence throughout the genome that is most concentrated in hypervariable regions (HVRs) of the capsid proteins (7–10). Epidemiological data indicate that all known serotypes are endemic to primates, although isolation of clinical isolates has been restricted to AAV2 and AAV3 from anal and throat swabs of human infants and AAV5 from a human condylomatous wart (11–14). No known clinical sequelae have been associated with AAV infection. Vectors based on replication-defective forms of AAV have been evaluated in preclinical and clinical models of gene therapy (2).
Materials and Methods
Nonhuman Primate (NHP) Animals.
Rhesus macaques from Penn colony were captive-bred and of Chinese or Indian origin. Tissues of rhesus macaques were kindly provided by Gary B. Baskin and Maurice J. Duplantis, III (Tulane University Regional Primate Research Center, Covington, LA). Several NHP centers or farms provided peripheral blood samples of other animals used in this study including New Iberia Research Center (New Iberia, LA), Covance (Alice, TX), Charles River Breeding Laboratories, Buckshire Farms (Perkasie, PA), Southwest Foundation of Biomedical Research (San Antonio, TX), Laboratories of Virginia (Yemassee, SC), and University of South Alabama (Mobile).
Detection and Recovery of AAV Sequences.
DNA was extracted and analyzed for the presence of AAV DNA by using a PCR strategy to amplify a 255-bp (15) fragment called the “signature region” by using conserved oligonucleotides. To directly amplify a 3.1-kb full-length Cap fragment from NHP tissue and blood DNAs, two other highly conserved regions were identified in AAV genomes for use in PCR amplification of large fragments. A primer within a conserved region located in the middle of the Rep gene was selected (AV1ns, 5′-GCTGCGTCAACTGGACCAATGAGAAC-3′) in combination with the 3′ primer located in another conserved region downstream of the Cap gene (AV2cas, 5′-CGCAGAGACCAAAGTTCAACTGAAACGA-3′) for amplification of full-length cap fragments. The PCR products were Topo-cloned (Invitrogen), and sequence analysis was performed by Qiagengenomics (Qiagengenomics, Seattle) with an accuracy of ≥99.9%. A total of 50 capsid clones were isolated and characterized. Among them, 37 clones were derived from rhesus macaque tissues (rh.1–rh.37), 6 clones from cynomologus macaques (cy.1–cy.6), 2 clones from baboons (bb.1 and bb.2), and 5 clones from chimpanzees (ch.1–ch.5).
Characterization of PCR for Artifacts.
To rule out the possibility that sequence diversity within the previously uncharacterized AAV family was not an artifact of the PCR, such as PCR-mediated gene splicing by overlap extension between different partial DNA templates with homologous sequences (16), or the result of a recombination process in bacteria, we performed a series of experiments under identical conditions for VP1 amplification using total cellular DNAs. First, we mixed intact AAV7 and AAV8 plasmids at an equal molar ratio followed by serial dilutions. The serially diluted mixtures were used as templates for PCR amplification of 3.1-kb VP1 fragments by using universal primers, and identical PCR conditions to that were used for DNA amplifications to determine whether any hybrid PCR products were generated. We also transformed the mixture into bacteria and isolated transformants to look for hybrid clones possibly derived from recombination process in bacterial cells. In a different experiment, we restricted AAV7 and AAV8 plasmids with MspI, AvaI, and HaeI, all of which cut both genomes multiple times at different positions, mixed the digestions in different combinations, and used them for PCR amplification of VP1 fragments under the same conditions to test whether any PCR products could be generated through overlap sequence extension of partial AAV sequences. In another experiment, a mixture of gel-purified 5′ 1.5-kb AAV7 VP1 and 3′ 1.7-kb AAV8 VP1 fragments with overlap in the signature region was serially diluted and used for PCR amplification in the presence and absence of 200 ng of cellular DNA extracted from a monkey cell line that was free of AAV sequences by TaqMan analysis. Under the conditions of the genomic DNA Cap amplification, none of these experiments generated products of PCR artifacts that would explain the described sequence heterogeneity (data not shown). As a further confirmation, we designed three pairs of primers that were located at different HVRs and were sequence-specific to the variants of clone 42s from rhesus macaque F953 in different combinations to amplify shorter fragments from mesenteric lymph node DNA from F953, from which clone 42s were isolated. All sequence variations identified in full-length Cap clones were found in these short fragments (data not shown).
Characterization of Previously Uncharacterized AAV Sequences.
NHP VP1 sequences were aligned by using CLUSTALX 1.8 (17) and analyzed further with the Vector NTI software package (Informax, Bethesda). HVRs were defined by means of the quality scores (Q) derived from the CLUSTALX alignment. In this analysis, sequence divergence is represented by a higher number. In short, an algorithm was developed that anchors an HVR to a residue with (100 − Q) ≥ 75 and then includes neighboring residues for which (100 − Q) ≥ 50. HVRs in close proximity were bridged. The space-filling model of the capsid protein structure (PDB ID code 1LP3) was made with MOLSCRIPT (18), and the ribbon representations were prepared within the SWISSPDB VIEWER program (19) and rendered with the Persistence of Vision Ray Tracer program (POV-RAY 2000 3.1G).
The grouping of structurally similar viruses was performed with the SPLITSTREE 3.1 program (20) making use of the split decomposition method (21) and the Hamming distance transformation. A formal test for recombination was performed by making use of SIMPLOT software (22). For the bootscanning phylogeny (23), a putative recombinant was analyzed with a number of reference sequences including the proposed parental clones. The neighbor-joining method based on the Kimura two-parameter distance model used with a transition/transversion bias of 2.0 was used after the generation of 100 bootstrap replicates.
Serological Analysis and Tissue Distribution of Proviral DNA.
Neutralizing antibodies were determined as described except the sensitivity of the assay was increased by decreasing the amount of vector for each serotype to 104 genome copies per cell (15).
Genome copies of AAV proviral sequences present in DNA samples extracted from 10 different tissues of rhesus macaques from two different colonies were quantified by real-time PCR (ABI PRISM 7700 sequence detector, Applied Biosystems) by using three sets of primers and probes to highly conserved regions of Cap, CapR2b (forward, 5′-CCGACGCCGAGTTTCA-3′; reverse, 5′-GCCTGGAAGACTGCTC-3′; probe, 6Fam-AAGATACGTCTTTTGGGGGCA-Tamra; amplicon size, 76 bp), CapR3b (forward, 5′-CCAATGGACAATAAC-3′; reverse, 5′-GACAGAGTCATCACCACCA-3′; probe, 6Tam-AATTGGCATTGCGATTCCACATG-Tamra), and RepR1b (forward, 5′-TCCAACACCAACATGT-3′; reverse, 5′-GGCTGCTGGTGCTCGAAGGT-3′; probe, 6Fam-CGCCGTGATTGACGGGAAC-Tamra). Animals were necropsied without prior experimentation or were being necropsied after prior treatment with adenoviral, AAV, or lentiviral vectors. We felt that including these latter animals was appropriate, from an animal-welfare standpoint, to maximize available tissues for analysis while minimizing necropsy of animals not enrolled in other studies.
Molecular and Cellular Analyses of AAV Sequences from Tissues.
In situ hybridization was performed per established procedures with optimization (24). Briefly, liver tissues were embedded in Tissue-Tek (Sakura Finetek, Torrance, CA) and sectioned to 10 μm, fixed in 4% paraformaldehyde, denatured, and hybridized to a digoxigenin-labeled probe. Hybridized probes were detected by using a sheep antidigoxigenin antibody (Roche Molecular Biochemicals) followed by Alexa fluor 488-labeled donkey anti-sheep antibody (Molecular Probes). Optical sections transversing the cells were collected at 0.5-μm intervals by using a Leica TCS SP2 confocal system and a Leica inverted DMIRBE microscope (Leica Microsystems, Exton, PA). DNA hybridization analysis of total cellular DNA was performed as described (25).
Results
Discovery of Diverse Populations of AAVs in NHP Populations.
In an attempt to better understand the biology of AAV, we used NHPs as models to characterize the sequelae of natural infections. Tissues from NHPs were screened for AAV sequences by using a PCR method based on oligonucleotides to highly conserved regions of known AAVs to amplify a 255-bp fragment in Cap called the signature region, which was shown later to span HVR3.
An initial survey of peripheral blood of a number of NHP species revealed detectable AAV in a subset of animals from species such as rhesus macaques, cynomologus macaques, chimpanzees, and baboons but no AAV in other species including Japanese macaques, pig-tailed macaques, and squirrel monkeys (Table 1). A more extensive analysis of vector distribution was conducted in tissues of rhesus macaques of the University of Pennsylvania and Tulane colonies recovered at necropsy, which revealed AAV sequence throughout a wide array of tissues (data not shown).
Table 1.
Serologic and molecular analysis of AAVs in NHP populations
| Animals (species/colony) | PCR (+) | Seropositive
|
|
|---|---|---|---|
| AAV7 | AAV8 | ||
| Rhesus macaque | |||
| University of Pennsylvania | 13/48 (27) | 45/56 (80) | 48/56 (85) |
| New Iberia | 2/10 (20) | 8/10 (80) | 10/10 (100) |
| Covance | 2/10 (20) | 5/10 (50) | 4/10 (40) |
| Cynomologus macaque | |||
| New Iberia | 1/10 (10) | 11/22 (50) | 13/22 (59) |
| Charles River | 6/30 (20) | 18/24 (75) | 16/24 (67) |
| Japanese macaque | |||
| ORPRC | 0/12 (0) | 2/12 (20) | 6/12 (50) |
| Pig-tailed macaque | |||
| LABS | 0/12 (0) | 10/12 (83) | 8/12 (67) |
| Squirrel macaque | |||
| University of South Alabama | 0/36 (0) | ND | ND |
| Chimpanzee | |||
| New Iberia | 7/10 (70) | 4/29 (14) | 5/29 (17) |
| Baboon | |||
| Buckshire Farms | 5/16 (31) | 5/12 (42) | 6/12 (50) |
| SFBR | 3/12 (25) | ND | ND |
Percentages are shown in parentheses. ORPRC, Oregon Regional Primate Center (Beaverton); LABS, Laboratories of Virginia; SFBR, Southwest Foundation for Biomedical Research; ND, not determined.
The amplified signature sequences were subcloned into plasmids, and individual transformants were subjected to sequence analysis. This revealed substantial variation in nucleotide sequence of clones derived from different animals. Variation in the signature sequence was also noted in clones obtained within individual animals. Tissues harvested from two animals in which unique signature sequences were identified (i.e., colon and heart) were characterized further by expanded PCR cloning and subsequent sequencing of the entire reconstructed virus genomes. These viral genomes differ from the other known AAVs, with the greatest sequence divergence noted in regions of the gene encoding VP1. In a previous study they were shown to be unique serotypes that we call AAV7 and AAV8 (15). The frequency of neutralizing antibodies to these previously uncharacterized serotypes in various NHP species is high (Table 1).
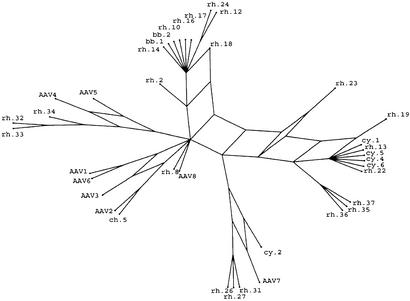
To further characterize the inter- and intraanimal variation of AAV signature sequence, selected tissues of different species were subjected to extended PCR to amplify entire VP1 ORFs and cloned, and individual transformants were isolated and sequenced fully. Of the 50 clones that were sequenced from 15 different animals, 30 were considered nonredundant based on the finding of at least 7-aa differences from one another. The VP1 clones are numbered sequentially as they were isolated, with a prefix indicating the species of NHP from which they were derived. The structural relationships between the 30 nonredundant clones and the previously described 8 AAV serotypes were determined by using the SPLITSTREE program (20) with implementation of the method of split decomposition (21) and shown in Fig. 1. The analysis depicts homoplasy between a set of sequences in a tree-like network rather than a bifurcating tree. The advantage is to enable detection of groupings that are the result of convergence and to exhibit phylogenetic relationships even when they are distorted by parallel events. Extensive phylogenetic research will be required to elucidate the AAV evolution, whereas the intention here is only to group the different clones by their sequence similarity.
Figure 1.
Split-decomposition analysis of 38 AAV sequences. The split-decomposition method performed as described in Materials and Methods represents the previously uncharacterized AAVs in relation to the known serotypes. This model is not on scale and magnifies network links from the analysis for better presentation.
To confirm that the previously uncharacterized VP1 sequences were derived from infectious viral genomes, cellular DNA from tissues with high abundance of viral DNA was restricted with an endonuclease that should not cleave within AAV and transfected into 293 cells together with an infectious molecular clone of adenovirus. This resulted in rescue and amplification of AAV genomes from DNA of tissues from three different animals (data not shown), which, when sequenced, were essentially identical to the clones rescued by PCR.
Sequence Variation in NHP AAVs Reside in HVRs on the Surface of the Virus.
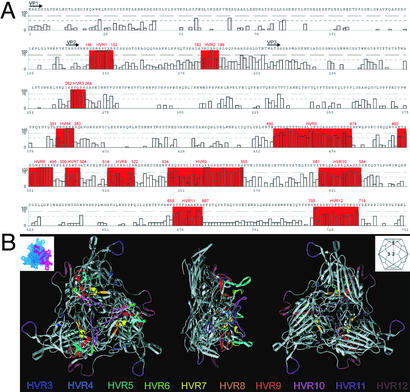
VP1 sequences of the previously uncharacterized AAVs were characterized further with respect to the nature and location of amino acid sequence variation. The 30 VP1 clones that were shown to differ from one another by >1% amino acid sequence were aligned and scored for variation at each residue as shown in Fig. 2A. An algorithm developed to determine areas of sequence divergence yielded 12 HVRs, of which 5 overlap or are part of the 4 previously described variable regions (7–9). The location of these HVRs 3–12 on the atomic structure of the AAV2 capsid protein on the atomic structure of VP3 of AAV2 is shown in Fig. 2B (26). The threefold proximal peaks contain most of the variability (HVRs 5–10). Interestingly, the loops located at the two- and fivefold axes show intense variation as well. HVRs 1 and 2 occur in the N-terminal portion of the capsid protein that is not resolved in the x-ray structure, suggesting that the N terminus of the VP1 protein is exposed on the surface of the virion. Conversely, certain domains that are implicated in preserving the structural integrity of the virion or infectivity such as the phospholipase A2 domain show a high degree of sequence conservation (27). Characterization of a more limited number of available Rep genes failed to reveal consistent regions of sequence variation (data not shown).
Figure 2.
Location of HVRs on both the primary and tertiary capsid structure of AAV2. (A) A total of 12 HVRs were identified and are highlighted in red on the figure in combination with their respective AAV2 coordinates. Plotted along the x axis are the coordinates of the 38 VP1 protein sequence alignments in combination with the AAV2 residue. Gaps in the AAV2 sequence are denoted with X. The y axis represents 100 − Q, an indicator of variation in the alignment. More variation is represented by a higher number. The black arrows indicate the N terminus of the different capsid proteins. (B) Ten HVRs (HVRs 3–12) of the AAV capsid protein are located on the resolved x-ray structure of the AAV2 capsid. Each capsid facet contains three subunits, one of which is highlighted (Upper Left; PDB ID code 1LP3). (Upper Right) A schematic shows the relative orientation of the 20 capsid facets and the five-, three-, and twofold symmetry elements. A ribbon drawing of facet with HVRs colored as viewed from the outside (Left), edge (Center), and inside (Right) of the virus. All these HVRs are located on the surface of the virion, and most are clustered in the threefold-related towers.
AAV Genomes Are Widely Distributed Throughout Tissues of NHPs.
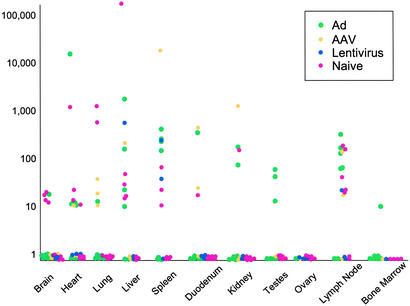
Real-time PCR was used to quantify AAV sequences from tissues of 21 rhesus macaques by using primers and probes to highly conserved regions of Rep (one set) and Cap (two sets) of known AAVs (Fig. 3). Each data point represents analysis from tissue DNA from an individual animal. This PCR experiment confirmed the wide distribution of AAV sequences, although the quantitative distribution differed between individual animals. The source of animals and previous history or treatments did not seem to influence distribution of AAV sequences in rhesus macaques. The three different sets of primers and probes used to quantify AAV yielded consistent results (Fig. 3 and data not shown). The highest levels of AAV were found in lymph node, spleen, and liver.
Figure 3.
Tissue distribution of AAV sequences in rhesus macaques. Genome copies of AAV sequences present in DNA samples extracted from 10 different tissues of 21 rhesus macaques that had not been enrolled in a previous study or had been administered an AAV, adenovirus (Ad), or lentiviral vector as part of another study were quantified by real-time PCR. Copy numbers of AAV genomes are reported as AAV copies per 100 ng of macaque genomic DNA detected in a 50-cycle amplification (y axis). The x axis represents different tissues surveyed in the study. The history of each animal is illustrated by the color of solid dots. The animals were naive at the time of necropsy (i.e., not on study, shown in pink) or were being necropsied after exposure to adenoviral (green), AAV (yellow), or lentiviral (blue) vectors.
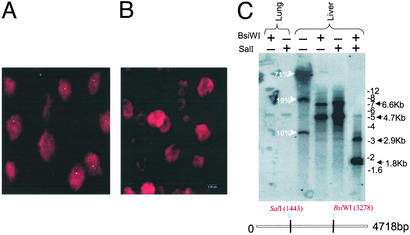
Liver tissues from a number of animals were evaluated for cellular distribution of virus DNA by in situ hybridization from cryosections with a probe to Rep and Cap of AAV8. Fig. 4A presents representative microscopic fields from the liver tissue of animal RQ4407 with a high virus-DNA content (i.e., 20 copies of virus per diploid genome), which showed strong foci of hybridization over the nuclei of 50% of cells with some cells showing multiple foci. No nuclear localized signal was detected in the liver tissue of another animal with substantially less viral DNA by TaqMan (Fig. 4B, V383).
Figure 4.
Molecular characterization and cellular distribution of AAV sequences in tissues. (A and B) In situ hybridization was performed on liver sections from rhesus macaques by using a digoxigenin-labeled AAV8 probe, and optical sections were collected at 0.5-μm intervals. A digital merge of individual green (digoxigenin) and red (propidium iodide) channel images from midplane sections of animals with >20 (RQ4407, A) and <0.1 (V383, B) proviral copies per diploid genome is shown. (C) Molecular state of AAV sequences in the cellular DNA from liver and lung tissues of rhesus macaque RQ4407 was analyzed by DNA hybridization. Total cellular DNA was analyzed by DNA hybridization as described in Materials and Methods. The enzymes used are indicated, and the probe was a mixture of Rep and Cap sequences.
Endogenous AAV Genomes Exist in a Variety of Molecular Forms.
Total Cellular DNA from liver (20 genomes per cell) and lung (0.1 genomes per cell) of a rhesus macaque was restricted with a variety of restriction endonucleases and analyzed by DNA hybridization with probes to both Rep and Cap. Fig. 4C shows the restriction sites in AAV genome selected to evaluate the molecular structure of the endogenous virus as well as a representative experiment. Analysis of uncut liver DNA indicated a broad diffuse band comigrating with chromosomal DNA, consistent with integrated viral DNA. In addition, discrete bands of 8- and 3.5-kb lengths were observed that could be explained only by the presence of substantial amounts of nonintegrated forms of the virus. More detailed structural analyses were performed from liver DNA with enzymes to consensus sites (SalI and BsiWI) that should be found only once in the AAV virus genome. Each enzyme released a predominant band of 4.7-kb length that can be formed by linearization of circular monomers or release of a unit virus length from integrated or nonintegrated head-to-tail concatamers. Larger fragments of ≈7.0-kb lengths were also observed in these digests, possibly reflecting fusion fragments of head-to-head or tail-to-tail concatamers. Digestion with both enzymes releases an internal fragment of 1.8-kb length, the size of which is independent of the molecular state of the virus genome. The abundance of virus DNA in lung of the same animal was 200-fold less, although enough was present to detect the 4.7-kb band after digestion with either single cutting enzyme. No hybridization was detected from DNA of a number of animals in which viral genomes were absent or substantially less as measured by PCR (data not shown). These studies provide indirect evidence for virus in high molecular weight fractions of DNA that could represent integrated forms as well as convincing evidence for persistent nonintegrated virus DNA. The presence of 20 copies of virus per diploid genome in liver with a limited number of hybridizing foci in hepatocytes, as demonstrated by in situ hybridization, supports the DNA hybridization experiments that suggest the presence of some concatamers of appreciable length.
Our results are consistent with previous studies of infectious and vector AAV proviral structures. These characterizations performed either in vitro or in mice showed that both monomers and concatamers were present as integrated provirus and episomes (28–36). Although the wild-type virus can integrate specifically at the chromosomal location 19q13.3-qter, the recombinant vector seemed to have lost that ability (37–40). Our results also correlate with previous findings of in vivo studies of both recombinant and wild-type AAV infection in rhesus macaques (41, 42).
In Vivo Homologous Recombination Contributes to AAV Diversity.
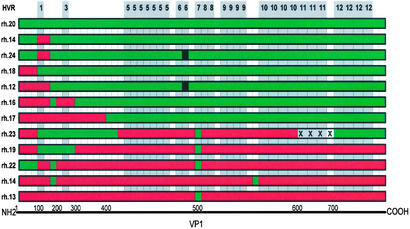
The heterogeneity in VP1 structure of the previously uncharacterized AAVs was most surprising in clones analyzed from the mesenteric lymph node of one single animal (Tulane/F953). A total of 9 distinct VP1 sequences were recovered from 15 characterized clones. A number of different controls were performed to rule out PCR artifact as an explanation for this heterogeneity (see Materials and Methods). Two of these clones, rhesus clones rh.13 and rh.20, were found to be nearly identical to cynomologus clone cy.6 and baboon clone bb.2, respectively, which are isolated from different animals from different colonies. The observation that viruses were linked by several pathways in the split-decomposition analysis in combination with the manual inspection of the amino acid sequences of these clones as shown in Fig. 5 suggested that they could have been derived by homologous recombination of these two putative parental clones.
Figure 5.
Structure of multiple AAV variants from a mesenteric lymph node of a rhesus macaque. The conserved regions were deleted from the alignment of the protein sequences of 12 AAV VP1 variants isolated from the mesenteric lymph node of animal Tulane/F953. The remaining variable residues were color-coded either red or green corresponding to their putative parental clone, rh.13 or rh.20, respectively. Location of the HVRs in relation to these differences are shaded in gray and numbered along the top. Each block represents a single residue or a group of neighboring residues. Black residues indicate a mutation that could not be brought back to any of the putative parents. X represents a deletion. Because of the nature of the presentation the coordinates corresponding to AAV2 VP1 amino acid residues are not linear.
A formal test of recombination from clones from this animal was obtained by using the bootscanning analysis (22, 23). Mosaicism was suggested when the putative recombinants were evaluated against the parental clones rh.13 and rh.20 as well as 14 other AAVs. Phylogenetic relatedness to the parents was proven to be different in adjacent fragments of the VP1 protein (data not shown). More limited sequence analysis of DNA isolated from other tissues from this animal confirmed the wide distribution of sequence variants throughout multiple organs. More detailed analysis of partial VP1 clones from one additional animal revealed sequence variation consistent with homologous recombination (data not shown).
The kind of sequence variation revealed in AAV proviral fragments isolated from different animals and within tissues of the same animals is reminiscent of the evolution that occurs for many RNA viruses during pandemics or even within the infection of an individual (43). In some situations the notion of a wild-type virus has been replaced by the existence of swarms of quasispecies that evolve as a result of rapid replication and mutations in the presence of selective pressure. One example is infection by HIV, which evolves in response to immunologic and pharmacologic pressure (44). Several mechanisms contribute to the high rate of mutations in RNA viruses including low fidelity and lack of proofreading capacity of reverse transcriptase and nonhomologous and homologous recombination (43).
Evidence for the formation of quasispecies of AAV was illustrated in our study by the systematic sequencing of multiple cloned virus genome fragments. An important mechanism for this evolution of sequence seems to be a high rate of homologous recombination between more limited numbers of parental viruses. The net result is extensive swapping of HVRs of the VP1 protein leading to an array of hybrids that could have different tropisms and serologic specificities (i.e., the ability to escape immunologic responses especially as it relates to neutralizing antibodies). Mechanisms by which homologous recombination could occur are unclear. One possibility is that positive and negative strands of different single-stranded AAV genomes anneal during replication, as has been described during high multiplicities of infection with AAV recombinants (35). In cotransfection experiments nonreplicative mutant AAV constructs have been shown to recombine with either another nonreplicative mutant or a wild-type AAV construct, generating viable virus after passages in the presence of adenovirus (45, 46). It was also proposed that AAV6 originated from a recombination event between AAV1 and AAV2 (10). The overall rate of mutation that occurs during AAV replication has been described to be relatively low, and our data do not suggest high frequencies of replication errors that led to nonsense mutations or frameshifts (47). However, substantial rearrangements of the AAV genome have been described during lytic infection leading to the formation of defective interfering particles (48). Irrespective of the mechanisms that lead to sequence divergence, with few exceptions, VP1 structures of the quasispecies remained intact without frameshifts or nonsense mutations, suggesting that competitive selection of viruses with the most favorable profile of fitness contributes to the population dynamics.
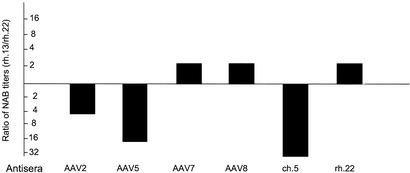
The location of most sequence divergence in the HVRs of capsid proteins suggests that immunologic pressure or modifications in vector tropism may contribute to selective pressure of variants. To test this hypothesis we evaluated the function of a number of previously uncharacterized AAVs by creating AAV pseudotypes based on AAV2 inverted terminal repeats that are packaged into the simian-derived capsids. These studies included VP1s from five viruses isolated from mesenteric lymph node of the rhesus macaque described in Fig. 5 and five viruses isolated from tissues of other animals. The vectors demonstrated a wide range of transduction efficiencies when equal quantities of GFP-expressing constructs were placed on 293 cells (data not shown). Function in vitro did not predict in vivo vector tropisms of the vectors that demonstrated 50- to 100-fold range of activity in lung and liver (data not shown). Remarkably small differences in capsid structures translated to substantial differences in the profiles of tropism. High-titer antibodies were generated to purified stocks of the vector pseudotypes and used to evaluate neutralizing activity against those in which GFP constructs were available. Distinct profiles of serologic activity were demonstrated in many of the previously uncharacterized constructs. Even minor changes in capsid structure that seem to have occurred through recombination in isolates rh.13 and rh.22 perturbed the profile of serologic reactivity (Fig. 6). These data support the hypothesis that the diversity in capsid structure observed in our studies translates to modifications in tropism and/or immune reactivity.
Figure 6.
Functional analysis of previously uncharacterized AAVs as pseudotypes. Example of the impact of capsid variation on serologic crossreactivity. Two pseudotypes from the mesenteric lymph nodes, which presumably are derived from recombination, were screened for neutralization against nonspecific antisera generated to a number of AAVs. Sera that show differences between pseudotypes based on rh.13 and rh.22 are presented as the ratios of neutralizing titers. Similar studies with sera from AAV1, rh.8, rh.10, rh.13, rh.21, and rh.24 showed no differences in neutralizing activity against rh.13 and rh.22. NAB, neutralizing antibody.
Discussion
These studies have implications in several areas of biology and medicine. The concept of rapid virus evolution, formerly thought to be a property restricted to RNA viruses, should be considered in DNA viruses, which classically have been characterized by serologic assays. It will be important in terms of parvoviruses to develop a new method for describing virus isolates that captures the complexity of its structure and biology, such as with HIV, which are categorized as general families of similar structure and function called clades. An alternative strategy is to continue to categorize isolates with respect to serologic specificity and develop criteria for describing variants within serologic groups. The implication of this work for applications of AAV in gene therapy is unclear, although the wide distribution of endogenous latent virus together with a propensity to rearrange might be the topic of further studies in the context of in vivo gene transfer in animals and humans.
Acknowledgments
Some tissue and blood samples were kindly provided by Dr. Gary B. Baskin and Mr. Maurice J. Duplantis III (Tulane University) and several other NHP research centers. Support from the Vector and Cell Morphology Cores at the University of Pennsylvania is appreciated. This work was supported by the Cystic Fibrosis Foundation, the Juvenile Diabetes Research Foundation, National Institutes of Health Grant P30 DK 47757-09, National Heart, Lung, and Blood Institute Grant P01 HL59407-03, and GlaxoSmithKline Pharmaceuticals. J.M.W. holds equity in Targeted Genetics, Corp.
Abbreviations
- AAV
adeno-associated virus
- HVR
hypervariable region
- NHP
nonhuman primate
Footnotes
References
- 1.Muzyczka N, Berns K I. In: Fields Virology. Knipe D M, Howley P M, editors. Vol. 2. Williams & Wilkins, Philadelphia: Lippincott; 2001. pp. 2327–2359. [Google Scholar]
- 2.Kotin R M. Hum Gene Ther. 1994;5:793–801. doi: 10.1089/hum.1994.5.7-793. [DOI] [PubMed] [Google Scholar]
- 3.Parks W P, Boucher D W, Melnick J L, Taber L H, Yow M D. Infect Immun. 1970;2:716–722. doi: 10.1128/iai.2.6.716-722.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atchison R W, Casto B C, Hammon W M. Science. 1965;149:754–756. doi: 10.1126/science.149.3685.754. [DOI] [PubMed] [Google Scholar]
- 5.Mayor H D, Melnick J L. Nature. 1966;210:331–332. doi: 10.1038/210331a0. [DOI] [PubMed] [Google Scholar]
- 6.Bantel-Schaal U, Zur Hausen H. Virology. 1984;134:52–63. doi: 10.1016/0042-6822(84)90271-x. [DOI] [PubMed] [Google Scholar]
- 7.Chiorini J A, Kim F, Yang L, Kotin R M. J Virol. 1999;73:1309–1319. doi: 10.1128/jvi.73.2.1309-1319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiorini J A, Yang L, Liu Y, Safer B, Kotin R M. J Virol. 1997;71:6823–6833. doi: 10.1128/jvi.71.9.6823-6833.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rutledge E A, Halbert C L, Russell D W. J Virol. 1998;72:309–319. doi: 10.1128/jvi.72.1.309-319.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao W, Chirmule N, Berta S C, McCullough B, Gao G, Wilson J M. J Virol. 1999;73:3994–4003. doi: 10.1128/jvi.73.5.3994-4003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blacklow N R, Hoggan M D, Rowe W P. Proc Natl Acad Sci USA. 1967;58:1410–1415. doi: 10.1073/pnas.58.4.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blacklow N R, Hoggan M D, Kapikian A Z, Austin J B, Rowe W P. Am J Epidemiol. 1968;88:368–378. doi: 10.1093/oxfordjournals.aje.a120897. [DOI] [PubMed] [Google Scholar]
- 13.Mayor H D, Ito M. Proc Soc Exp Biol Med. 1967;126:723–725. [Google Scholar]
- 14.Blacklow N R, Hoggan M D, Rowe W P. J Natl Cancer Inst. 1968;40:319–327. [PubMed] [Google Scholar]
- 15.Gao G P, Alvira M, Wang L, Calcedo R, Johnston J, Wilson J M. Proc Natl Acad Sci USA. 2002;99:11854–11859. doi: 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horton R M. Mol Biotechnol. 1995;3:93–99. doi: 10.1007/BF02789105. [DOI] [PubMed] [Google Scholar]
- 17.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 19.Guex N, Peitsch M C. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 20.Huson D H. Bioinformatics. 1998;14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- 21.Bandelt H J, Dress A W. Mol Phylogenet Evol. 1992;1:242–252. doi: 10.1016/1055-7903(92)90021-8. [DOI] [PubMed] [Google Scholar]
- 22.Lole K S, Bollinger R C, Paranjape R S, Gadkari D, Kulkarni S S, Novak N G, Ingersoll R, Sheppard H W, Ray S C. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salminen M O, Carr J K, Burke D S, McCutchan F E. AIDS Res Hum Retroviruses. 1995;11:1423–1425. doi: 10.1089/aid.1995.11.1423. [DOI] [PubMed] [Google Scholar]
- 24.Gowans E J, Blight K, Arthur J, Higgins G D. Methods Mol Biol. 1994;33:243–256. doi: 10.1385/0-89603-280-9:243. [DOI] [PubMed] [Google Scholar]
- 25.Gao G P, Lu F, Sanmiguel J C, Tran P T, Abbas Z, Lynd K S, Marsh J, Spinner N B, Wilson J M. Mol Ther. 2002;5:644–649. doi: 10.1006/mthe.2001.0591. [DOI] [PubMed] [Google Scholar]
- 26.Xie Q, Bu W, Bhatia S, Hare J, Somasundaram T, Azzi A, Chapman M S. Proc Natl Acad Sci USA. 2002;99:10405–10410. doi: 10.1073/pnas.162250899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Girod A, Wobus C E, Zadori Z, Ried M, Leike K, Tijssen P, Kleinschmidt J A, Hallek M. J Gen Virol. 2002;83:973–978. doi: 10.1099/0022-1317-83-5-973. [DOI] [PubMed] [Google Scholar]
- 28.Cheung A K, Hoggan M D, Hauswirth W W, Berns K I. J Virol. 1980;33:739–748. doi: 10.1128/jvi.33.2.739-748.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duan D, Sharma P, Yang J, Yue Y, Dudus L, Zhang Y, Fisher K J, Engelhardt J F. J Virol. 1998;72:8568–8577. doi: 10.1128/jvi.72.11.8568-8577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laughlin C A, Cardellichio C B, Coon H C. J Virol. 1986;60:515–524. doi: 10.1128/jvi.60.2.515-524.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLaughlin S K, Collis P, Hermonat P L, Muzyczka N. J Virol. 1988;62:1963–1973. doi: 10.1128/jvi.62.6.1963-1973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, Zhou W, Zhang Y, Zidon T, Ritchie T, Engelhardt J F. J Virol. 1999;73:9468–9477. doi: 10.1128/jvi.73.11.9468-9477.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang C C, Xiao X, Zhu X, Ansardi D C, Epstein N D, Frey M R, Matera A G, Samulski R J. J Virol. 1997;71:9231–9247. doi: 10.1128/jvi.71.12.9231-9247.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakai H, Yant S R, Storm T A, Fuess S, Meuse L, Kay M A. J Virol. 2001;75:6969–6976. doi: 10.1128/JVI.75.15.6969-6976.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakai H, Storm T A, Kay M A. J Virol. 2000;74:9451–9463. doi: 10.1128/jvi.74.20.9451-9463.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fisher K J, Jooss K, Alston J, Yang Y, Haecker S E, High K, Pathak R, Raper S E, Wilson J M. Nat Med. 1997;3:306–312. doi: 10.1038/nm0397-306. [DOI] [PubMed] [Google Scholar]
- 37.Giraud C, Winocour E, Berns K I. J Virol. 1995;69:6917–6924. doi: 10.1128/jvi.69.11.6917-6924.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duan D, Fisher K J, Burda J F, Engelhardt J F. Virus Res. 1997;48:41–56. doi: 10.1016/s0168-1702(96)01425-6. [DOI] [PubMed] [Google Scholar]
- 39.Mendelson E, Smith M G, Carter B J. Virology. 1988;166:154–165. doi: 10.1016/0042-6822(88)90157-2. [DOI] [PubMed] [Google Scholar]
- 40.Nakai H, Iwaki Y, Kay M A, Couto L B. J Virol. 1999;73:5438–5447. doi: 10.1128/jvi.73.7.5438-5447.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hernandez Y J, Wang J, Kearns W G, Loiler S, Poirier A, Flotte T R. J Virol. 1999;73:8549–8558. doi: 10.1128/jvi.73.10.8549-8558.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Afione S A, Conrad C K, Kearns W G, Chunduru S, Adams R, Reynolds T C, Guggino W B, Cutting G R, Carter B J, Flotte T R. J Virol. 1996;70:3235–3241. doi: 10.1128/jvi.70.5.3235-3241.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Domingo E, Holland J J. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 44.Domingo E, Mas A, Yuste E, Pariente N, Sierra S, Gutierrez-Riva M, Menendez-Arias L. Prog Drug Res. 2001;57:77–115. doi: 10.1007/978-3-0348-8308-5_2. [DOI] [PubMed] [Google Scholar]
- 45.Senapathy P, Carter B J. J Biol Chem. 1984;259:4661–4666. [PubMed] [Google Scholar]
- 46.Bowles D E, Rabinowitz J E, Samulski R J. J Virol. 2003;77:423–432. doi: 10.1128/JVI.77.1.423-432.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smuda J W, Carter B J. Virology. 1991;184:310–318. doi: 10.1016/0042-6822(91)90847-5. [DOI] [PubMed] [Google Scholar]
- 48.Faust E A, Hogan A. In: Handbook of Parvoviruses. Tijssen P, editor. Vol. 1. Boca Raton, FL: CRC; 1990. pp. 91–107. [Google Scholar]